Abstract
Developments in the use of genomics to guide natural product discovery and a recent emphasis on understanding the molecular mechanisms of microbiota-host interactions have converged on the discovery of natural products from the human microbiome. Here, we review what is known about small molecules produced by the human microbiota. Numerous molecules representing each of the major metabolite classes have been found that have a variety of biological activities, including immune modulation and antibiosis. We discuss technologies that will affect how microbiota-derived molecules are discovered in the future, and consider the challenges inherent in finding specific molecules that are critical for driving microbe-host and microbe-microbe interactions and their biological relevance.
INTRODUCTION
Symbiotic relationships – including mutualism, commensalism, and parasitism – are ubiquitous in nature (1). Some of the best known symbioses are between a microorganism and a multicellular host; in these inter-kingdom relationships, the fitness of the microbe-host system (the holobiont) often relies on a diverse set of molecular interactions between the symbiotic partners (2, 3). Examples include food digestion, nitrogen and carbon fixation, oxidation and reduction of inorganic molecules, and the synthesis of essential amino acids and cofactors (2, 4–6). In light of the critical role of a molecular dialog in maintaining a productive mutualism, the community of researchers studying the symbiosis between humans and their microbiota has begun moving from a focus on ‘who’s there’ to ‘what are they doing’. The accompanying emphasis on molecular mechanism has sparked a concerted hunt for the mediators of microbe-host interactions, including microbiota-derived small molecules.
It is now possible to identify biosynthetic genes in bacterial genome sequences and, in some cases, predict the chemical structure of their small molecule products. This genome mining has led to the discovery of a growing number of molecules, and recently developed algorithms (7–9) have not only automated biosynthetic gene cluster identification, but also have led to the unexpected discovery of numerous biosynthetic gene clusters in genomes of the human microbiota (10) . In addition, a wealth of natural products have been discovered from bacterial and fungal symbionts of insects, nematodes, sponges and ascidians, and plants (11–15). The many known examples of microbe-host mutualisms in which the microbe synthesizes a metabolite important for the ecology of the pair raise an intriguing question: To what extent are mammals, including humans, a part of this paradigm?
In this review, we review what is known about natural products from the human microbiota, examining in depth the diverse chemistries and biological functions of these molecules. We focus predominantly on commensal bacterial species, although we include a few notable examples of small molecules from bacterial pathogens. We then discuss recent insights into the metabolic potential of the human microbiota from computational analyses, and conclude by considering four approaches to identify and discover the function of ‘important’ molecules out of a complex cellular and molecular milieu. Some prominent microbiota-derived metabolite classes are not covered here, including short- chain fatty acids (SCFAs) and trimethylamine-N-oxide (TMAO); for an in-depth discussion of their role in microbe-host interactions in the gut, see (16).
THE MENU OF MOLECULES
A wide range of natural products have been isolated from human-associated bacteria. These molecules cover the entire spectrum of chemical classes that have been isolated from terrestrial and aquatic bacterial species and include well-characterized mediators of microbe-host and microbe-microbe interactions. Exploring their chemistry and function provides an entry point for understanding the effects of the microbiota on human health and disease.
Ribosomally synthesized, post-translationally modified peptides (RiPPs)
Most human-associated bacteria live in complex communities and compete with other species for resource-limited niches. RiPP natural products, which are unusually prevalent among the microbiota (10) , are often active against a limited set of species closely related to the producer (i.e., narrow-spectrum) and are therefore thought to determine which strain of a bacterial species (or species of a genus) colonizes a niche. RiPPs are divided into numerous subclasses (17) , five of which include members that have been isolated from human-associated bacteria: lantibiotics, bacteriocins, microcins, thiazole/oxazole-modified microcins (TOMMs), and thiopeptides (see Outlook section for a discussion about thiopeptides).
Lantibiotics and bacteriocins
Lantibiotics and bacteriocins are the most commonly isolated RiPPs from members of the human microbiota; dozens have been found to date. Lantibiotics are short peptides (<40 amino acids) with chemical crosslinks formed post-translationally by connecting the terminal thiol of a cysteine residue to a dehydrated serine or threonine. The resulting ‘lanthionine’ contains a thioether bond, which is typically more redox-stable than a disulfide. In contrast, bacteriocins are longer peptides (>40 amino acids) and are usually unmodified. Microbiota-derived lantibiotics are predominantly produced by the Firmicutes, and are usually active against a narrow spectrum of Gram-positive bacteria that are closely related to the producing strain.
Some microbiota-derived lantibiotics are produced by commensals (18), including the salivaricins from oral resident Streptococcus salivarus (19–22), a cocktail of five lantibiotics from the skin commensal Staphylococcus epidermidis (23–27), and ruminococcin A from the gut commensals Ruminococcus gnavus and Clostridium nexile (Table 1) (28, 29). Each of these molecules inhibits pathogens that are closely related to the producer. Lantibiotics have also been isolated from human pathogens: staphylococcin Au-26 (also known as Bsa) from Staphylococcus aureus (30, 31), SA-FF22 from Streptoccous pyogenes (32, 33), and the two- component lantibiotic cytolysin from Enterococcus faecalis (34) exert antibacterial activity against a range of common human commensals, indicating that lantibiotic production are used by commensals and pathogens to compete and establish resilient colonization.
Table 1.
Selected small molecules from the human microbiota
| Class | Compound | Producer (example) | Phylum | Host Site | Known/Predicted Activity |
|---|---|---|---|---|---|
| RiPP (thiopeptide) | lactocillin | Lactobacillus gasseri | Firmicutes | Vagina | Antibiotic |
| RiPP (lantibiotic) | epidermin | Staphylococcus epidermidis | Firmicutes | Skin | Antibiotic |
| RiPP (lantibiotic) | salivaricin A2 and B | Streptococcus salivarius | Firmicutes | Mouth | Antibiotic |
| RiPP (lantibiotic) | cytolysin | Enterococcus faecalis* | Firmicutes | Gut | Antibiotic, Cytotoxic |
| RiPP (lantibiotic) | ruminococcin A | Ruminococcus gnavus | Firmicutes | Gut | Antibiotic |
| RiPP (lantibiotic) | staphylococcin Au-26 (Bsa) | Staphylococcus aureus | Firmicutes | Skin | Antibiotic |
| RiPP (lantibiotic) | SA-FF22 | Streptoccous pyogenes | Firmicutes | Oral/Skin | Antibiotic |
| RiPP (bacteriocin) | ruminococcin C | Ruminococcus gnavus | Firmicutes | Gut | Antibiotic |
| RiPP (microcin) | microcin C7/C51 | Escherichia coli | Proteobacteria | Gut | Antibiotic |
| RiPP (microcin) | microcin B17 | Escherichia coli | Proteobacteria | Gut | Antibiotic |
| RiPP (microcin) | microcin J25 | Escherichia coli | Proteobacteria | Gut | Antibiotic |
| RiPP (microcin) | microcin H47 | Escherichia coli | Proteobacteria | Gut | Antibiotic |
| RiPP (TOMM) | streptolysin S | Streptoccous pyogenes | Firmicutes | Oral/Skin | Cytotoxic |
| RiPP (TOMM) | clostridiolysin S | Clostridium sporogenes | Firmicutes | Gut | Unknown |
| RiPP (TOMM) | listeriolysin S | Listeria monocytogenes | Firmicutes | Gut | Unknown |
| RiPP | heat-stable enterotoxin | Escherichia coli | Proteobacteria | Gut | GI motility (guanylate cyclase 2C) |
| Amino acid metabolite | indolepropionic acid | Clostridium sporogenes | Firmicutes | Gut | Immunomodulatory |
| Amino acid metabolite | indole | Unknown | Unknown | Gut | Converted to indoxyl sulfate |
| Amino acid metabolite | skatole | Clostridium spp. | Firmicutes | Gut | Unknown |
| Amino acid metabolite | tryptamine | Ruminococcus gnavus | Firmicutes | Gut | Neurotransmitter |
| Amino acid metabolite | phenyllactic acid | Bifidobacterium spp. | Actinobacteria | Gut | Unknown |
| Amino acid metabolite | phenethylamine | Lactobacillus spp. | Firmicutes | Gut | Neurotransmitter |
| Amino acid metabolite | δ-aminovaleric acid | Clostridium spp. | Firmicutes | Gut | Unknown |
| Amino acid metabolite | GABA | Unknown | Unknown | Gut | Unknown |
| Amino acid metabolite | α-aminobutyric acid | Unknown | Unknown | Gut | Unknown |
| Amino acid metabolite | 3-aminoisobutyric acid | Clostridium spp. | Firmicutes | Gut | Unknown |
| Amino acid metabolite | p-cresol | Clostridium spp. | Firmicutes | Gut | Unknown |
| Acid (short-chain) | propionic acid | Bacteroides spp. | Bacteroidetes | Gut | Immunomodulatory (GPR43) |
| Oligosaccharide | polysaccharide A | Bacteroides fragilis | Bacteroidetes | Gut | Immunomodulatory (TLR2) |
| Oligosaccharide | capsular polysaccharide | Streptococcus pneumoniae | Firmicutes | Airways | Immunomodulatory |
| Glycolipid | α-galactosylceramide | Bacteroides fragilis | Bacteroidetes | Gut | Immunomodulatory (CD1d) |
| Glycolipid | corynomycolic acid | Corynebacterium spp. | Actinobacteria | Skin | Unknown |
| Glycolipid | mycolic acid | Mycobacterium spp. | Actinobacteria | Airways | Immunomodulatory (CD1b) |
| Glycopeptide | muramyl di- and tripeptides | Fusobacterium nucleatum | Fusobacteria | Oral | Immunomodulatory (NOD1, NOD2) |
| Terpenoid | staphyloxanthin | Staphylococcus aureus | Firmicutes | Skin | Unknown (antioxidant?) |
| Terpenoid | bile acids (e.g., deoxycholic acid) | Clostridium spp. | Firmicutes | Gut | Metabomodulatory (TGR5, FXR, VDR) |
| NRP | phevalin | Staphylococcus aureus | Firmicutes | Skin | Unknown (virulence inducer?) |
| NRP | cereulide | Bacillus cereus* | Firmicutes | Gut | Cytotoxic, Immunomodulatory |
| NRP | yersiniabactin | Yersinia pestis* | Proteobacteria | Bloodstream | Siderophore |
| NRP | corynebactin | Corynebacterium spp. | Actinobacteria | Skin | Siderophore |
| NRP | tilivalline | Klebsiella oxytoca* | Proteobacteria | Gut | Cytotoxic |
| NRP-PK | zwittermicin | Bacillus cereus* | Firmicutes | Gut* | Antimicrobial |
| NRP-PK | mutanobactin | Streptococcus mutans | Firmicutes | Mouth | Unknown |
| NRP-PK | colibactin | Escherichia coli | Proteobacteria | Gut | Cytotoxic |
| PK | mycolactone | Mycobacterium ulcerans* | Actinobacteria | Skin | Immunomodulatory |
| Porphyrin | coproporphyrin III | Propionibacterium acnes | Actinobacteria | Skin | Unknown |
| Citrate amide | staphyloferrin B | Staphylococcus aureus | Firmicutes | Skin | Siderophore |
Microcins and thiazole/oxazole modified microcins (TOMMs)
Microcins are prototypical narrow-spectrum antibacterials. They contain a wide range of unusual post-translational modifications including, the conversion of cysteine and serine residues to thiazoles and oxazoles (microcin B17), the addition of adenosine monophosphate (microcin C7) or a siderophore to the C-terminus (microcin E492, Figure 2) and internal amide crosslinking forming a lasso-like topology (microcin J25) (35–38) . As they derive exclusively from enterobacteria and have potent (typically nM) antibacterial activity against close relatives of the producer (35) , their role in the Gram-negative microbiota is analogous to that of lantibiotics in the Gram-positive microbiota. Most microcins have been isolated from E. coli strains and are widely distributed in both commensal and pathogenic enterobacteria (35, 39–41).
Figure 2. Small-molecule mediated microbe-host and microbe-microbe interactions.
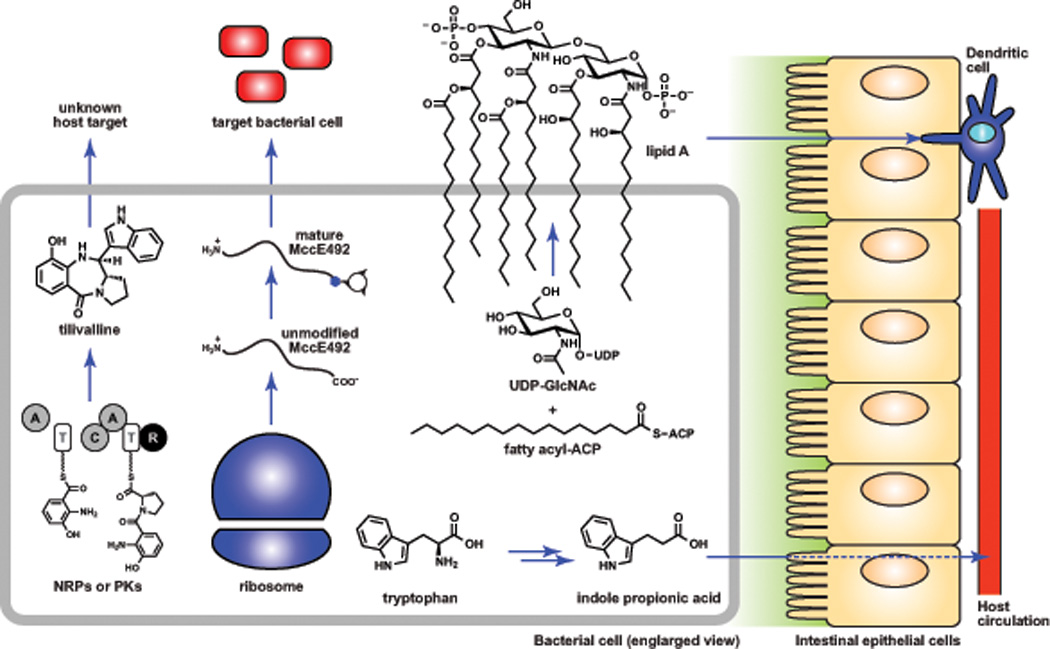
The microbiota produce a range of small molecules from various classes with distinct targets. Four examples are shown: the nonribosomal peptide tilivalline, whose host target is unknown; the ribosomally synthesized and post-translationally modified peptide microcin E492 (MccE492), a narrow spectrum antibacterial; lipid A, the glycolipid core of lipopolysaccharide, which targets TLR4 in host immune cells; and indole propionic acid, a reductive metabolite of tryptophan that enters host circulation but whose biological activity is poorly understood. These metabolites are each produced by different species of the microbiota, but are shown here in a single cell for schematic purposes. The following are abbreviations for domains in the nonribosomal peptide synthetase that produces tilivalline: A = adenylation domain, T = thiolation domain, C = condensation domain, R = terminal reductase domain.
TOMMs are similar to microcin B17 in their biosynthesis and post-translational modifications, but encompass a larger family of natural products from both Gram-positive and Gram-negative bacteria (17, 42). The best-studied example is streptolysin S from the human pathogen Streptococcus pyogenes (43) . Despite intensive efforts for almost a century, the precise chemical structure and mechanism of action of streptolysin S have not been fully determined (43). However, streptolysin S contains multiple oxazole and thiazole residues that are required for its hemolytic activity (44, 45). Related biosynthetic gene clusters from other human pathogens and commensals have been characterized (43), including listeriolysin S from Listeria monocytogenes (46) and clostridiolysin S from Clostridium botulinum and Clostridium sporogenes (47) .
Heat-stable enterotoxin
While most RiPPs from the human microbiota are thought to mediate microbe-microbe interactions, heat-stable enterotoxin is a RiPP produced by strains of E. coli associated with diarrheal disease and has a well-characterized host target. It is a 14-amino acid peptide stabilized by three internal disulfide bonds (48) and mimics the effect of the host peptide hormones guanylin and uroguanylin by agonizing guanylate cyclase 2C, a transmembrane protein expressed in intestinal epithelial cells with an extracellular ligand binding domain and a cytoplasmic catalytic domain (49) . Guanylate cyclase 2C generates cGMP to stimulate electrolyte secretion into the gut lumen. A single amino acid variant of heat-stable enterotoxin was approved by the FDA in 2012 for the treatment of constipation ssociated with irritable bowel syndrome (linaclotide, Figure 1) (50) . The enzyme that introduces disulfide crosslinks post-translationally into heat-stable enterotoxin is not encoded in its biosynthetic gene cluster, raising the question of whether the endogenous disulfide bond formation (Dsb) system is responsible, and whether other small peptides from enterobacteria undergo similar post-translational processing. Importantly, heat-stable enterotoxin indicates that a modified peptide can survive the proteolytic milieu of the gut lumen and target a host receptor expressed in intestinal epithelial cells, delivering a potent biological activity without absorption into host circulation.
Figure 1. Structurally diverse small molecules from the human microbiota.
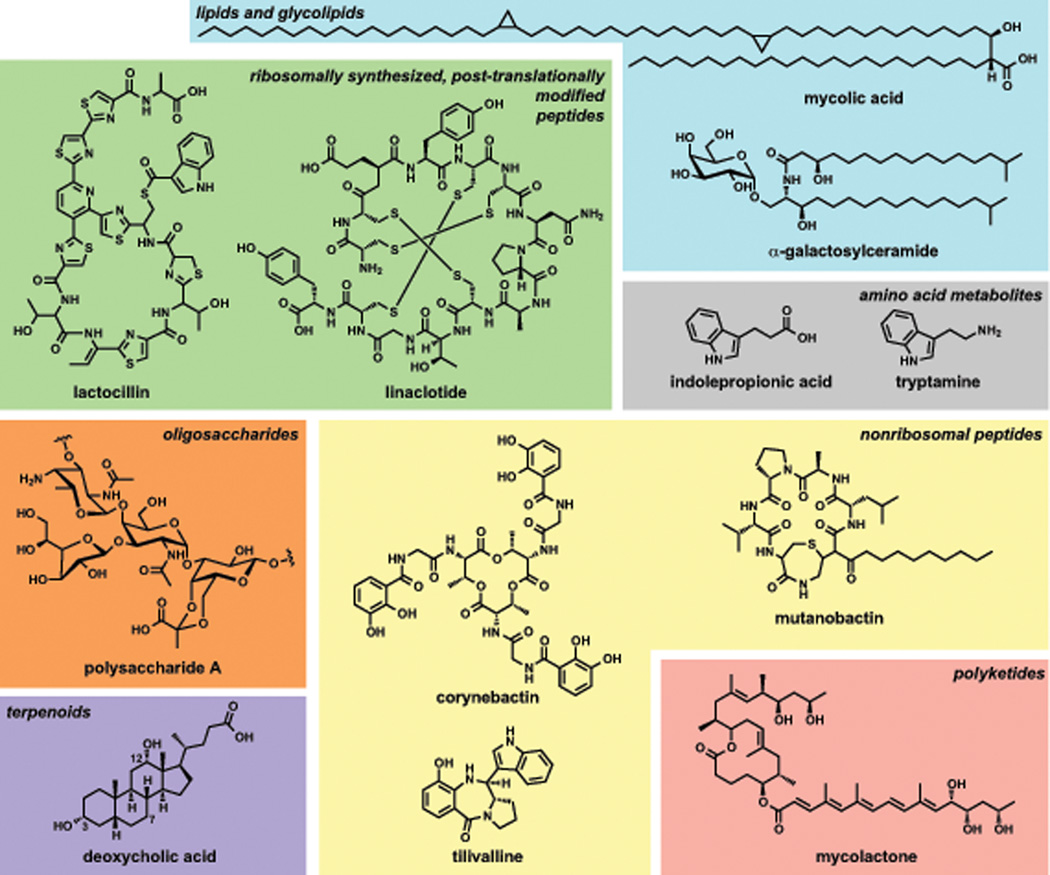
The diversity of chemical classes produced by the human microbiota rivals that of microorganisms from any ecological niche. Representative molecules are shown for each of the major molecular classes discussed: the RiPPs lactocillin and linaclotide; the amino acid metabolites indolepropionic acid and tryptamine; the oligosaccharide polysaccharide A; the lipids/glycolipids mycolic acid and α-galactosylceramide; the terpenoid deoxycholic acid, in which carbons 3, 7, and 12 of the bile acid scaffold are labeled; the nonribosomal peptides corynebactin, tilivalline, and mutanobactin; and the polyketide mycolactone.
Products of amino acid metabolism
Gut bacteria living in an anaerobic environment require an electron acceptor to drive fermentation or an anaerobic electron transport chain (51) . Commonly amino acids are used as electron acceptors, resulting in the production by gut bacteria of high levels (in some cases, >100 mg/day) of reductive amino acid metabolites, such as phenylpropionic acid and phenylacetic acid - molecules that are not found in most other habitats. Importantly, the amounts of these metabolites produced vary widely among individuals and, unlike RiPPs, they are generally permeable and accumulate systemically in the host (52). For example, humans with comparable levels of dietary tryptophan but distinct gut bacterial communities can end up with markedly different profiles of gut metabolites following microbiotal metabolism. One prominent tryptophan metabolite, indole, is derived from tryptophan by as-yet-unidentified enzyme(s) that are presumably homologs of tryptophanases seen in other bacterial species. In its unmodified form, indole serves as a signalling agent in bacterial communities (53). In addition, following absorption through the intestinal epithelium, indole is 3-hydroxylated and O-sulfated in the liver to become indoxyl sulfate, a well-known uremic toxin that is known from germ-free rodent studies to be derived entirely from the GI microbiota (54) . Indoxyl sulfate is present at a wide range of concentrations in human urine (10–200 mg/day), likely reflecting differences among individuals in diet and in the level of indole-producing bacterial species in the gut community (55) . A second reductive tryptophan metabolite, indolepropionic acid (Figure 2), is found in mouse serum if Clostridium sporogenes is present in the gut (52) . Although the function of this molecule is unknown, several other Clostridium spp. producers have been identified. A third tryptophan metabolite, the decarboxylation product tryptamine, is a biogenic amine neurotransmitter that is synthesized by a variety of gut bacteria (56) and has been linked to signaling in the enteric nervous system (57), one of several findings that reveal a role for the microbiota in the gut-brain axis (58–61). Thus, tryptophan can be diverted to end products with distinct biological activities depending on the composition of the gut community.
The metabolic products of aliphatic amino acids are equally prominent but less well characterized. Notable examples include δ-aminovaleric acid, which derives from arginine, proline, and ornithine, and acts as an electron source for secondary fermenters (62) ; and α-aminobutyric acid, which derives from threonine or methionine. Notably, the neurotransmitter γ-aminobutyric acid (GABA), the decarboxylation product of glutamate, is both produced and consumed by various species of gut bacteria (62) , although its potential role in microbe-host signaling remains unexplored. Many of the less common short-chain fatty acids, including isobutyric, valeric, 2- and 3-methylbutyric, caproic, and isocapropic acids, are also the products of reductive amino acid metabolism, but it is not known whether their signaling properties differ from those of the better known SCFAs (62) .
Oligosaccharides
Oligosaccharides provide some of the best-characterized examples of how small molecules from the human microbiota can mediate microbe-host interactions. Diffusible oligosaccharides are well known in the natural products community (63, 64) but the best-studied oligosaccharides from the human microbiota are cell-associated. Capsular polysaccharides from Bacteroides and Streptococcus show that capsular polysaccharides are not simply structural or nonspecifically adhesive, they can have highly specific ligand-receptor interactions that result in immune modulation, similarly to glycolipids (see below).
Species of Bacteroides, the most abundant bacterial genus in the human gut, produce an array of capsular polysaccharides, the best characterized of which is polysaccharide A from Bacteroides fragilis. Polysaccharide A is an oligomer in which the tetrasaccharide repeating unit consists of four derivatives of galactose: galactofuranose, N-acetylgalactosamine (GalNAc), 4,6- pyruvoylgalactose, and 4-amino-6-deoxy-GalNAc. Although the biosynthesis of polysaccharide A has just begun to be explored (65, 66) , its biological activity has been investigated in detail. Polysaccharide A signals through TLR2 to induce regulatory T cells to produce the tolerogenic cytokine IL-10. This signaling event restricts the activity of Th17 cells, promotes B. fragilis colonization and suppresses Helicobacter hepaticus-induced colitis (67, 68) . Remarkably little is known about the structures and biological activities of the tens to hundreds of other Bacteroides capsular polysaccharides, although they are likely to be the most abundant natural products in the human gut (10) .
Cell-associated oligosaccharides can also play a defensive role, as is the case with the richly diverse capsular polysaccharides elaborated by species of Streptococcus, including a family of capsular polysaccharides from group B Streptococcus, a pathogen (69) . Although the repeating unit varies in size, composition, and connectivity among group B Streptococcus serotypes, a common chemical feature is a terminal sialic acid, which blocks phagocytosis by inhibiting the deposition of the complement component C3b (69) .
An area of great promise in the microbiome is the identification of microbially derived ligands for host receptors. In addition to recent studies on the role of short-chain fatty acids and G-protein-coupled receptor 43 (GPR43) in regulatory T-cell function (70) , glycolipids and saccharides are significant candidate ligands. One example is muramyl dipeptide, a glycopeptide fragment of the repeating unit of peptidoglycan, which is a ligand for nucleotide-binding oligomerization domain-containing protein 2 (NOD2) and forms the scaffold for the immune -stimulatory osteosarcoma drug mifamurtide (Table 2) (71, 72) . Additionally, the large number of uncharacterized oligosaccharide biosynthetic loci in the human microbiome are particularly interesting in light of the C-type lectin receptors in the dectin/langerin/DC-SIGN family, which are known to bind oligosaccharides and modulate immune cell function, but for which few convincing ligands have been discovered (73) .
Glycolipids and Terpenoids
Perhaps the best known microbiota-derived molecule is lipopolysaccharide (LPS), a glycolipid that is a major component of the outer membrane of Gram-negative bacteria (Figure 2). LPS is the ligand for the innate immune receptor TLR4 and has been reviewed extensively elsewhere (74); here, we focus on two other families of bacterial glycolipids with similar immunomodulatory activities: the Bacteroides glycosphingolipid α-galactosylceramide and the mycolic acids of Mycobacterium and Corynebacterium.
Glycolipids
α-Galactosylceramide is a glycosphingolipid that was originally discovered nearly two decades ago as a natural product from a sponge and was later found to be a potent ligand for CD1d- restricted natural killer T (NKT) cells (75). Even though >1000 papers have been published on synthetic derivatives of this sphingolipid and the identification and stimulation of NKT cells, the source of the ‘native’ ligand for CD1d has remained a mystery (76). Recently, Bacteroides fragilis, a common gut commensal, was discovered to produce α-galactosylceramide (77) . Colonization of germ-free mice as neonates by wild-type B. fragilis suppress NKT cells in the gut and block oxazolone-induced colitis (78) . These findings suggest that the ‘native’ ligand for the highly conserved mammalian receptor CD1d might originate in the microbiota rather than the host. This may be true for other ‘orphan’ receptors expressed in immune and epithelial cells.
Mycolic acids are distinctive components of the cell wall of Mycobacterium (mostly pathogens) and Corynebacterium (both pathogens and skin and oral commensals). They consist of an all-carbon backbone with two lipid tails, one of which can be 40–60 carbons long in Mycobacterium, and are present both as free carboxylic acids and as esters of cell-wall polysaccharides. In addition to being important structural components of the capsule (79) , glucose monomycolate serves as a ligand for CD1b-restricted T cells, eliciting a specific immune response against infection (80–83) . Another glycolipid derivative of mycolic acid, trehalose-6,6-dimycolate, is a potent immune elicitor that binds to the C-type lectin Mincle to induce macrophage activation and a T-cell response characteristic of vaccination (84) . Mycolic acid has many chemical modifications, including methylation and cyclopropanation, both of which appear to shield it from immune detection: the MmaA4-catalyzed methylation of mycolic acid by M. tuberculosis blocks IL-12 production and prevents detection and elimination by macrophages (85) , and an M. tuberculosis mutant that is deficient in mycolic acid cyclopropanation is attenuated and hyperinflammatory in a mouse model of infection (86) .
Terpenoids
The major microbiota-derived terpenoids are not synthesized de novo by the microbiota; they are secondary bile acids, derivatives of the host-derived primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) (Figure 1). CA and CDCA are biosynthesized in the human liver, conjugated to taurine or glycine, and then excreted in bile; although 90% of the bile acid pool is absorbed in the terminal ileum, the remaining 10% enters the large intestine (87). Here, the bile acid pool reaches concentrations of ~1 mM and varies widely in composition among healthy humans. Numerous biochemical transformations of CA and CDCA are performed by gut bacteria, including deconjugation from taurine and glycine, oxidation and subsequent epimerization of the hydroxyl groups at C3, C7 and C12, dehydration and subsequent reduction of the hydroxyl group at C7, and esterification with ethanol at the C24 carboxylate. Among these, one modification – dehydroxylation at C7 – has been characterized most extensively (87–89).
Several species of Firmicutes (e.g., Clostridium scindens and Clostridium hylemonae) dehydroxylate CA and CDCA at the C7 position to form deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. The high flux of this biochemical transformation results in DCA and LCA making up nearly two-thirds of the fecal bile acid pool (87) . Both DCA and LCA are toxic to human cells and have been implicated in hepatoxicity and colon cancer (90). 7-Dehydroxylation is carried out by the 8-gene bai operon. This gene cluster is thought to perform eight successive chemical transformations in a pathway for which the early oxidative steps have been characterized biochemically, but the later, reductive steps remain speculative (89) . Little is known about the biosynthetic genes for other secondary bile acids, although there is preliminary evidence that some bile acid pathways might be collaborative, in the sense that they involve transformations by more than one gut bacterial species.
The carotenoids are terpenoids, exemplified by staphyloxanthin, the golden-coloured pigment for which Staphylococcus aureus is named. Staphyloxanthin is composed of a glucose residue that is esterified with both a fatty acid (12-methyltetradecanoate) at the C6” position and a carotenoid (4,4’-diaponeurosporen-4-oate) at the C1” position (91) . The core structure of staphyloxanthin is assembled by two biosynthetic enzymes: a glycosyltransferase, which esterifies the C1” position of glucose, and an acyltransferase, which esterifies its C6” position. The unusual 4,4’-diaponeurosporen-4-oate originates from dehydrosqualene by further dehydrogenation and oxidation steps (92, 93) . The conjugated double bonds of staphyloxanthin’s carotenoid tail serve as a ‘sponge’ for oxygen radicals, protecting S. aureus against killing by hydrogen peroxide, superoxide and hydroxyl radical, which are produced by host neutrophils and macrophages (94, 95) .
Polyketides and Nonribosomal Peptides
Although polyketides (PKs) and nonribosomal peptides (NRPs) are among the largest classes of natural products in soil and aquatic bacteria, relatively few are known from human-associated bacteria. Two recently discovered examples come from the common pathobionts Staphylococcus aureus and Streptococcus mutans (Table 1 and Figure 1). A conserved NRPS gene cluster in S. aureus encodes a family of pyrazinones, which are derivatives of the ubiquitous diketopiperazines (96, 97). Although the role of the pyrazinones in regulating the expression of S. aureus virulence factors remains unclear (96, 98), they are unlikely to function exclusively in the context of pathogenesis since they are also produced by the skin commensal Staphylococcus epidermidis (97) . Isolates of S. mutans, the leading cause of dental caries, harbor a genomic island encoding hybrid PKS/NRPS pathways (99) . The product of one of these pathways, mutanobactin, contains an unusual 1,4-thiazepan-5-one ring, which originates from the cyclization of a cysteine and a glycine residue. The biological activity of the mutanobactins has not been fully determined, but may involve modulating growth and biofilm formation by the fungal pathogen Candida albicans (100–102) .
Four pathogen-derived NRPs and PKs cause disease: cereulide, mycolactone, colibactin, and tilivalline. Cereulide, a dodecadepsipeptide toxin, is responsible for the emetic effects of the food-poisoning pathogen Bacillus cereus (103, 104). The ester bonds in cereulide’s alternating ester/amide backbone enable the molecule to have high affinity for potassium ions, which causes mitochondrial toxicity and results in uncoupling of oxidative phosphorylation (105, 106).
Mycolactones are PK toxins produced by the causative agent of Buruli ulcer, Mycobacterium ulcerans (107, 108). These molecules, which cause the necrosis, ulceration, and immune suppression associated with this disease, are encoded by a >100 kb type I PKS biosynthetic gene cluster that includes two genes >40 kb. Interestingly, a heterogeneous suite of mycolactone derivatives is produced by different strains of M. ulcerans, which may explain the variation observed in the virulence of the strains and their geographical distribution among affected countries (107, 109–112).
Colibactin is produced by a subset of enterobacteria, including strains of E. coli B2, Enterobacter aerogenes, Klebsiella pneumoniae, and Citrobacter koseri (113, 114) . Exposure of mammalian cells to colibactin producing E. coli and K. pneumoniae induces DNA damage in vitro and in vivo. Surprisingly, the colibactin gene cluster is present in one of the most commonly used probiotic E. coli strains (E. coli Nissle 1917 or EcN) (114–118). Considerable efforts have been made to study the biosynthesis of colibactin (119–121), and its chemical structure has recently been characterized, revealing a unique spirocyclopropane warhead that crosslinks DNA (122). It is not yet clear what role colibactin plays in the ecology of the interaction between E. coli and the host, and how the genotoxic activity of colibactin benefits its producer.
Tilivalline is an NRP toxin produced by colitogenic strains of the pathobiont Klebsiella oxytoca (Figure 2) (123) . Importantly, tilivalline is essential for the inflammatory pathology characteristic of antibiotic-associated hemorrhagic colitis, and induces apoptosis in cultured human epithelial cells. Although the discovery of tilivalline sheds light on one mechanism of antibiotic-induced colitis, there are likely to be alternative mechanisms, since K. oxytoca is present in, at most, 10% of the healthy human population (124).
Much is known about the biosynthesis of NRPS-derived and NRPS-independent siderophores in a broad range of human pathogens, and their role in iron acquisition as an essential component of bacterial pathogenesis, both of which have been reviewed extensively elsewhere (125–127). In contrast, very little is known about the mechanisms by which commensals acquire iron and, to our knowledge, no iron acquisition system has ever been shown to be required for colonization by a commensal.
OUTLOOK
We have used ClusterFinder (7) to identify biosynthetic gene clusters in the human microbiome as a way to assess the metabolic potential of the human microbiota (10). Of >14,000 putative natural product biosynthetic gene clusters identified in human-associated bacterial genomes, 3,118 were present in one or more of the 752 whole-genome shotgun metagenomic sequence samples from the NIH Human Microbiome Project (HMP). Although each of the major natural product classes is produced by the human microbiota, oligosaccharide and RiPP gene clusters are predominant, underscoring the need to improve analytical chemical techniques to purify and assay these molecules. Nearly all of the gene clusters that were present in >10% of the subjects in the study are uncharacterized, highlighting the potential of studying these genetic elements and the small molecules they encode.
There are two central challenges facing the field: First, from the wealth of microbiota-derived molecules, which ones are the functionally ‘important’ ones? Second, what experimental systems are appropriate for testing the activity of an individual molecule from a complex milieu?
Identifying significant microbiota-derived molecules
Human-associated microbial communities can consist of hundreds of abundant bacterial species and perhaps thousands of molecules at physiologically relevant concentrations. Figuring out which of these molecules drive a phenotype and how they act requires new computational and experimental approaches.
Initial mapping of metagenomic sequence data onto coarse KEGG or COG gene categories provides a way of seeing coarse changes, e.g., a shift from oligosaccharide toward amino acid catabolism in a community. Resolution is not yet high enough to make reliable predictions about specific biosynthetic pathways or products. Methods that predict the gene content of a sample from 16S data can predict pathways that are present or absent in every member of an operational taxonomic unit (OTU) (128) , but have limited utility for biosynthetic pathways, which are highly variable even among closely related strains of a bacterial species (129, 130) .
Methods are needed that take genomic and metagenomic sequence data as an input and use it to predict, at high resolution, pathways for specific molecules. Multiple algorithms will likely be needed: some for identifying clustered biosynthetic pathways, characteristic of a conventional secondary metabolite, and others for predicting unclustered pathways more commonly found in primary metabolism. The dearth of knowledge about primary metabolic pathways in anaerobes from the gut community is a critical gap in current knowledge, and addressing this problem will be a major achievement.
Nicholson, Holmes and colleagues have pioneered the use of metabolomics to profile microbiota-derived metabolites in a variety of sample types and disease models, developing powerful and widely applicable analytical pipelines (131, 132) (Figure 3a). Using similar approaches, a range of microbiota-derived molecules have been connected to specific bacterial species through the metabolomic profiling of various artificial and disease- associated communities from mice (52, 59, 133, 134). However, although metabolomic profiling is capable of measuring hundreds to thousands of known metabolites in a single run, its application (known as untargeted metabolomics) to discovering molecules of interest is more laborious and generally requires purification and structural characterization of milligram quantities of compound.
Figure 3. Approaches to discovering small molecules from the microbiota.
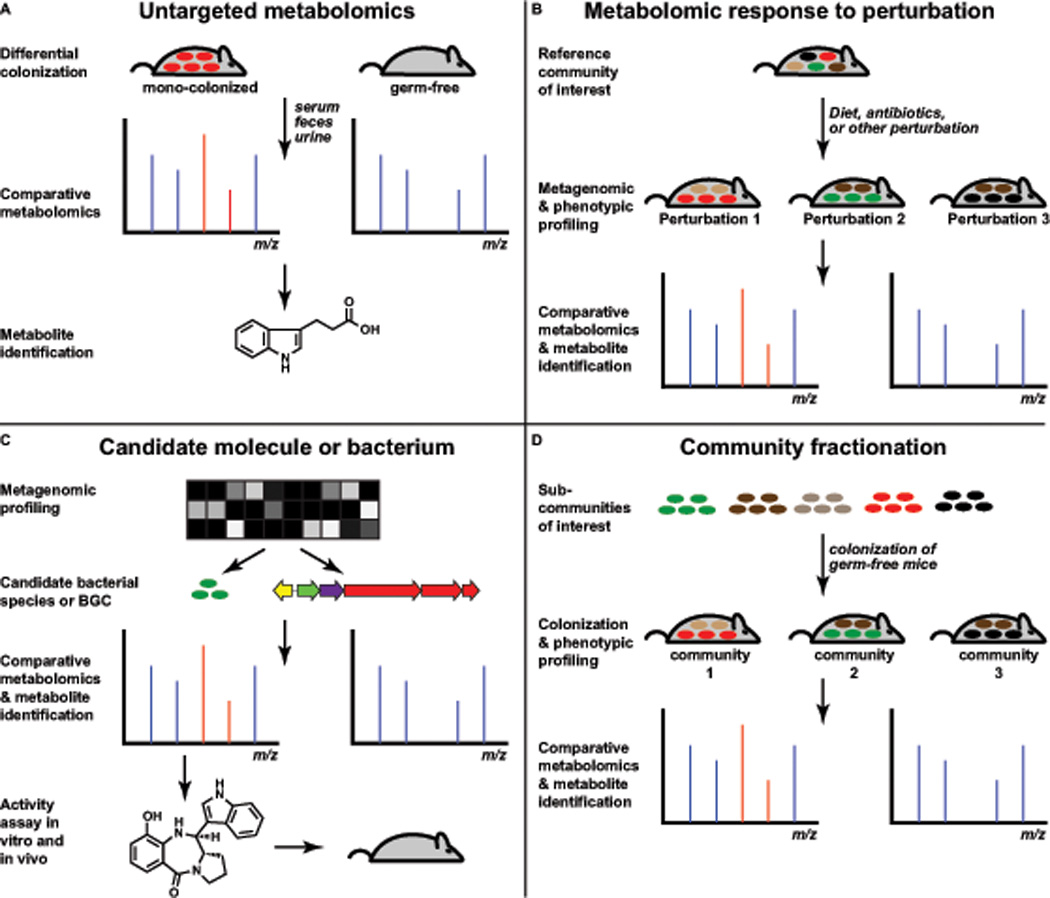
(A) Samples from germ-free and colonized mice can be analyzed by untargeted metabolomics to identify molecules that are present in a microbiota-dependent fashion. (B) A mouse harboring a reference gut community can be subjected to antibiotic treatment, a dietary shift, or another perturbation. Comparative metabolomics can be used to identify microbiota-derived molecules whose abundance changes as a consequence of the perturbation. (C) Candidate bacterial gene clusters or bacterial species can be selected by metagenomic profiling (e.g., for gene clusters or species that are widely distributed, or differ in abundance between cases and controls). Comparative metabolomics can then be used to identify molecules produced by a gene cluster or bacterial species of interest. (D) Subsets of bacteria from a fractionated complex community or designed synthetic communities can be used to colonize mice in order to identify specific bacterial species whose presence correlates with the production of a molecule of interest.
Although bioassay-guided fractionation is immensely powerful, it is painstaking and difficult to scale, so it is better suited for unusually important phenotypes of interest, including the search for ligands for orphan G-protein coupled receptors (GPCRs) expressed in the gut (Figure 3b). A derivative of this approach in which microbes, not molecules, are “fractionated” was recently used to identify a cocktail of 17 gut bacterial strains that induce regulatory T cells and attenuate colitis. This activity was further correlated with short-chain fatty acids produced by this anti-inflammatory cocktail of bacteria (135, 136) .
An alternative to bioassay-guided fractionation is the candidate molecule approach (Figure 3C). We used this method for the discovery of the potent thiopeptide antibiotic, lactocillin, from Lactobacillus gasseri, a prominent member of the vaginal community (10, 137). Initially, we made a systematic analysis of all biosynthetic gene clusters for small molecules in genomes of human-associated bacteria and identified 13 novel thiopeptide gene clusters, four of which are present in >20% of the HMP samples at one of the body sites. Thiopeptides are a class of antibiotics with potent activity against Gram-positive bacteria that bind to a site on the 50S subunit of the bacterial ribosome. One member of this class, LFF571, is currently in a phase II clinical trial (138). Lactocillin has been purified, structurally characterized, and shown to have low-to-mid nanomolar antibiotic activity against vaginal pathogens but not against vaginal commensals.
Another class of molecules ideally suited to the candidate molecule approach is secondary bile acids: bile acids, and the sterol scaffold more generally, are rich in biological activity and there are numerous host receptors for these molecules (87). The levels of secondary bile acids vary widely among individuals, although the bile acid pool in the gut lumen is held at a physiologically relevant concentrations. Although dozens of secondary bile acids are known, very few have been assigned a biological activity or have known biosynthetic genes, making this a promising area for detailed experimental investigation.
Studying individual molecules from a pool
The discoveries of highest impact will come not from simply cataloging new microbiota- derived molecules, but from studying the biological activities of individual molecules in a complex molecular milieu. With the exception of fecal metabolite profiling, few microbiota-derived molecules have been detected in host-derived samples. More sensitive analytical and synthetic techniques (e.g., nanospray desorption electrospray ionization (nanoDESI) mass spectrometry) are needed to verify the production of a molecule in the skin, oral, and vaginal communities. An alternative approach – the detection of RNA transcripts for a particular bacterial gene cluster in metatranscriptomic data – has been used to profile the expression of the cluster in a native sample. This approach is consistent with, but not proof of, small molecule production in the ‘native’ habitat of the microbiome (10).
Although preliminary studies to study the biological effects of target molecules have been carried out for three microbiota-derived RiPPs (139–143), their biological relevance have not yet been tested experimentally. Although colonization studies in gem-free mice have the advantage of simplicity, there are drawbacks: (i) a molecule derived from a mono-colonist could be produced at super-physiological levels, leading to a false signal; (ii) the activity of some molecules, e.g., immune modulators, may require the activity of co-stimulatory signals from other bacterial species; and (iii) the biological relevance of certain molecules, e.g. antibacterials, may only be observed when other members of the microbiota are present. Faith et al. have overcome some of these challenges in studies on regulatory T cells in the gut, by using small subsets of gut bacterial strains to colonize mice (Figure 3d) (144) .
Similar experimental systems have recently been developed for the skin. Conventional mice can be colonized by individual skin commensals and the T-cell response can be tracked over long time periods (145) . By contrast, few experimental systems are available for interrogating the role of small- molecule-mediated interactions in the community structure (146) and dynamics (137) of oral and vaginal communities.
Many molecules of interest are produced by bacterial species that are difficult to manipulate genetically, such as the anaerobic Firmicutes (Clostridium and its relatives). Two technologies would be transformative for their study: gene knock out and expression (147) , and synthetic-biology-based approaches to ‘refactor’ biosynthetic gene clusters and express them in a more genetically tractable host (148) .
Conclusion
Much is known about which bacterial species are most abundant in human-associated communities and how they vary among individuals. Yet comparatively little is known about the most abundant bacterially derived or modified small molecules in the gut, despite the fact that these molecules are present at high micromolar concentration, their levels can vary greatly among individuals, and the human host is chronically exposed to them for decades. Certain low- abundance molecules with potent biological activities may also be significant. Against this backdrop, it seems likely that in the near future the suite of microbiota- derived molecules in an individual’s gut community will not be left to chance. Pharmaceutical companies go to great lengths to get a single molecule into the human gut at a comparable concentration. Discovering the most abundant, widely (or variably) distributed, and biologically active molecules produced by the microbiota – and connecting them to the genes that encode them – are critical first steps in understanding which molecules have desired effects and which are deleterious, what receptors they target, and how therapeutic communities of microorganisms can be designed in which the production and non-production of molecules can be genetically specified.
ONE-PAGE SUMMARY.
Background
Two developments in distinct fields are converging to create interest in discovering natural products from the human microbiome. First, the use of genomics to guide natural product discovery has led to the unexpected discovery of numerous biosynthetic gene clusters in genomes of the human microbiota. Second, the microbiome research community is moving from a focus on ‘who’s there’ to ‘what are they doing’, with an accompanying emphasis on understanding microbiota-host interactions at the level of molecular mechanism. This merger has sparked a concerted hunt for the mediators of microbe-host and microbe-microbe interactions, including microbiota-derived small molecules.
Advances
Numerous small molecules are known that are produced by the human microbiota. The microbiota-derived ribosomally synthesized, post-translationally modified peptides (RiPPs) include widely distributed lantibiotics and microcins with narrow-spectrum activity that are presumptive mediators of interactions among closely related species, and Escherichia coli heat-stable enterotoxin, a guanylate cyclase 2C agonist from which the recently approved GI motility drug linaclotide was derived. Fewer amino acid metabolites are synthesized by the microbiota, but they are produced at very high levels that vary widely among individuals (e.g., indoxyl sulfate at 10–200 mg/day). Gut bacterial species convert common dietary amino acids into distinct end products, such as tryptophan to indoxyl sulfate, indole propionic acid and tryptamine – indicating that humans with the same diet but different gut colonists can have widely varying gut metabolic profiles. Microbially produced oligosaccharides differ from other natural products because they are cell-associated (i.e., non-diffusible) and because many more biosynthetic loci exist for them than for other small molecule classes. Well-characterized examples, such as Bacteroides polysaccharide A, show that oligosaccharides may not simply play a structural role or mediate adhesion; rather, they can be involved in highly specific ligand-receptor interactions that result in immune modulation. Similarly, the (glyco)lipids α-galactosylceramide and mycolic acid can play roles in immune signaling. The most prominent microbiota-derived terpenoids are microbial conversion products of the cholic acid and chenodeoxycholic acid in host bile. These secondary bile acids can reach high concentration (mM) in the gut and vary widely in composition among individuals. Several canonical virulence factors from pathogens are derived from nonribosomal peptides (NRPs) and polyketides (PKs), but less is known about NRPs and PKs from the commensal microbiota. A recent computational effort has identified ~14,000 biosynthetic gene clusters in sequenced genomes from the human microbiota, 3,118 of which were present in one or more of the 752 metagenomic sequence samples from the NIH Human Microbiome Project. Nearly all of the gene clusters that were present in >10% of the samples from the body site of origin are uncharacterized, highlighting the potential for identifying the molecules they encode and studying their biological activities.
Outlook
There are two central challenges facing the field. The first is to distinguish, from among thousands of microbiota-derived molecules, which are the ‘important’ ones, i.e., the ones that drive a key phenotype at physiologically relevant concentrations? Second, which experimental systems are appropriate for testing the activity of an individual molecule from a complex milieu? Meeting these challenges will require the development of new computational and experimental technologies, including a capacity to identify biosynthetic genes and predict the structure and target of their biological activity, and new systems in which germ-free mice are colonized by mock communities that differ only by the presence/absence of a biosynthetic gene cluster.
Acknowledgements
We thank members of the Fischbach and Donia groups for helpful discussions. Work in the authors’ laboratories is supported by Princeton University (M.S.D), a Medical Research Program Grant from the W.M. Keck Foundation (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.), an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Foundation (M.A.F.), DARPA award HR0011-12-C-0067 (M.A.F.), the Program for Breakthrough Biomedical Research (M.A.F.), and NIH grants OD007290, AI101018, GM081879, and DK101674 (M.A.F.). M.A.F. is on the scientific advisory boards of NGM Biopharmaceuticals and Warp Drive Bio.
References and Notes
- 1.Margulis L, Fester R. Symbiosis as a source of evolutionary innovation : speciation and morphogenesis. Cambridge, Mass.: MIT Press; 1991. pp. xii–454. [PubMed] [Google Scholar]
- 2.Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Annual review of genetics. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 3.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annual review of genetics. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 4.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature reviews. Microbiology. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 5.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woyke T, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 7.Cimermancic P, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blin K, et al. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic acids research. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic acids research. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long SR. Genes and signals in the rhizobium-legume symbiosis. Plant physiology. 2001;125:69–72. doi: 10.1104/pp.125.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Ruyter-Spira C, Bouwmeester HJ. Engineering the plant rhizosphere. Current opinion in biotechnology. 2015;32:136–142. doi: 10.1016/j.copbio.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Piel J. Metabolites from symbiotic bacteria. Natural product reports. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 14.Brachmann AO, Bode HB. Identification and bioanalysis of natural products from insect symbionts and pathogens. Advances in biochemical engineering/biotechnology. 2013;135:123–155. doi: 10.1007/10_2013_192. [DOI] [PubMed] [Google Scholar]
- 15.Crawford JM, Clardy J. Bacterial symbionts and natural products. Chemical communications. 2011;47:7559–7566. doi: 10.1039/c1cc11574j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nature chemical biology. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 17.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Natural product reports. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Heel AJ, Montalban-Lopez M, Kuipers OP. Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert opinion on drug metabolism & toxicology. 2011;7:675–680. doi: 10.1517/17425255.2011.573478. [DOI] [PubMed] [Google Scholar]
- 19.Barbour A, Philip K, Muniandy S. Enhanced production, purification, characterization and mechanism of action of salivaricin 9 lantibiotic produced by Streptococcus salivarius NU10. PloS one. 2013;8:e77751. doi: 10.1371/journal.pone.0077751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross KF, Ronson CW, Tagg JR. Isolation and Characterization of the Lantibiotic Salivaricin A and its Structural Gene salA from Streptococcus salivarius 20P3. Applied and environmental microbiology. 1993;59:2014–2021. doi: 10.1128/aem.59.7.2014-2021.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wescombe PA, et al. Salivaricin G32, a Homolog of the Prototype Streptococcus pyogenes Nisin-Like Lantibiotic SA-FF22, Produced by the Commensal Species Streptococcus salivarius. International journal of microbiology. 2012;2012:738503. doi: 10.1155/2012/738503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wescombe PA, et al. Production of the lantibiotic salivaricin A and its variants by oral streptococci and use of a specific induction assay to detect their presence in human saliva. Applied and environmental microbiology. 2006;72:1459–1466. doi: 10.1128/AEM.72.2.1459-1466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nascimento JS, et al. Bacteriocins as alternative agents for control of multiresistant staphylococcal strains. Letters in applied microbiology. 2006;42:215–221. doi: 10.1111/j.1472-765X.2005.01832.x. [DOI] [PubMed] [Google Scholar]
- 24.Ekkelenkamp MB, et al. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS letters. 2005;579:1917–1922. doi: 10.1016/j.febslet.2005.01.083. [DOI] [PubMed] [Google Scholar]
- 25.Heidrich C, et al. Isolation, Characterization, and Heterologous Expression of the Novel Lantibiotic Epicidin 280 and Analysis of Its Biosynthetic Gene Cluster. Applied and environmental microbiology. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahl HG, Brandis H. Production, Purification and Chemical Properties of an Antistaphylococcal Agent Produced by Staphylococcus epidemtidis. J Gen Microbiol. 1981;127:377–384. doi: 10.1099/00221287-127-2-377. [DOI] [PubMed] [Google Scholar]
- 27.Velasquez JE, Zhang X, van der Donk WA. Biosynthesis of the antimicrobial peptide epilancin 15X and its N-terminal lactate. Chemistry & biology. 2011;18:857–867. doi: 10.1016/j.chembiol.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dabard J, Ruminococcin A, et al. a New Lantibiotic Produced by a Ruminococcus gnavus Strain Isolated from Human Feces. Applied and environmental microbiology. 2001;67:4111–4118. doi: 10.1128/AEM.67.9.4111-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcille F, et al. Distribution of Genes Encoding the Trypsin-Dependent Lantibiotic Ruminococcin A among Bacteria Isolated from Human Fecal Microbiota. Applied and environmental microbiology. 2002;68:3424–3431. doi: 10.1128/AEM.68.7.3424-3431.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott JC, Sahl HG, Carne A, Tagg JR. Lantibiotic-mediated anti-lactobacillus activity of a vaginal Staphylococcus aureus isolate. FEMS Microbiol Lett. 1992;93:97–102. doi: 10.1016/0378-1097(92)90496-b. [DOI] [PubMed] [Google Scholar]
- 31.Daly KM, et al. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. Journal of bacteriology. 2010;192:1131–1142. doi: 10.1128/JB.01375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jack RW, Elucidation of the structure of SA-FF22, et al. a lanthionine-containing antibacterial peptide produced by Streptococcus pyogenes strain FF22. Eur J Biochem. 1994;220:455–462. doi: 10.1111/j.1432-1033.1994.tb18643.x. [DOI] [PubMed] [Google Scholar]
- 33.Tagg JR, Wannamaker LW. Streptococcin A-FF22: Nisin-Like Antibiotic Substance Produced by a Group A Streptococcus. Antimicrobial agents and chemotherapy. 1978;14:31–39. doi: 10.1128/aac.14.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coburn PS, Gilmore MS. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cellular Microbiology. 2003;5:661–669. doi: 10.1046/j.1462-5822.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 35.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Natural product reports. 2007;24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 36.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 37.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. Journal of the American Chemical Society. 2008;130:3603–3609. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 38.Nolan EM, Fischbach MA, Koglin A, Walsh CT. Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. Journal of the American Chemical Society. 2007;129:14336–14347. doi: 10.1021/ja074650f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Micenkova L, et al. Bacteriocin-encoding genes and ExPEC virulence determinants are associated in human fecal Escherichia coli strains. BMC microbiology. 2014;14:109. doi: 10.1186/1471-2180-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smajs D, et al. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC microbiology. 2010;10:288. doi: 10.1186/1471-2180-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zschuttig A, et al. Identification and characterization of microcin S, a new antibacterial peptide produced by probiotic Escherichia coli G3/10. PloS one. 2012;7:e33351. doi: 10.1371/journal.pone.0033351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Current opinion in chemical biology. 2011;15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molloy EM, Cotter PD, Hill C, Mitchell DA, Ross RP. Streptolysin S-like virulence factors: the continuing sagA. Nature reviews. Microbiology. 2011;9:670–681. doi: 10.1038/nrmicro2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SW, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell DA, et al. Structural and functional dissection of the heterocyclic peptide cytotoxin streptolysin S. The Journal of biological chemistry. 2009;284:13004–13012. doi: 10.1074/jbc.M900802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cotter PD, Listeriolysin S, et al. a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS pathogens. 2008;4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez DJ, Clostridiolysin S, et al. a post-translationally modified biotoxin from Clostridium botulinum. The Journal of biological chemistry. 2010;285:28220–28228. doi: 10.1074/jbc.M110.118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozaki H, et al. Molecular structure of the toxin domain of heat-stable enterotoxin produced by a pathogenic strain of Escherichia coli. A putative binding site for a binding protein on rat intestinal epithelial cell membranes. The Journal of biological chemistry. 1991;266:5934–5941. [PubMed] [Google Scholar]
- 49.Schulz S, Green CK, Yuen PS, Garbers DL. Guanylyl cyclase is a heat-stable enterotoxin receptor. Cell. 1990;63:941–948. doi: 10.1016/0092-8674(90)90497-3. [DOI] [PubMed] [Google Scholar]
- 50.McWilliams V, Whiteside G, McKeage K. Linaclotide: first global approval. Drugs. 2012;72:2167–2175. doi: 10.2165/11470590-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 51.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. Fems Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 54.Aronov PA, et al. Colonic contribution to uremic solutes. Journal of the American Society of Nephrology : JASN. 2011;22:1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel KP, Luo FJ, Plummer NS, Hostetter TH, Meyer TW. The production of p-cresol sulfate and indoxyl sulfate in vegetarians versus omnivores. Clinical journal of the American Society of Nephrology : CJASN. 2012;7:982–988. doi: 10.2215/CJN.12491211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams BB, et al. Discovery and characterization of two tryptamine-producing decarboxylases from the gut microbiota. Cell host & microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takaki M, Mawe GM, Barasch JM, Gershon MD, Gershon MD. Physiological responses of guinea-pig myenteric neurons secondary to the release of endogenous serotonin by tryptamine. Neuroscience. 1985;16:223–240. doi: 10.1016/0306-4522(85)90059-4. [DOI] [PubMed] [Google Scholar]
- 58.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. The Journal of clinical investigation. 2015;125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burokas A, Moloney RD, Dinan TG, Cryan JF. Microbiota regulation of the Mammalian gut-brain axis. Advances in applied microbiology. 2015;91:1–62. doi: 10.1016/bs.aambs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 62.Barker HA. Amino acid degradation by anaerobic bacteria. Annual review of biochemistry. 1981;50:23–40. doi: 10.1146/annurev.bi.50.070181.000323. [DOI] [PubMed] [Google Scholar]
- 63.Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Natural product reports. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 64.McCranie EK, Bachmann BO. Bioactive oligosaccharide natural products. Natural product reports. 2014;31:1026–1042. doi: 10.1039/c3np70128j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mostafavi AZ, Troutman JM. Biosynthetic assembly of the Bacteroides fragilis capsular polysaccharide A precursor bactoprenyl diphosphate-linked acetamido-4-amino-6-deoxygalactopyranose. Biochemistry. 2013;52:1939–1949. doi: 10.1021/bi400126w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Troutman JM, Sharma S, Erickson KM, Martinez CD. Functional identification of a galactosyltransferase critical to Bacteroides fragilis Capsular Polysaccharide A biosynthesis. Carbohydrate research. 2014;395C:19–28. doi: 10.1016/j.carres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 68.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cieslewicz MJ, et al. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infection and immunity. 2005;73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Girardin SE, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 72.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. The Journal of biological chemistry. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 73.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nature reviews. Immunology. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annual review of biochemistry. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 75.Akimoto K, Natori T, Morita M. Synthesis and stereochemistry of agelasphin-9b. Tetrahedron Lett. 1993;34:5593–5596. [Google Scholar]
- 76.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annual review of immunology. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 77.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS biology. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marrakchi H, Laneelle MA, Daffe M. Mycolic acids: structures, biosynthesis, and beyond. Chemistry & biology. 2014;21:67–85. doi: 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 80.Layre E, et al. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chemistry & biology. 2009;16:82–92. doi: 10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 81.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nature immunology. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 82.Moody DB, et al. Structural requirements for glycolipid antigen recognition by CD1b–restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 83.Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nature immunology. 2013;14:706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. The Journal of experimental medicine. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dao DN, et al. Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS pathogens. 2008;4:e1000081. doi: 10.1371/journal.ppat.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barkan D, Hedhli D, Yan HG, Huygen K, Glickman MS. Mycobacterium tuberculosis lacking all mycolic acid cyclopropanation is viable but highly attenuated and hyperinflammatory in mice. Infection and immunity. 2012;80:1958–1968. doi: 10.1128/IAI.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. Journal of lipid research. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 88.Gérard P. Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathogens. 2013;3:14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ridlon JM, Kang DJ, Hylemon PB. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marshall JH, Wilmoth GJ, Pigments of Staphylococcus aureus. a series of triterpenoid carotenoids. Journal of bacteriology. 1981;147:900–913. doi: 10.1128/jb.147.3.900-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pelz A, et al. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. The Journal of biological chemistry. 2005;280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 93.Wieland B, et al. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4’-diaponeurosporene of Staphylococcus aureus. Journal of bacteriology. 1994;176:7719–7726. doi: 10.1128/jb.176.24.7719-7726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clauditz A, Resch A, Wieland KP, Peschel A, Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infection and immunity. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Olivier AC, Lemaire S, Van Bambeke F, Tulkens PM, Oldfield E. Role of rsbU and staphyloxanthin in phagocytosis and intracellular growth of Staphylococcus aureus in human macrophages and endothelial cells. The Journal of infectious diseases. 2009;200:1367–1370. doi: 10.1086/606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wyatt MA, et al. Staphylococcus aureus nonribosomal peptide secondary metabolites regulate virulence. Science. 2010;329:294–296. doi: 10.1126/science.1188888. [DOI] [PubMed] [Google Scholar]
- 97.Zimmermann M, Fischbach MA. A family of pyrazinone natural products from a conserved nonribosomal peptide synthetase in Staphylococcus aureus. Chemistry & biology. 2010;17:925–930. doi: 10.1016/j.chembiol.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 98.Sun F, et al. Aureusimines in Staphylococcus aureus are not involved in virulence. PloS one. 2010;5:e15703. doi: 10.1371/journal.pone.0015703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu C, et al. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Applied and environmental microbiology. 2010;76:5815–5826. doi: 10.1128/AEM.03079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Joyner PM, et al. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Organic & biomolecular chemistry. 2010;8:5486–5489. doi: 10.1039/c0ob00579g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sztajer H, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. The ISME journal. 2014;8:2256–2271. doi: 10.1038/ismej.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X, Du L, You J, King JB, Cichewicz RH. Fungal biofilm inhibitors from a human oral microbiome-derived bacterium. Organic & biomolecular chemistry. 2012;10:2044–2050. doi: 10.1039/c2ob06856g. [DOI] [PubMed] [Google Scholar]
- 103.Agata N, Ohta M, Mori M, Isobe M. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol Lett. 1995;129:17–20. doi: 10.1016/0378-1097(95)00119-P. [DOI] [PubMed] [Google Scholar]
- 104.Suwan S, Structure of cereulide, et al. a cyclic dodecadepsipeptide toxin from Bacillus cereus and studies on NMR characteristics of its alkali metal complexes including a conformational structure of the K+ complex. J Chem Soc Perkin Trans. 1995;1:765–775. [Google Scholar]
- 105.Ehling-Schulz M, et al. Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC microbiology. 2006;6:20. doi: 10.1186/1471-2180-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teplova VV, Mikkola R, Tonshin AA, Saris NE, Salkinoja-Salonen MS. The higher toxicity of cereulide relative to valinomycin is due to its higher affinity for potassium at physiological plasma concentration. Toxicology and applied pharmacology. 2006;210:39–46. doi: 10.1016/j.taap.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 107.Kishi Y, Chemistry of mycolactones. the causative toxins of Buruli ulcer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6703–6708. doi: 10.1073/pnas.1015252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sarfo FS, et al. Kinetics of mycolactone in human subcutaneous tissue during antibiotic therapy for Mycobacterium ulcerans disease. BMC infectious diseases. 2014;14:202. doi: 10.1186/1471-2334-14-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mve-Obiang A, Lee RE, Portaels F, Small PLC. Heterogeneity of Mycolactones Produced by Clinical Isolates of Mycobacterium ulcerans: Implications for Virulence. Infection and immunity. 2003;71:774–783. doi: 10.1128/IAI.71.2.774-783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chany AC, Tresse C, Casarotto V, Blanchard N. History, biology and chemistry of Mycobacterium ulcerans infections (Buruli ulcer disease) Natural product reports. 2013;30:1527–1567. doi: 10.1039/c3np70068b. [DOI] [PubMed] [Google Scholar]
- 111.Hong H, Demangel C, Pidot SJ, Leadlay PF, Stinear T. Mycolactones: immunosuppressive and cytotoxic polyketides produced by aquatic mycobacteria. Natural product reports. 2008;25:447–454. doi: 10.1039/b803101k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marion E, et al. Mycobacterial toxin induces analgesia in buruli ulcer by targeting the Angiotensin pathways. Cell. 2014;157:1565–1576. doi: 10.1016/j.cell.2014.04.040. [DOI] [PubMed] [Google Scholar]
- 113.Putze J, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infection and immunity. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 115.Cuevas-Ramos G, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cougnoux A, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 117.Secher T, Samba-Louaka A, Oswald E, Nougayrede JP. Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PloS one. 2013;8:e77157. doi: 10.1371/journal.pone.0077157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lai YC, et al. Genotoxic Klebsiella pneumoniae in Taiwan. PloS one. 2014;9:e96292. doi: 10.1371/journal.pone.0096292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bian X, et al. In vivo evidence for a prodrug activation mechanism during colibactin maturation. Chembiochem : a European journal of chemical biology. 2013;14:1194–1197. doi: 10.1002/cbic.201300208. [DOI] [PubMed] [Google Scholar]
- 120.Vizcaino MI, Engel P, Trautman E, Crawford JM. Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. Journal of the American Chemical Society. 2014;136:9244–9247. doi: 10.1021/ja503450q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brotherton CA, Balskus EP. A prodrug resistance mechanism is involved in colibactin biosynthesis and cytotoxicity. Journal of the American Chemical Society. 2013;135:3359–3362. doi: 10.1021/ja312154m. [DOI] [PubMed] [Google Scholar]
- 122.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nature chemistry. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schneditz G, et al. Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13181–13186. doi: 10.1073/pnas.1403274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hogenauer C, et al. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. The New England journal of medicine. 2006;355:2418–2426. doi: 10.1056/NEJMoa054765. [DOI] [PubMed] [Google Scholar]
- 125.Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem : a European journal of chemical biology. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 126.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiology and molecular biology reviews : MMBR. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nature chemical biology. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 128.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Donia MS, et al. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1423–E1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ziemert N, et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Holmes E, et al. Therapeutic modulation of microbiota-host metabolic interactions. Science translational medicine. 2012;4:137rv136. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- 132.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 133.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 134.Marcobal A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. The ISME journal. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 137.Gajer P, et al. Temporal dynamics of the human vaginal microbiota. Science translational medicine. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.LaMarche MJ, et al. Discovery of LFF571: an investigational agent for Clostridium difficile infection. Journal of medicinal chemistry. 2012;55:2376–2387. doi: 10.1021/jm201685h. [DOI] [PubMed] [Google Scholar]
- 139.Crost EH, Ruminococcin C, et al. a new anti-Clostridium perfringens bacteriocin produced in the gut by the commensal bacterium Ruminococcus gnavus E1. Biochimie. 2011;93:1487–1494. doi: 10.1016/j.biochi.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 140.Pujol A, et al. Characterization and distribution of the gene cluster encoding RumC, an anti-Clostridium perfringens bacteriocin produced in the gut. FEMS microbiology ecology. 2011;78:405–415. doi: 10.1111/j.1574-6941.2011.01176.x. [DOI] [PubMed] [Google Scholar]
- 141.Ramare F, et al. Trypsin-dependent production of an antibacterial substance by a human Peptostreptococcus strain in gnotobiotic rats and in vitro. Applied and environmental microbiology. 1993;59:2876–2883. doi: 10.1128/aem.59.9.2876-2883.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cursino L, et al. Exoproducts of the Escherichia coli strain H22 inhibiting some enteric pathogens both in vitro and in vivo. Journal of applied microbiology. 2006;100:821–829. doi: 10.1111/j.1365-2672.2006.02834.x. [DOI] [PubMed] [Google Scholar]
- 143.Wooley RE, Gibbs PS, Shotts EB. Inhibition of Salmonella typhimurium in the Chicken Intestinal Tract by a Transformed Avirulent Avian Escherichia coli. Avian Diseases. 1999;43:245. [PubMed] [Google Scholar]
- 144.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Science translational medicine. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Naik S, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nature reviews. Microbiology. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 147.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Temme K, Zhao D, Voigt CA. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7085–7090. doi: 10.1073/pnas.1120788109. [DOI] [PMC free article] [PubMed] [Google Scholar]


