Abstract
Driver tyrosine kinase mutations are rare in sarcomas, and patterns of tyrosine phosphorylation are poorly understood. To better understand the signaling pathways active in sarcoma, we examined global tyrosine phosphorylation in sarcoma cell lines and human tumor samples. Anti-phosphotyrosine antibodies were used to purify tyrosine phosphorylated peptides, which were then identified by liquid chromatography and tandem mass spectrometry. The findings were validated with RNA interference, rescue, and small-molecule tyrosine kinase inhibitors. We identified 1,936 unique tyrosine phosphorylated peptides, corresponding to 844 unique phosphotyrosine proteins. In sarcoma cells alone, peptides corresponding to 39 tyrosine kinases were found. Four of 10 cell lines showed dependence on tyrosine kinases for growth and/or survival, including platelet-derived growth factor receptor (PDGFR)α, MET, insulin receptor/insulin-like growth factor receptor signaling, and SRC family kinase signaling. Rhabdomyosarcoma samples showed overexpression of PDGFRα in 13% of examined cases, and sarcomas showed abundant tyrosine phosphorylation and expression of a number of tyrosine phosphorylated tyrosine kinases, including DDR2, EphB4, TYR2, AXL, SRC, LYN, and FAK. Together, our findings suggest that integrating global phosphoproteomics with functional analyses with kinase inhibitors can identify drivers of sarcoma growth and survival.
Introduction
Sarcomas are rare and diverse malignancies that arise from mesenchymal derived connective tissues. Advances in understanding the genetic nature of cancer have led to the development of new treatment options for sarcoma. For example, gastrointestinal stromal tumors (GIST) that harbor activating mutations in the c-KIT gene are sensitive to treatment with imatinib mesylate, a tyrosine kinase inhibitor, whereas those without c-KIT mutations are less sensitive (1). Patients with advanced GIST who have progressed on imatinib treatment were subsequently shown to benefit when treated with sunitinib malate, a broad spectrum, orally available multitargeted tyrosine kinase inhibitor of VEGF receptor, platelet-derived growth factor receptor (PDGFR), c-KIT, and FLT-3 kinases (2). The example of GIST is encouraging and hopefully will prove to be a model for developing new agents for the other sarcoma subtypes. Furthermore, many sarcomas harbor balanced translocations that result in unique fusion proteins that have been shown to deregulate various kinases (3).
Despite advances in GIST, effective treatment options for metastatic soft tissue sarcomas and osteosarcoma have yet to be shown. In addition to c-KIT in GIST, a number of other tyrosine kinases (TK) have been suggested to be important as drivers of oncogenesis in sarcoma (reviewed in ref. 4). These include PDGFs and their tyrosine kinase receptors (PDGFR), the epidermal growth factor receptor (EGFR), HER-2, VEGF and its receptors, and the insulin-like growth factor receptor (IGF1R). Despite encouraging preclinical studies and studies showing receptor expression in sarcoma tumor specimens, activity of tyrosine kinase inhibitors (TKI) in patients with advanced sarcoma has been limited. For example, phase II studies with EGFR TKI in sarcoma have disappointingly shown no clinical activity (5). There are a number of potential reasons for lack of efficacy of TKI in sarcoma. These include not enriching for patients whose tumor depends on the particular tyrosine kinase for growth/survival and a lack of assays that detect an activated tyrosine kinase that predicts drug sensitivity. In addition, it is possible that other driver tyrosine kinases are coexpressed in sarcoma cells and maintain signaling despite inhibition of one particular tyrosine kinase (6). Thus, for true efficacy, combinations of different TKI may be required.
One technique that may be helpful to identifying tumor cells dependent on kinases for growth and/or survival, as well as charting the landscape of activated tyrosine kinases in tumor cells, is mass spectrometry (MS)–based phosphoproteomics (7). The technique has been limited because phosphorylated tyrosine residues (pY) represent only 0.5% of the total phosphoamino acids within a cell (8). However, more sensitive mass spectrometers have been coupled with anti-pY antibodies to purify either proteins or enzymatically digested peptides for analysis. This approach has been used to characterize protein networks and pathways downstream of oncogenic HER2, BCR-ABL, and SRC (9–12). These methods can also be used to identify novel tyrosine phosphorylation sites and identify oncogenic proteins resulting from activating mutations in protein tyrosine kinases (10, 11, 13, 14). The data can then be used in either expert literature curation or machine learning techniques to synthesize network models that can be further evaluated (9). The methodologies can be coupled with TKI or other compounds to further understand their effect on protein networks. Identification of critical tyrosine kinase proteins in an important oncogenic network may also suggest “druggable” targets that can be entered into therapeutic discovery research.
We hypothesized that a phosphoproteomics strategy in sarcoma cells and tumors could (i) identify tyrosine kinases and substrate proteins important in the malignant process, (ii) define functional tyrosine kinases driving sarcoma cell growth and survival, (iii) suggest studies in human tumors for activated kinases and kinase substrates. We used multiple validation strategies, including RNA interference, use of small-molecule tyrosine kinase inhibitors, and rescue strategies to define MET, PDGFR, SRC, and IGF1R/insulin receptor (INSR) signaling as important in individual sarcoma cell lines. Finally, we conducted pilot experiments using primary explant models of sarcoma tissues to explore tyrosine kinase signaling in human tumor tissues.
Materials and Methods
Full descriptions of all materials and methods can be found under Supplementary Methods and Materials.
Cell lines
Cell lines A204 (HTB-82), Saos2 (HTB-85), MG63 (CRL-1427), MNNG/HOS Cl#5 (CRL-1547), A673 (CRL-1598), RD-ES(HTB-166), U2OS (HTB-96), SK-LMS1 (HTB-88), and HT1080 (CCL-221) were purchased from American Type Culture Collection (ATCC) and subcultured with their recommended conditions. ATCC conducted short tandem repeat (STR) authentication on cells, which were used within 6 months of purchase. RD18 cells were from Dr. Jack Pledger (Moffitt Cancer Center, Tampa, FL) and have not been authenticated secondary to lack of reference STR.
Phosphopeptide immunoprecipitation, analysis, and data processing
Phosphopeptide immunoprecipitation and purification were carried out and analyzed by nano-liquid chromatography/tandem mass spectrometry (LC/MS-MS) as described previously with a total of 2 × 108 cells of each cell line (12) and 30 to 80 mg of extracted proteins from tissue samples. The eluted peptide mixtures were analyzed by a nanoflow liquid chromatograph (U3000; Dionex) coupled to an LTQ-Orbitrip hybrid mass spectrometer (Thermo Scientific) in a data-dependent manner for tandem MS peptide sequencing experiments. MASCOT and SEQUEST searches were carried out against the Swiss-Prot human database downloaded on April 03, 2008. The results were summarized with Scaffold 2.0 software. We counted the number of spectra observed for each peptide in 2 total MS-MS runs (1 biological and 2 technical) per cell line. To calculate a protein spectrum count, we summed the numbers for all the peptides assigned to each protein in that run.
Kinase inhibitors
Erlotinib was provided by Genentech, dasatinib by Bristol Myers Oncology, PHA665752 by Pfizer, OSI868 by OSI Pharmaceuticals, and imatinib by Novartis. JAK inhibitor and SU5402 were purchased from EMD. ZD6474 (vandetanib) was purchased by Huskerchem Inc.. Viability was assessed after 72 hours treatment with MTT assays (MTT; Roche). The IC50 was defined as the drug concentration that induced 50% cell viability in comparison with dimethyl sulfoxide (DMSO) controls and was calculated by nonlinear regression analysis (Prism, GraphPad 5.0).
Phospho-receptor tyrosine kinase array assay
Forty-two human receptor tyrosine kinases phosphorylation level determination in sarcoma cell lines was carried out with the Proteome Profiler Array Kit (R&D Systems).
Protein expression analysis
Western blotting was conducted as previously described (12). Primary antibodies used in these studies consisted of PDGFRα, pIGF1R/INSR-Tyr1131/1146, IGF1R, INSR, pEGFR-Tyr1068, EGFR, pMET-Tyr1234/1235, MET, pAKT-Ser473, AKT, pERK, ERK, FYN, LYN, and PARP (Cell Signaling), pPDGFRα-Tyr742 (Invitrogen), as well as β-actin (Sigma).
Transfection of siRNA
The siRNAs ON-TARGETplus SMARTpool were used along with ON-TARGET plus nontargeting pool as a negative control obtained from Dharmacon. Transfection was carried out with Lipofectamine RNAiMAX from Invitrogen with reverse transfection procedure as recommended by manufacturer.
Rescue experiments with gatekeeper mutant version of tyrosine kinases
Rescue experiments were carried out as previously described (12). Briefly, cells were seeded in black wall 96-well plate from NUNC. After overnight incubation, cells were infected with 30 μL of viruses plus polybrene per well for 48 hours, and then treated with a series dilution of dasatinib for 120 hours for CellTiter-Glo Cell Viability Assay from Promega.
Human tumor tissues
Patients with the diagnosis of rhabdomyosarcoma and archived tissue block were identified under an Institutional Review Board–approved protocol. Formalin-fixed and paraffin-embedded tumor tissues were stained with a Ventana Discovery XT automated system (Ventana Medical Systems) as per manufacture’s protocol with proprietary reagents. The detection system used was the Ventana ChromoMap DAB Kit, and slides were then counterstained with hematoxylin. Primary sarcoma explant models were generated as described under a University of South Florida Institutional Animal Care and Use Committee (IACUC) approved protocol (15).
Statistical analysis
Significant differences between values obtained in control groups and different treatment groups were determined by the Student t test. P < 0.05 was assigned significance.
Results
Expression profiling of tyrosine phosphorylated peptides in sarcoma cell lines identifies novel tyrosine kinases
We employed phosphotyrosine (pY) peptide immunoprecipitation followed by LC/MS-MS to identify pY proteins in a collection of sarcoma cell lines. Briefly, pY containing peptides are isolated directly from protease-digested cellular protein extracts with a pY-specific antibody and are identified by tandem MS peptide sequencing (11). Applying this approach to several cell systems, including cancer cell lines, has shown that it can identify activated protein kinases and their phosphorylated substrates without prior knowledge of the active signaling networks (12). Ten sarcoma cell lines originating from distinct histologies were characterized. These include osteosarcoma (MNNG, U2OS, MG63, and Saos2), rhabdomyosarcoma (RD18 and A204), leiomyosarcoma (SK-LMS1), fibrosarcoma (HT1080), and Ewing’s sarcoma (RD-ES and A673). Tyrosine phosphorylated peptides were purified and analyzed by nanoflow LC coupled to a hybrid linear ion trap mass spectrometer (LTQ-Orbitrap).
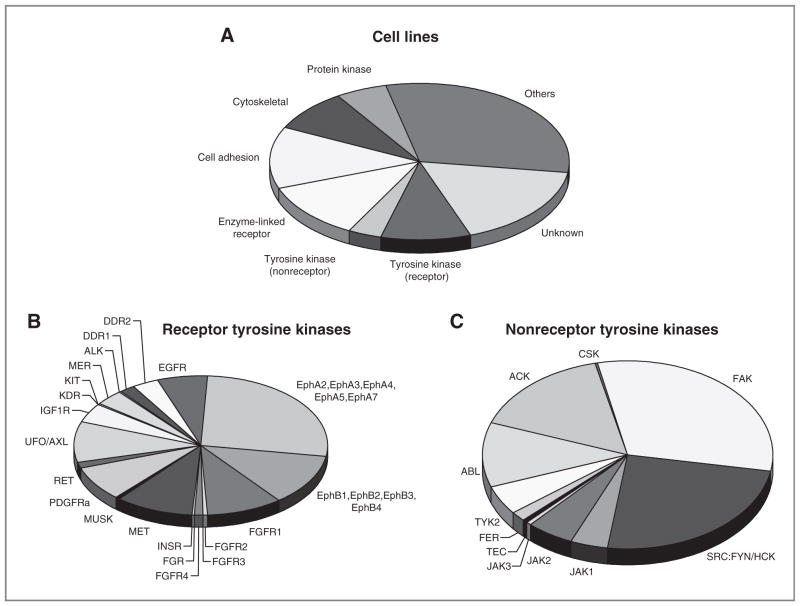
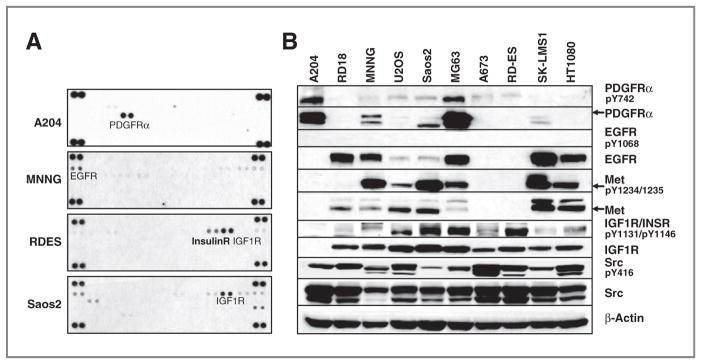
We identified 3,878 (1,936 unique) tyrosine phosphorylated peptides corresponding to 844 unique phosphotyrosine proteins (Supplementary Tables S1 and S2). Of these proteins, 39 represent tyrosine kinases (Fig. 1A and Table 1). Therefore, of the 99 tyrosine kinases present in the human genome, we identified peptides corresponding to nearly 40% of the tyrosine kinome (16). Some peptides correspond to known sites of autoactivation (autophosphorylation) suggesting that the kinase is active, while other peptides correspond to sites important in protein–protein interactions, enzymatic activity, or of unknown function. Importantly, we identify numerous tyrosine kinases that can be important in signaling transduction in human sarcomas, could drive the natural progression of sarcoma, and could be targeted by small-molecule inhibitors to alter that progression. A number of receptor tyrosine kinases were identified including EGFR, multiple Ephrin receptors, multiple FGFR receptors, IGF1R, KIT, MET, and PDGFRα (Fig. 1B). A number of nonreceptor tyrosine kinases were identified including FAK, ACK, FYN, SRC family kinases (SFK), JAK members, and ABL (Fig. 1C). Important signaling proteins such as p130CAS, paxillin, Stat3, and ERK1/2 that are activated by tyrosine phosphorylation were also identified. We carried out confirmatory assays using either receptor tyrosine kinase (RTK) arrays or Western blotting for a subset of these targets (Fig. 2A and B and Supplementary Fig. S1A–S1C). Phosphorylation of PDGFRα was abundant in A204 cells, whereas higher levels of IGF1R were more prominent in Saos2 and RD-ES cells (Fig. 2A). MET phosphorylation was abundant in MNNG and Saos2 cells and to a lesser extent in U2OS, MG63, SK-LMS1, and H1080 (Fig. 2B). PDGFRα protein expression was more abundant in A204 and MG63 cells. Overall, these results were consistent with observations from the pY MS data.
Figure 1.
Identification of tyrosine kinases in 10 sarcoma cell lines. A, distribution of phosphoprotein types. Each observed phosphoprotein was assigned a protein category from the PhosphoSite ontology. The numbers of unique proteins in each category, as a fraction of the total, are represented by the wedges in the pie. Distribution of spectral counts among receptor tyrosine kinases (B) or nonreceptor tyrosine kinases (C).
Table 1.
Tyrosine kinases identified by phosphoproteomics in sarcoma cell lines
| A204 | RD18 | MNNG | U2OS | Saos2 | MG63 | A673 | RD-ES | SK-LMS1 | HT1080 | |
|---|---|---|---|---|---|---|---|---|---|---|
| ABL | 6 | 11 | 8 | 16 | 4 | 2 | 7 | 3 | 5 | 11 |
| ACK | 15 | 11 | 4 | 7 | 24 | 15 | 3 | 3 | 8 | 15 |
| ALK | 1 | 1 | ||||||||
| CSK | 3 | |||||||||
| DDR1 | 1 | 12 | 8 | |||||||
| DDR2 | 13 | 3 | 1 | 2 | 2 | 6 | 8 | 2 | 2 | |
| EGFR | 3 | 11 | 6 | 3 | 10 | 9 | 15 | 12 | ||
| EphA2 | 34 | 18 | 10 | 12 | 23 | 13 | 23 | 5 | 11 | 22 |
| EphA3 | 1 | 4 | 4 | 4 | 15 | 4 | 3 | 4 | ||
| EphA4 | 7 | 8 | 4 | |||||||
| EphA5 | 18 | 13 | 6 | |||||||
| EphA7 | 4 | 17 | 2 | 2 | ||||||
| EphB1 | 7 | 4 | 2 | 2 | 3 | 2 | ||||
| EphB2 | 16 | 3 | 6 | |||||||
| EphB3 | 4 | 10 | 9 | 6 | 3 | |||||
| EphB4 | 11 | 5 | 5 | 12 | 1 | 15 | 4 | 2 | ||
| FAK | 19 | 11 | 25 | 28 | 32 | 28 | 39 | 2 | 7 | 14 |
| FER | 2 | 3 | 2 | 4 | ||||||
| FGFR1 | 28 | 8 | 4 | 11 | 8 | 24 | 6 | 3 | 13 | |
| FGFR2 | 2 | 3 | ||||||||
| FGFR3 | 1 | |||||||||
| FGFR4 | 3 | 4 | 6 | |||||||
| FGR | 1 | 1 | ||||||||
| SRC:FYN/HCK | 24 | 17 | 14 | 17 | 10 | 15 | 21 | 17 | 10 | 17 |
| IGF1R | 2 | 9 | 8 | 10 | 5 | 5 | 2 | 1 | 6 | |
| INSR | 1 | |||||||||
| JAK1 | 3 | 4 | 2 | 1 | 5 | 1 | 7 | 4 | ||
| JAK2 | 11 | 3 | 3 | 4 | 1 | 5 | 4 | 4 | ||
| JAK3 | 1 | 1 | 1 | 2 | ||||||
| KDR | 1 | |||||||||
| KIT | 3 | 1 | ||||||||
| MER | 6 | 2 | 2 | 4 | 4 | 11 | 4 | 3 | 3 | |
| MET | 14 | 23 | 7 | 20 | 8 | 22 | 15 | |||
| MUSK | 4 | |||||||||
| PDGFRα | 33 | 10 | 1 | 10 | 23 | 13 | ||||
| RET | 14 | 1 | ||||||||
| TEC | 2 | 1 | ||||||||
| TYK2 | 4 | 4 | 2 | 3 | 5 | 2 | 3 | 4 | 3 | 3 |
| UFO/AXL | 6 | 4 | 5 | 13 | 19 | 24 | 3 | 13 | 15 |
NOTE: Ten sarcoma cell pellets were lysed and phosphopeptides were purified and identified by phosphopeptide immunoprecipitation and LC/MS-MS. pY sites associated with the corresponding tyrosine kinases were summed and total spectral counts from one biological sample and 2 MS-MS runs are listed.
Figure 2.
Receptor tyrosine kinase phosphorylation and total expression in the different sarcoma cell lines. A, A204, Saos2, MNNG, and RD-ES cells were lysed and the relative level of 42 phosphorylated human receptor tyrosine kinases were determined by human Phospho-RTK Array Kit. A darker exposure is shown in Supplementary Fig. S1B. B, indicated sarcoma cells were lysed and protein lysates were run on SDS-PAGE and exposed to indicated antibodies. Equal protein loading was confirmed by evaluation of β-actin.
Effects of TKIs on sarcoma cell line proliferation
Which, if any, of the roughly 40 TKs we identified in tumor cell lines act as driver kinases to regulate tumor cell growth and survival? This is important to help prioritize which targets to examine further in human specimens. For example, some tyrosine kinase may be expressed and be signaling, yet are redundant given the coexpression of other tyrosine kinases. To help investigate this further, we screened the same sarcoma cell lines against inhibitors of EGFR (erlotinib), JAK (P6), FGFR (SU5402), MET (PHA665752), IGF1R (OSI868), RET (ZD6474), PDGFR (imatinib and dasatinib), and SRC (dasatinib; refs. 17, 18). These compounds were selected based on the tyrosine kinases observed in the MS data to gain good coverage of kinases and identify sensitive cells for further validation experiments. We also used results of chemical proteomics reports that more accurately map targets of these kinases and could point to further validation studies (17, 19–23). These data are shown in Table 2 and suggest that some kinases may in fact be drivers of sarcoma cell growth. MNNG cells were sensitive to MET TKI (PHA) with IC50 = 330 nmol/L while A204 cells were sensitive to both dasatinib and imatinib with IC50 = 34 and 135 nmol/L, respectively. RD-ES cells were sensitive to IGF1R TKI OSI868 with IC50 = 96 nmol/L. Finally, Saos2 cells were sensitive to dasatinib with IC50 = 390 nmol/L. Next, we examined these sensitive cell lines for effects of downstream signaling as well as used another series of validation experiments to validate drivers of sarcoma cell growth and survival.
Table 2.
Kinase inhibitor sensitivity
| A204 | RD18 | MNNG | U2OS | Saos2 | MG63 | A673 | RD-ES | SK-LMS1 | HT1080 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Erlotinib | >10 | >10 | >10 | >10 | >10 | >10 | >10 | >10 | >10 | 9.63 |
| Dasatinib | 0.03 | 6.07 | >10 | >10 | 0.39 | 0.80 | 6.75 | >10 | >10 | 1.62 |
| Imatinib | 0.13 | >10 | >10 | >10 | >10 | >10 | >10 | >10 | >10 | >10 |
| PHA665752 | 4.20 | 2.87 | 0.33 | 8.03 | 3.03 | 2.50 | 3.33 | 6.31 | >10 | 3.10 |
| OSI868 | 9.80 | >10 | >10 | >10 | 2.24 | 4.83 | 4.26 | 0.10 | >10 | 4.12 |
| SU5402 | 15.24 | >40 | >40 | >40 | >40 | 34.08 | >40 | >40 | >40 | >40 |
| JAK | 9.23 | >10 | 3.47 | >10 | >10 | >10 | >10 | 8.36 | >10 | 0.91 |
| ZD6474 | 5.77 | 9.33 | 6.37 | >10 | >10 | 8.28 | 5.37 | 5.78 | >10 | 9.04 |
NOTE: IC50 values for each indicated compound tested in the 10 sarcoma cell lines. Values are given in μmol/L.
Identification of sarcoma cells dependent on PDGFR signaling
We noted that the A204 rhabdomyosarcoma cell line had highly abundant pY peptides corresponding to PDGFRα. Validation experiments with RTK array confirmed tyrosine phosphorylated PDGFRα, and Western blotting showed overexpression of PDGFRα compared with other cell lines studied (see Fig. 2A and B). We find that A204 cells are very sensitive to both dasatinib and imatinib, both of which can inhibit PDGFRα kinase activity (17, 18). A closer examination of the data presented in Table 1 finds that A204 expressed abundant levels of pY peptides corresponding to PDGFRα, a known target of imatinib/dasatinib, while no such PDGFRα pY peptides were found in A673 or RD18 cells, which are resistant to both dasatinib and imatinib. For these reasons, we believed that PDGFRα may be an important target in some rhabdomyosarcoma cells and tumors.
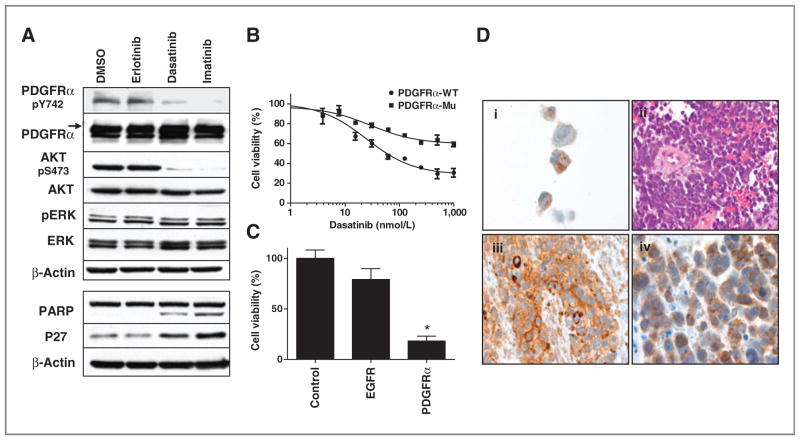
Next, we examined downstream signaling in A204 cells. We find that both dasatinib and imatinib inhibit the AKT pathway and induce apoptosis (PARP cleavage) as well as cell-cycle arrest (p27 elevation). No obvious changes were observed in ERK phosphorylation, and erlotinib had no effects (Fig. 3A). We engineered A204 cells to express a mutant form of PDGFRα expressing a gatekeeper mutation that prevents TKI binding to the ATP binding pocket (12). As shown in Fig. 3B, cells expressing this mutant form of PDGFRα were rescued from the effects of dasatinib compared with cells expressing wild-type PDGFRα (Fig. 3B). We also transfected A204 cells with siRNA against PDGFRα and found significant effects of cell viability (Fig. 3C). Target inhibition with siRNA was confirmed with Western blotting (Supplementary Fig. S2A). We attempted to determine the mechanism of PDGFRα over-expression and drug sensitivity in the A204 cells. We found no evidence of PDGFRα gene amplification nor did we find evidence of gene mutation (data not shown). In addition, we found no evidence of PAX3:FKHR fusion in these cells that has been reported to drive overexpression of PDGFRα. During the course of our work it was discovered that A204 cells secrete abundant amounts of PDGF that produce autocrine stimulation of PDGFRα (24).
Figure 3.
A204 cell line dependent on PDGFRα signaling. A, signaling changed by TKIs in A204 cells. A204 cells were exposed to DMSO, erlotinib 1,000 nmol/L, dasatinib 1,000 nmol/L, and imatinib 1,000 nmol/L for 3 hours (top) or 24 hours (bottom) after which total proteins were run on SDS-PAGE and exposed to indicated antibodies. Equal protein loading was confirmed by evaluation of β-actin. B, rescue effects of dasatinib target PDGFRα gatekeeper mutants on cell viability. A204 cells were infected with lentivirus expressing wild-type and mutant gatekeeper form of PDGFRα for 48 hours. Subsequently cells were exposed to increasing concentrations of dasatinib for 120 hours after which cell viability was assessed. Cell viability as a function of dasatinib concentration is plotted with error bars representing SD and results normalized to DMSO-treated cells. C, effects of siRNA-mediated knockdown of PDGFRα on cell viability in A204 cells. A204 cells were transfected with 20 nmol/L of indicated siRNA. Cell viability was assessed after 5 days transfection and normalized to the control siRNA-transfected cells. Each bar represents mean ± SD of 2 separate experiments. *, P < 0.01 represents the results comparing each siRNA group with control siRNA group with the Student t test. D, PDGFRα expression in human sarcoma tissue: (i) immunohistochemical stain of the cellblock prepared from A204 cell line (magnification, ×100); (ii) hematoxylin and eosin stain of undifferentiated rhabdomyosarcoma cells with small blue, round cell morphology (magnification, ×60); (iii) immunohistochemical stain of the above tumor myoglobin expression (magnification, ×60); (iv) immunohistochemical stains of the above tumor with PDGFRα immunoreactivity in the cytoplasm and the membrane (magnification, ×100).
Using the same antibody used in Western blotting experiments and using A204 as a positive control, we established an immunohistochemistry assay to assay PDGFRα in rhabdomyosarcoma tumors obtained from patients. Of 23 specimens, 3 had 2+ to 3+ staining for PDGFRα and were counted as positive (3 of 23, 13%; Fig. 3D and Supplementary Table S3); 2 had focal or weak staining (1+) and were counted as negative with the remainders (19 of 23, 87%). Two of 3 cases are primary tumors and the other was a brain metastasis. The metastatic lesion had duplicate blocks that were both positive for PDGFRα; however, multiple blocks from other metastatic sites of the same case (lung and subcutaneous) were negative for PDGFRα indicating heterogeneity of the tumor. Primary tumor was not available from this case. Collectively, our MS and tumor data along with data from other groups suggested PDGFRα as a potential target that is overexpressed in a small group of rhabdomyosarcoma tumors (25, 26).
Identification of sarcoma cells dependent on MET signaling
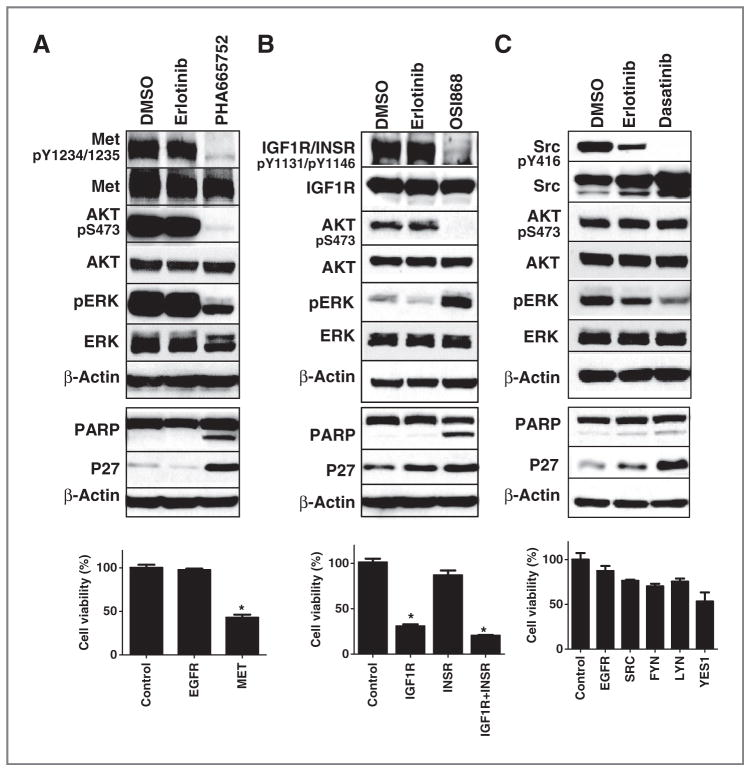
We identified pY peptides corresponding to MET in 7 of the 10 cells with higher spectral counts (a semiquantitative measure of peptide abundance) in MNNG, SK-LMS1, and Saos2 cells. We identified MNNG cells as being sensitive to PHA665752, an inhibitor of MET, while the other cells were largely resistant (IC50 > 1 μmol/L; refs. 17, 18). PHA resulted in strong inhibition of pAKT and pERK in MNNG cells, while no effects were observed with erlotinib as a negative control (Fig. 4A, top). PHA resulted in increased PARP cleavage and increased p27 levels (Fig. 4A, middle), suggestive of cell apoptosis and cell-cycle arrest, respectively. MET siRNA had strong effects on cell viability in these cells while siRNA against EGFR had no effects (Fig. 4A, bottom, and Supplementary Fig. S2B). These results together suggest MET is a driver kinase responsible for cell growth and survival in this particular cell line. Interestingly, the MNNG cells are known to harbor a gene fusion between TPR and MET leading to constitutive MET signaling and transformation (27).
Figure 4.
MNNG, RD-ES, and Saos2 cell lines dependent on MET, IGF1R/INSR, and SFK signaling respectively. A, MNNG cells dependent on MET signaling. MNNG cells were exposed to DMSO, erlotinib 1,000 nmol/L, and PHA665752 500 nmol/L for 3 (top) or 24 hours (middle) and protein lysates used for Western analysis with indicated antibodies. Bottom, MNNG cells were exposed to 20 nmol/L of siRNA and cell viability was assessed after 120 hours. Results were normalized to control siRNA-treated cells and error bars represent SD. *, P < 0.01 represents the results comparing each siRNA group with control siRNA group using the Student t test. B, RD-ES cells dependent on IGF1R/INSR signaling. RDES cells were exposed to DMSO, erlotinib 1,000 nmol/L, and OSI868 1,000 nmol/L for 3 (top) or 24 hours (middle) and protein lysates used for Western analysis with indicated antibodies. Bottom, RD-ES cells were transfected with 40 nmol/L of siRNA and cell viability was assessed after 120 hours. Results were normalized to control siRNA-treated cells and error bars represent SD. *, P < 0.01 comparing siRNA groups with control siRNA group using the Student t test. C, Saos2 cells dependent on SFK signaling. Saos2 cells were exposed to DMSO, erlotinib 1,000 nmol/L, and dasatinib 1,000 nmol/L for 3 (top) or 24 hours (middle) and protein lysates used for Western analysis with indicated antibodies. Bottom, Saos2 cells were exposed to 20 nmol/L of siRNA and effects on cell viability was assessed after 120 hours. Results were normalized to control siRNA-treated cells and error bars represent SD. D, rescue the effect of dasatinib in Saos2 cells. Saos2 cells were infected with lentivirus expressing wild-type and gatekeeper mutant form of indicated kinases for 48 hours, and then treated with indicated concentrations of dasatinib for 120 hours. Cell viability was assessed by CellTiter-Glo and normalized to DMSO-treated cells. Cell viability curves were plotted by GraphPad Prism 5. Each data point represents mean ± SD.
Identification of sarcoma cells dependent on IGF1R/INSR signaling
pY peptides corresponding to IGF1R were found in 9 of the 10 cell lines but only RD-ES was sensitive to OSI868, an IGF1R TKI, with IC50 = 96 nmol/L. OSI868 strongly inhibited pAKT and paradoxically increased levels of pERK, perhaps through a cross-talk or adaptive mechanisms (Fig. 4B, top). OSI868 also increased PARP cleavage indicative of cell apoptosis with similar evidence of p27 elevation (Fig. 4B, middle). To examine the central targets of OSI868 in these cells, we transfected cells with siRNA against IGF1R, INSR, and the combination of IGF1R and INSR. We observed that dual inhibition of IGF1R and INSR causes pronounced inhibition of RD-ES cell viability (Fig. 4B, bottom). Interestingly, we observed no effects of INSR inhibition on RD-ES cell viability. Effects of siRNA on IGF1R and INSR protein levels showed substantial target inhibition, yet we were unable to identify an siRNA that only affected IGF1R but not INSR (Supplementary Fig. S2C). In addition to these RNAi experiments, we also examined the effect of another IGF1R/INSR inhibitor BMS-754807 (28). RD-ES cells exhibited the highest sensitivity to this compound compared with the other 9 sarcoma cell lines (Supplementary Fig. S3A and S3B), and we observed similar effects of BMS-754807 compared with OSI868 on RD-ES cells (Supplementary Fig. S3C). Overall, our results are consistent with preclinical and, more recently, clinical trial data suggesting subsets of Ewing sarcoma cells can be sensitive to IGF1R/INSR signaling (29–32).
Identification of sarcoma cells dependent on SRC signaling
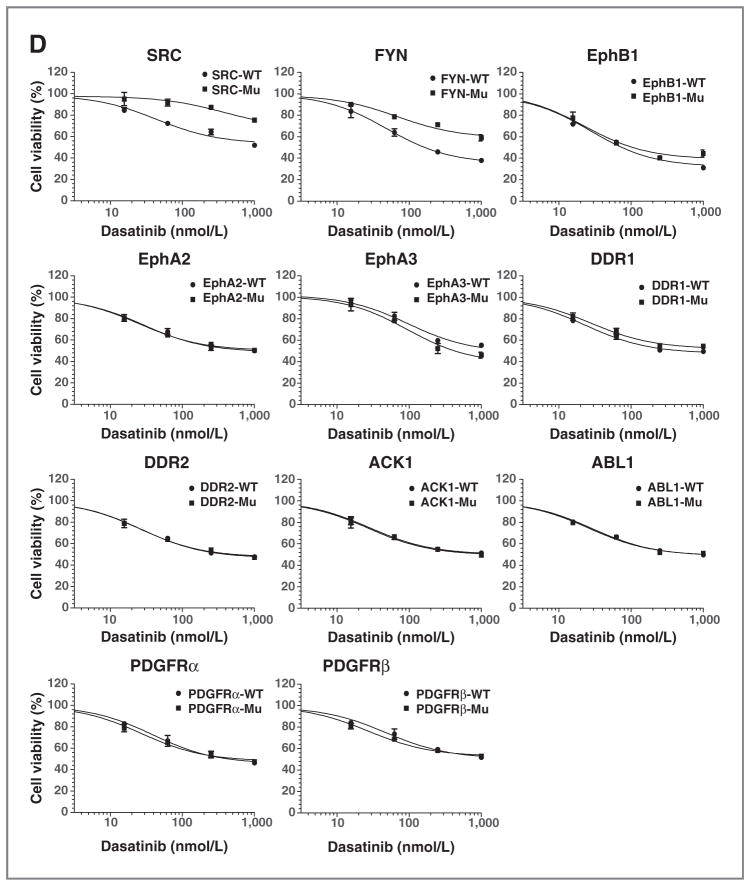
Saos2 cells were identified to be sensitive to dasatinib and had multiple pY peptides corresponding to known targets of dasatinib (SFK, Ephrin receptors, ABL, etc). Dasatinib had minimal effects on downstream pAKT with some modest effects on pERK (Fig. 4C, top). It also had minimal effects on PARP cleavage, yet had effects on p27 although similar to those observed with erlotinib (Fig. 4C, middle). We did not examine imatinib as the cells were highly resistant. We attempted to use siRNA for SFK and found no effects on cell viability despite good target inhibition (Fig. 4C, bottom and Supplementary Fig. S2D). This is not particularly surprising because a great degree of redundancy exists for SFK signaling. To address this, we next engineered Saos2 cells to express drug-resistant alleles of tyrosine kinases targeted by dasatinib. Here, we expressed wild-type and mutant (drug-resistant gatekeeper mutant) forms of 11 dasatinib targets in Saos2 cells and then examined for their effect on dasatinib sensitivity. We specifically examined c-SRC, FYN, EphB1, EphA2, EphA3, DDR1, DDR2, ACK1, PDGFRα, PDGFRβ, and ABL1 (Fig. 4D). A number of important conclusions are derived from this experiment. First, we do in fact find compensation by alternative SFK, in particular both c-SRC and FYN drug-resistant alleles can rescue cells from dasatinib. Second, we found no effect of Ephrins or other dasatinib targets on cell sensitivity. These new data show that dasatinib effects result from SFK inhibition. From these experiments, we conclude that SFK are important drivers of cell growth in Saos2 cells through downstream signaling pathways independent of AKT and ERK.
Phosphotyrosine profiling of human sarcoma tissues
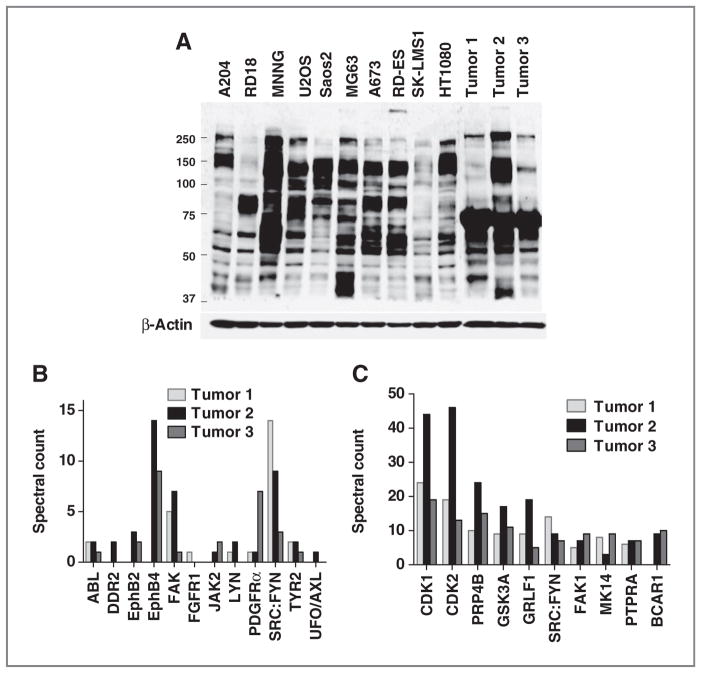
We carried out experiments with LC/MS-MS and phosphopeptide enrichment in snap-frozen human sarcoma specimens by employing primary explant models of sarcoma (15). In these studies, tumor tissue taken directly from patients is implanted into mice and grown as xenografts. These systems are thought to better recapitulate human tumors and are amenable to signaling studies as the excision from mice is rapid and human surgery variables (time of operation, arterial ligations, time to freezing) are mitigated. Tumors included liposarcoma, small round blue tumor, and malignant peripheral nerve sheath tumor histology (Supplementary Table S4). We observed comparable amounts of tyrosine phosphorylation in these tumors compared with cell lines indicating retention of tyrosine phosphorylation in these tumor models (Fig. 5A). Overall, in these 3 tumor samples, we observed 220 unique pY peptides corresponding to 170 unique proteins (Supplementary Tables S2 and S5). A number of tyrosine kinases were again observed including the collagen receptor tyrosine kinase DDR2, multiple Ephrin receptors, FAK, and SFK (Fig. 5B). Highly abundant pY proteins are listed in Fig. 5C and include serine/threonine kinases (CDK1/2, PRP4 kinase, GSK3A, and MK14), tyrosine kinases (SFK and FAK), receptor-type tyrosine-protein phosphatase alpha (PTPRA), and the scaffold protein p130CAS. These results suggest that pY profiling on tumor tissue is feasible and patterns of pY activation in human sarcoma tumor samples could be useful for identification of additional targets, especially with broader histologic types of tumors and in conjunction with drug therapy experiments in mice.
Figure 5.
Phosphotyrosine profiling of xenograft human sarcoma tissues. A, phosphotyrosine expression in cell lines and human tumor tissues. Lysates from cells and tumor tissues were subjected to SDS-PAGE gel, and phosphotyrosine expression was detected by Western blotting. Equal protein loading was confirmed by evaluation of β-actin. B and C, tyrosine kinases and top phosphotyrosine proteins identified in sarcoma tumor tissue. Three xenograft human sarcoma tumors were lysed and phosphopeptides were purified and identified by phosphopeptide immunoprecipitation and LC/MS-MS. All tyrosine kinases (B) and top 10 proteins with most abundant phosphotyrosine peptides (C) are indicated at x-axis. pY sites associated with the corresponding protein were summed, and total spectral counts from one biological sample and 2 MS-MS runs are shown at y-axis.
Discussion
Phosphotyrosine profiling with affinity capture and LC/MS-MS is a powerful tool for elucidating signaling mechanisms in cancer and monitoring changes during TKI treatment (33). Tyrosine kinases play an important role in controlling hallmarks of cancer and have a proven track record as drug-gable targets for the treatment of cancer (34). Screens for tyrosine kinase signaling with anti-phosphotyrosine antibodies to purify tyrosine phosphorylated peptides with identification using LC/MS-MS have identified novel tyrosine kinases that drive cancer cell growth, including ALK and PDGFR in lung cancer cell lines and JAK in leukemia (35, 36). However, interpretation of these data sets can be difficult and identifying driver kinases can be problematic. Combining genomic or proteomic screens with functional screens has the ability to provide additional evidence for a particular target’s importance in cancer signaling. The main goal of the phosphotyrosine MS experiments reported here was to produce a landscape of tyrosine phosphorylated proteins and tyrosine kinases prioritized for subsequent functional validation. We carried out this analysis by a battery of sarcoma cell lines with representative cells from different histologic subtypes. Results of our study uncovered sarcoma cells dependent on MET, SFK, IGF1R/INSR, and PDGFRα tyrosine kinases. This approach was able to identify drivers either through genetic changes (TPR-MET in MNNG) or autocrine ligand production (PDGF in A204 cells). The results also uncovered kinases in sarcoma cells that deserve further study, such as the nonreceptor tyrosine kinase ACK and the AXL receptor tyrosine kinase. Our results suggest that further studies on human sarcoma tissues, either directly removed from patients or propagated in primary mouse explant models, could be informative. Integration of phosphoproteomic data with either genomic studies or drug response data could further define functional kinases in human sarcomas. Our results suggest the overall applicability of the approach as well as suggest further studies on these targets in preclinical models and in clinical trials.
Our studies revealed that sarcoma cells and tissues express multiple tyrosine kinases and this may ultimately limit the effectiveness of targeted agents. Merely observing phosphorylated peptides corresponding to tyrosine kinases does not imply functional relevance of the kinases. For example, MG63 cells have a high expression of PDGFRα and phosphotyrosine PDGFRα, yet are insensitive to agents inhibiting PDGFRα. In addition, despite EGFR phosphopeptides observed in multiple cell lines, all the cell lines tested are highly resistant to EGFR TKI. Thus, in the context of analyzing global phosphoproteomics data, it is important to have integrated strategies with other approaches to identify driver kinases as opposed to bystander (irrelevant) kinases. It is currently difficult to infer the driver kinases based solely on a phosphotyrosine MS data set. Rather, we have used these MS experiments to provide hypothesis-generating data sets that require further functional characterization. Future use of mathematical modeling tools, coupled with drug response data, could potentially reveal the important pathways driving sarcoma growth and survival by the entire pY peptide/protein list. Such future studies will need to have more accurate quantitative analysis and possibly perturbation data to enable the modeling. Combining phosphoproteomic data sets with proteomic data sets that map protein–protein interactions could also be useful to better define signaling architecture and dependence on particular kinases. Similar studies characterizing global phosphorylation with immobilized metal affinity chromatography could expand beyond tyrosine kinase signaling to examine serine and threonine kinase signaling in sarcoma. Such studies could also enable combination pathway inhibition therapeutic strategies, which may be important in complex tumors with multiple activated pathways driving tumor growth and survival.
Supplementary Material
Acknowledgments
The authors thank Drs. Sam Agresta (Agios; Cambridge, MA) and Tony Conley (Moffitt Cancer Center, FL) for helpful discussions, Jack Pledger for support and guidance, Chris Cubitt for assistance with cell lines, Uwe Rix for providing compounds and Patricia Johnston for administrative assistance.
Grant Support
This work was supported by grants from the V Foundation and the National Functional Genomics Center; the Proteomics Core, Molecular Genomics Core, Tissue Core, and Cancer Informatics Core at the H. Lee Moffitt Cancer Center & Research Institute (Tampa, FL).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/)
Disclosure of Potential Conflicts of Interest
E.B. Haura has received commercial research support from Bristol-Myers Squibb Oncology. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: M. Bui, E.B. Haura
Development of methodology: Y. Bai, J. Li, B. Fang, M. Bui, S. Eschrich, E.B. Haura
Acquisition of data: B. Fang, M. Bui, S. Altiok, J. Koomen, E.B. Haura
Analysis and interpretation of data: Y. Bai, J. Li, B. Fang, A. Edwards, G. Zhang, M. Bui, S. Eschrich, J. Koomen, E.B. Haura
Writing, review, and/or revision of the manuscript: Y. Bai, J. Li, B. Fang, M. Bui, J. Koomen, E.B. Haura
Administrative, technical, or material support: S. Altiok, E.B. Haura
Study supervision: E.B. Haura
References
- 1.Joensuu H, Roberts PJ, Sarlomo-Rikala M, Andersson LC, Tervahartiala P, Tuveson D, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001;344:1052–6. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- 2.Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368:1329–38. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 3.Crose LE, Linardic CM. Receptor tyrosine kinases as therapeutic targets in rhabdomyosarcoma. Sarcoma. 2011;2011:756982. doi: 10.1155/2011/756982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shor AC, Agresta SV, D’Amato GZ, Sondak VK. Therapeutic potential of directed tyrosine kinase inhibitor therapy in sarcomas. Cancer Control. 2008;15:47–54. doi: 10.1177/107327480801500106. [DOI] [PubMed] [Google Scholar]
- 5.Ray-Coquard I, Le Cesne A, Whelan JS, Schoffski P, Bui BN, Verweij J, et al. A phase II study of gefitinib for patients with advanced HER-1 expressing synovial sarcoma refractory to doxorubicin-containing regimens. Oncologist. 2008;13:467–73. doi: 10.1634/theoncologist.2008-0065. [DOI] [PubMed] [Google Scholar]
- 6.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 7.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 8.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–80. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 9.Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, Bader JS, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci U S A. 2006;103:9773–8. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goss VL, Lee KA, Moritz A, Nardone J, Spek EJ, MacNeill J, et al. A common phosphotyrosine signature for the Bcr-Abl kinase. Blood. 2006;107:4888–97. doi: 10.1182/blood-2005-08-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–9. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters DK, Goss VL, Stoffregen EP, Gu TL, Lee K, Nardone J, et al. Phosphoproteomic analysis of AML cell lines identifies leukemic oncogenes. Leuk Res. 2006;30:1097–104. doi: 10.1016/j.leukres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Walters DK, Mercher T, Gu TL, O’Hare T, Tyner JW, Loriaux M, et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell. 2006;10:65–75. doi: 10.1016/j.ccr.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Kreahling JM, Gemmer JY, Reed D, Letson D, Bui M, Altiok S. MK1775, a selective Wee1 inhibitor, shows single-agent antitumor activity against sarcoma cells. Mol Cancer Ther. 2012;11:174–82. doi: 10.1158/1535-7163.MCT-11-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 17.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–36. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 18.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 19.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci U S A. 2005;102:11011–6. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hantschel O, Rix U, Superti-Furga G. Target spectrum of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib. Leuk Lymphoma. 2008;49:615–9. doi: 10.1080/10428190801896103. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–9. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Remsing Rix LL, Rix U, Colinge J, Hantschel O, Bennett KL, Stranzl T, et al. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–85. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 23.Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–63. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 24.McDermott U, Ames RY, Iafrate AJ, Maheswaran S, Stubbs H, Greninger P, et al. Ligand-dependent platelet-derived growth factor receptor (PDGFR)-alpha activation sensitizes rare lung cancer and sarcoma cells to PDGFR kinase inhibitors. Cancer Res. 2009;69:3937–46. doi: 10.1158/0008-5472.CAN-08-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armistead PM, Salganick J, Roh JS, Steinert DM, Patel S, Munsell M, et al. Expression of receptor tyrosine kinases and apoptotic molecules in rhabdomyosarcoma: correlation with overall survival in 105 patients. Cancer. 2007;110:2293–303. doi: 10.1002/cncr.23038. [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi E, Nishijo K, McCleish AT, Michalek JE, Grayson MH, Infante AJ, et al. PDGFR-A is a therapeutic target in alveolar rhabdomyosarcoma. Oncogene. 2008;27:6550–60. doi: 10.1038/onc.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park M, Dean M, Cooper CS, Schmidt M, O’Brien SJ, Blair DG, et al. Mechanism of met oncogene activation. Cell. 1986;45:895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 28.Carboni JM, Wittman M, Yang Z, Lee F, Greer A, Hurlburt W, et al. BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Mol Cancer Ther. 2009;8:3341–9. doi: 10.1158/1535-7163.MCT-09-0499. [DOI] [PubMed] [Google Scholar]
- 29.Buck E, Gokhale PC, Koujak S, Brown E, Eyzaguirre A, Tao N, et al. Compensatory insulin receptor (IR) activation on inhibition of insulin-like growth factor-1 receptor (IGF-1R): rationale for cotargeting IGF-1R and IR in cancer. Mol Cancer Ther. 2010;9:2652–64. doi: 10.1158/1535-7163.MCT-10-0318. [DOI] [PubMed] [Google Scholar]
- 30.Olmos D, Martins AS, Jones RL, Alam S, Scurr M, Judson IR. Targeting the insulin-like growth factor 1 receptor in Ewing’s Sarcoma: reality and expectations. Sarcoma. 2011;2011:402508. doi: 10.1155/2011/402508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juergens H, Daw NC, Geoerger B, Ferrari S, Villarroel M, Aerts I, et al. Preliminary efficacy of the anti-insulin-like growth factor type 1 receptor antibody figitumumab in patients with refractory Ewing sarcoma. J Clin Oncol. 2011;29:4534–40. doi: 10.1200/JCO.2010.33.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappo AS, Patel SR, Crowley J, Reinke DK, Kuenkele KP, Chawla SP, et al. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J Clin Oncol. 2011;29:4541–7. doi: 10.1200/JCO.2010.34.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Rosario AM, White FM. Quantifying oncogenic phosphotyrosine signaling networks through systems biology. Curr Opin Genet Dev. 2010;20:23–30. doi: 10.1016/j.gde.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–8. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 35.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Walters DK, Goss VL, Stoffregen EP, Gu TL, Lee K, Nardone J, et al. Phosphoproteomic analysis of AML cell lines identifies leukemic oncogenes. Leukemia Res. 2006;30:1097–104. doi: 10.1016/j.leukres.2006.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.