Abstract
Carbohydrates, in addition to their metabolic functions, serve important roles as receptors, ligands, and structural molecules for diverse biological processes. Insight into carbohydrate biology and mechanisms has been aided by metabolic oligosaccharide engineering (MOE). In MOE, unnatural carbohydrate analogs with novel functional groups are incorporated into cellular glycoconjugates and used to probe biological systems. While MOE has expanded knowledge of carbohydrate biology, limited metabolism of unnatural carbohydrate analogs restricts its use. Here we assess metabolism of SiaDAz, a diazirine-modified analog of sialic acid, and its cell-permeable precursor, Ac4ManNDAz. We show that the efficiency of Ac4ManNDAz and SiaDAz metabolism depends on cell type. Our results indicate that different cell lines can have different metabolic roadblocks in the synthesis of cell surface SiaDAz. These findings point to roles for promiscuous intracellular esterases, kinases, and phosphatases during unnatural sugar metabolism and provide guidance for ways to improve MOE.
Keywords: sialic acid, metabolic engineering, diazirine, photocrosslinking, esterase
INTRODUCTION
Sialic acids are typically located at the non-reducing termini of glycans. When attached to cell surface proteins, this location positions sialic acid distal from the cell surface, an ideal position to modulate extracellular recognition events. Indeed, sialic acids are implicated in a wide variety of physiological processes such as neural development and leukocyte homing, as well as pathological processes such as viral infections and cancer.[1] In select cases, the sialylated molecules that mediate these recognition events are known,[2] but in many instances the identities of the relevant interaction partners remain poorly defined. In efforts to probe the functional roles of sialylated molecules, metabolic oligosaccharide engineering (MOE) has emerged as an important tool.[3]
MOE refers to the use of variant monosaccharide analogs that can be metabolized by living cells and incorporated into glycoconjugates in place of the naturally occurring sugars.[4] Variant monosaccharides differ from the natural sugars by addition of steric bulk, or by modification with bioorthogonal or other functional groups. While MOE has been used to replace a variety of naturally occurring monosaccharides, the earliest and widest successes have come in the area of sialic acid, where dozens of analogs have been shown to be metabolically incorporated into mammalian glycoconjugates.[3, 5] Specifically, the metabolic incorporation of modified sialic acid analogs can be achieved by simply culturing cells with unnatural sialic acid analogs. Alternatively, it is possible to take advantage of the sialic acid biosynthetic pathway of mammalian cells in which sialic acids are produced from N-acetylmannosamine (ManNAc). In this approach, cells can be cultured with unnatural ManNAc analogs, which are metabolized to their sialic acid counterparts and then incorporated into glycoconjugates.
One application of MOE is in the metabolic incorporation of photocrosslinking sialic acid analogs. These reagents have been developed to identify binding interactions that depend on sialic acid. Both metabolically incorporated photocrosslinking sialic acid analogs and ManNAc precursors have been described. [6–9] Photocrosslinking sialic acid analogs can be activated by UV irradiation, producing a reactive species that forms a covalent adduct with neighboring molecules.[10] The resulting crosslinked complexes can be analyzed to identify sialic acid-dependent binding interactions. Indeed, one photocrosslinking sialic acid analog, 9-aryl azide NeuAc (9-AAz-NeuAc), enabled elucidation of the binding partners of CD22, a sialic acid-recognizing protein that regulates B-cell activation.[7, 11]
While MOE is a powerful approach, some limitations exist. In particular, the utility of MOE is limited by the efficiency with which the unnatural analog replaces the natural monosaccharide in glycoconjugates.[12] For instance, achieving efficient incorporation of a photocrosslinking sialic acid into cell surface glycoconjugates would increase the likelihood of crosslinking to a binding partner and provide more material for biochemical evaluation (i.e. immunoblot or mass spectrometry analysis). Both sialic acid and ManNAc analogs are polar molecules that cross the plasma membrane inefficiently, so these compounds typically must be used at millimolar concentrations to achieve adequate incorporation. Thus, large quantities of the sugar analogs are required. Alternatively, compounds may be protected by peracylation to yield more lipophilic molecules that can readily traverse the plasma membrane.[13, 14] Inside cells, the acyl protecting groups are postulated to be removed by endogenous esterases, yielding free sugar analogs that can be metabolized to glycoconjugates. In some cases, efficient analog incorporation can be achieved with protected sugars used at concentrations nearly three orders of magnitude lower than the corresponding free sugars.[12]
The utility of MOE is also limited by the variable ability of different cell lines to metabolize sialic acid analogs and their precursors. However, quantification of cell line-dependent differences in sialic acid analog metabolism has been limited.[15–18] Here we describe experiments in which metabolism of diazirine-modified sialic acid and ManNAc analogs is quantitatively evaluated in multiple human cell lines. The results of these experiments suggest that different cell lines harbor metabolic barriers at different steps in analog metabolism. Based on these findings, we propose that cell line-specific expression of promiscuous enzymes, including esterases, kinases, and phosphatases, may affect the efficiency of MOE. Taken together, we expect that these results will inspire strategies to improve the efficiency and generality of MOE.
MATERIALS AND METHODS
Cell lines and culturing conditions
All cell lines were cultured at 37 °C, 5% CO2 in a water-saturated environment. BJAB K88 and BJAB K20 were obtained from Michael Pawlita (German Cancer Research Center) and James Paulson (The Scripps Research Institute). Cells were cultured in RPMI 1640 containing 2 mM glutamine, 10% fetal bovine serum, and 1% Pen/Strep (serum normal) or in 1% Nutridoma-SP (11011375001 Roche) and 0.5% Pen/Strep (serum free). Sugars were added to culture media for 48 h. Jurkat cells were obtained from Kim Orth (UT Southwestern) and were cultured in RPMI 1640 containing 2 mM glutamine and 10% fetal bovine serum and 1% Pen/Strep. Sugars were added to culture media for 48 h for Table 1, or as indicated for Figures 5 and 7. Daudi cells were obtained from Ellen Vitetta (UT Southwestern) and were cultured in RPMI 1640 media containing 2 mM glutamine and supplemented with 10% fetal bovine serum and 1% Pen/Strep. Sugars were added to culture media for 72 h. HeLa and SW48 cells were obtained from the ATCC and were cultured in DMEM containing 4.5 g/L D-glucose, 110mg/L sodium pyruvate, 4 mM L-glutamine, 10% FBS, and 1% Pen/Strep. Sugars were added to culture media for 48 h for Table 1, or as indicated for Figure 4. K562 cells were obtained from ATCC and were cultured in RPMI 1640 medium containing 2 mM glutamine, 10% FBS, and 1% Pen/Strep. Sugars were added to culture media for 48 h. HEK293T cells were obtained from the ATCC and cultured in DMEM containing 4.5 g/L D-glucose, 110 mg/L sodium pyruvate, 4 mM L-glutamine, 10% FBS, and 1% Pen/Strep. Sugars were added to culture media for 48 h. HEK293 cells were obtained from Chou-Long Huang (UT Southwestern) and cultured in DMEM containing 4.5 g/L D-glucose, 110 mg/L sodium pyruvate, 4 mM L-glutamine, 10% FBS, and 1% Pen/Strep. Sugars were added to culture media for 72 h. T84 cells were obtained from the ATCC and cultured in DMEM/F-12 with L-glutamine and HEPES (Gibco #11330-032) supplemented with 5% FBS and 1% Pen/Strep. Sugars were added to culture media for 72 h. Caco-2 cells were obtained from the ATCC and cultured in MEM with L-glutamine (Gibco #11095-080) supplemented with 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 10 mM HEPES, 10% FBS, and 1% Pen/Strep. Sugars were added to culture media for 72 h. MDA-MB-231 cells were obtained from the ATCC and cultured in DMEM containing 10% FBS and 1% Pen/Strep. Sugars were added to culture media for 48 h. SK-N-SH cells were obtained from the ATCC and cultured in alpha MEM containing 10% fetal bovine serum. Sugars were added to culture media for 72 h. SH-SY cells were obtained from Melanie Cobb (UT Southwestern) and cultured in a 1:1 mixture of ATCC-formulated Eagle’s MEM and F12 medium including 10% FBS. Sugars were added to culture media for 72 h. PC-3 cells were obtained from Jer-Tsong Hsieh (UT Southwestern) and cultured in DMEM containing 4.5 g/L D-glucose, 110 mg/L sodium pyruvate, 4 mM L-glutamine, 10% FBS, and 1% Pen/Strep. Sugars were added to culture media for 48 h.
Table 1. Cell surface incorporation of SiaDAz.
Cells were cultured with 100 μM Ac4ManNDAz. Membrane-associated Neu5Ac and SiaDAz were quantified. The fraction [SiaDAz]/([SiaDAz] +[Neu5Ac]) is reported as percent cell surface incorporation of SiaDAz.
| Cell Line | Cell type | Media | % cell surface incorporation of SiaDAz |
|---|---|---|---|
| BJAB K88 | B cell lymphoma | Serum Normal | 34 |
| BJAB K20 | B cell lymphoma | Serum Normal | 50 |
| BJAB K20 | B cell lymphoma | Serum Free | 100 |
| Jurkat | T cell lymphoma | Serum Normal | 65 |
| Daudi | B cell lymphoma | Serum Normal | 36 |
| HeLa | cervical carcinoma | Serum Normal | 0 |
| SW48 | colorectal carcinoma | Serum Normal | 10 |
| K562 | myelogenous leukemia | Serum Normal | 0 |
| HEK293T | kidney adenocarcinoma | Serum Normal | 13 |
| HEK293 | kidney adenocarcinoma | Serum Normal | 30 |
| T84 | colorectal carcinoma | Serum Normal | 7 |
| T84 | colorectal carcinoma | Serum Free | 15 |
| Caco-2 | colorectal carcinoma | Serum Normal | 39 |
| MDA-MB-231 | mammary carcinoma | Serum Normal | 42 |
| SK-N-SH | neuroblastoma | Serum Normal | 15 |
| SH-SY | neuroblastoma | Serum Normal | 0 |
| PC-3 | prostate cancer | Serum Normal | 0 |
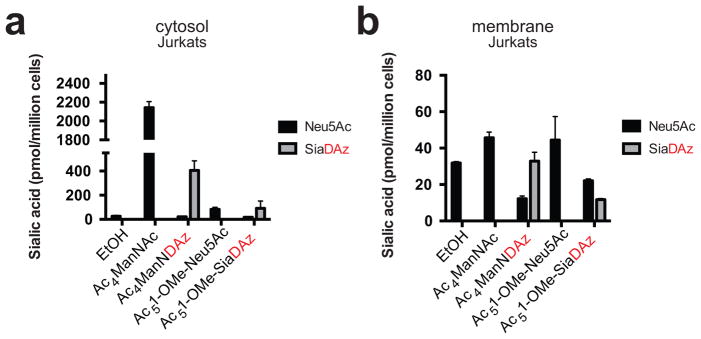
Figure 5. Ac5-1-OMe-SiaDAz is poorly metabolized by Jurkat cells.
Jurkat cells were cultured with ethanol, 100 μM Ac4ManNAc, 100 μM Ac4ManNDAz, 100 μM Ac5-1-OMe-Neu5Ac, or 100 μM Ac5-1-OMe-SiaDAz. Intracellular (a) and membrane (b) sialic acids were derivitized with DMB and quantified by fluorescence HPLC. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean.
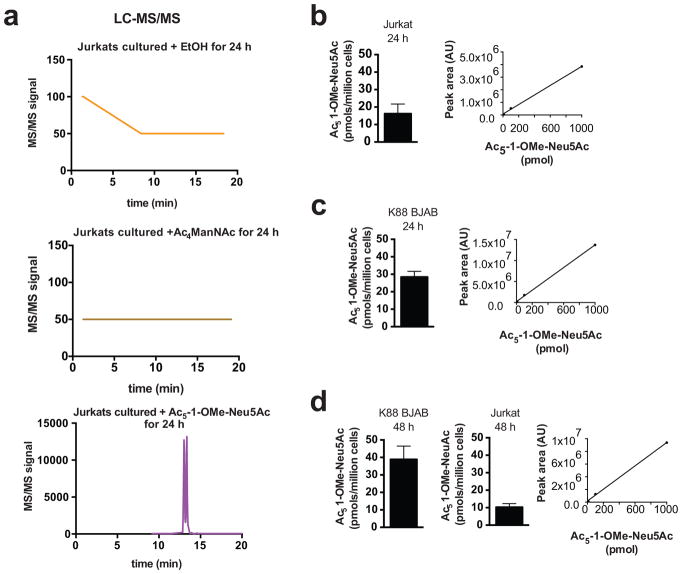
Figure 7. Ac5-1-OMe-Neu5Ac persists intracellularly.
(a) LC-MS/MS detection of Ac5-1-OMe-Neu5Ac in Jurkat cells cultured with ethanol control, Ac4ManNDAz, or Ac5-1-OMe-Neu5Ac for 24 h. BJAB K88 or Jurkat cells were cultured with Ac5-1-OMe-Neu5Ac for 24 h (b, c) or 48 h (d). Intracellular Ac5-1-OMe-Neu5Ac was quantified by MRM with LC-MS/MS. Insets show calibration curves for synthetic Ac5-1-OMe-Neu5Ac. Data presented are the average of duplicate trials with error bars depicting the standard deviation.
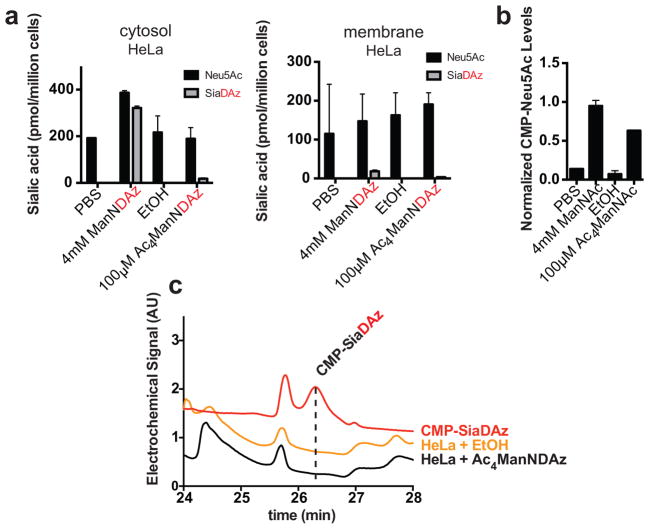
Figure 4. Metabolism of ManNDAz and Ac4ManNDAz by HeLa cells.
(a) HeLa cells were cultured with 4 mM ManNDAz or 100 μM Ac4ManNDAz for 48 h. Intracellular and membrane sialic acid were measured following fractionation, DMB derivatization, and HPLC analysis with fluorescence detection. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean. (b) HeLa cells were cultured with 4 mM ManNAc or 100 μM Ac4ManNAc for 24 h. CMP-Neu5Ac levels were quantified by HPAEC analysis with UV detection. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean. (c) HeLa cells were cultured with 100 μM Ac4ManNDAz for 48 h. Intracellular metabolites were harvested and analyzed by HPAEC-PAD. No CMP-SiaDAz production was observed. Data presented are a single trial representative of replicate experiments.
ManNAc was obtained from Sigma-Aldrich and Neu5Ac was purchased from Carbosynth. Ac4ManNAc, Ac5-1-OMe-NeuAc, Ac5-1-OMe-SiaDAz, Ac4ManNDAz were synthesized as described previously.[19] Synthesis of ManNDAz is presented in the electronic supplementary material (Online Resource 1). Briefly, we previously reported the synthesis of ManNDAz by amide coupling of 4,4-azo-pentanoic acid and mannosamine via activating the acid by a carbodiimide reagent and HOBt.[19] Although ManNDAz could not be completely separated from HOBt at that point, acetylation of the mixture enabled isolation of peracetylated ManNDAz (Ac4ManNDAz) for cell culture experiments. In order to isolate free ManNDAz for this study, we therefore started from pure Ac4ManNDAz prepared as reported previously. Basic cleavage of the ester bonds afforded ManNDAz in good purity with a yield of 52 %. See Online Resource 1 for full characterization of ManNDAz. Peracetylated sugars were stored in ethanol at a concentration of 10 mM. Free sugars were dissolved in phosphate-buffered saline (PBS) at a concentration of 400 mM and filtered prior to storage at −20 °C.
Generation of SW48 cells overexpressing ST6GALI
The human ST6GAL1 gene was amplified by PCR from pCMV-Sport6-ST6GAL1 (MHS1010-97227857, Thermo Scientific). The PCR product was digested and ligated into the lentiviral destination vector, pSIN4-EF1a-mGli2-IRES-Puro (a gift from Dr. Jiang Wu, University of Texas Southwestern Medical Center).[20] The plasmid was verified by DNA sequencing (UT Southwestern DNA Sequencing Core Facility).
HEK293T cells were transfected with pSIN4-ST6GAL1 or pSIN4-eGFP, accompanied with psPAX2 (12260, Addgene) and pMD2.G (C12259, Addgene) in the presence of Fugene 6 (E2691, Promega) to generate lentivirus. Media was replaced after 20 h. After 2 days, supernatant containing lentivirus was harvested. SW48 cells were seeded into 6-well tissue culture wells for 18 h. A 1:1 lentivirus stock solution diluted with DMEM was added and supplemented with 4 μg/mL polybrene (AL-118, Sigma-Aldrich) to enhance infection efficiency. After 24 h, supernatant was removed by aspiration and fresh DMEM was added to the cells. After an additional 24 h, cells were transferred to a 10 cm diameter tissue culture dish and cultured for two weeks in the presence of 1.5 μg/mL puromycin to select for infected clones.
Flow cytometry analysis
BJAB K20 cells were collected and washed twice with Dulbecco’s phosphate buffered saline (PBS), then resuspended in PBS at 2.0 × 106 cells/mL. Then, 200 μl of cell suspension was transferred to V-bottom 96-well plate and centrifuged to pellet cells. Cell pellets were resuspended in 100 μl of PBS + 0.1% BSA (w/v) containing 0.01 mg/mL MALII-biotin (Vector) for 60 min at 4 °C, then washed with PBS three times. Cells were then incubated in PBS containing 0.1% BSA and 0.005 mg/mL allophycocyanin-streptavidin (APC-streptavidin; S-868, Life Technologies) for 45 min at 4 °C. Fluorescence was analyzed by flow cytometry on a FACSCalibur instrument (BD Biosciences) equipped with dual lasers at 488 nm and 635 nm.
Cell fractionation
Freshly harvested cells were fractionated as described previously.[21] Briefly, adherent cells were removed from tissue culture plates by incubation with 1 mM in PBS and then lysed, or lysed directly in hypotonic lysis buffer containing protease inhibitors for 15 min on ice. Non-adherent cells were counted and harvested by centrifugation and then suspended in hypotonic lysis buffer containing protease inhibitors for 15 min on ice. Cells were lysed by extrusion through a 25 gauge needle. Nuclei and unbroken cells were removed from the post-nuclear supernatant by two rounds of centrifugation at 1000g for 15 min at 4 °C. The post-nuclear supernatant was transferred to heavy-walled polycarbonate tubes and centrifuged at 100,000g for 1 h in a Beckman TLA 100.3 or Thermo Sorvall MX120 ultracentrifuge rotor. The resulting supernatant was designated the cytosolic fraction. The pellet was washed twice with 400 μL cold hypotonic lysis buffer followed by centrifugation at 100,000g for 1 h after each wash. The remaining pellet was designated the membrane fraction. Samples were flash frozen in liquid nitrogen and the solvent removed by vacuum overnight.
Quantification of sialic acids by DMB derivatization
To release sialic acids, 50 μL 2.0 M acetic acid was added to each membrane (M) sample and and 100 μL 2.0 M acetic acid was added to each cytosolic (C) sample. After incubation at 80 °C for 2 h, samples were cooled to room temperature, then 1,2-diamino-4,5-methylenedioxybenzene (DMB) reaction solution (7.0 mM DMB, 1.4 M acetic acid, 750 mM 2-mercaptoethanol, 18 mM Na2S2O3) was added to each sample (M: 40 μL and C: 80 μL). After a 2 h incubation at 50 °C, 200 mM NaOH was added to each sample (M: 10 μL and C: 20 μL). Samples were filtered through 10 kDa MWCO filters and the resulting flow-through was stored at −20 °C in the dark until analysis. Generally, 10 μL of derivatized material was diluted with 90 μL ddH2O and analyzed by fluorescence HPLC. To do this, samples were injected onto a using Dionex Acclaim® Polar Advantage C16 5μm, 4.6 × 250 mm column attached to a Dionex Ultimate 3000 HPLC with fluorescence detector. Separation was performed using a gradient of 2–90% acetonitrile (ddH2O) and detected by fluorescence (ex. 373 nm, em. 448 nm). The integrated areas of both DMB-Neu5Ac and DMB-SiaDAz peaks were determined. DMB-Neu5Ac and DMB-SiaDAz yield slightly different fluorescence signals, so the areas were normalized by linear regression analysis using calibration curves prepared by injecting between 100 to 600 fmol of DMB-Neu5Ac and 275 fmol to 11000 fmol of DMB-SiaDAz. The fluorescence signals of DMB-Neu5Ac and DMB-SiaDAz were linear in these ranges. For DMB-SiaDAz, signals corresponding to amounts below 100 fmol were considered to be below the detection limit. Experiments were performed in biological duplicate.
Analysis of intracellular metabolites by HPAEC and LC-MS/MS
Cells were cultured with sugars or sugar analogs as indicated. Typically ~2 × 106 cells were harvested by centrifugation, then washed with cold DPBS twice and flash frozen in liquid nitrogen. Cells were lysed with 80% “super-cold” methanol (on dry ice).[22] Lysates were centrifuged at 2,000g for 15 min at 4 °C. The supernatant was flash frozen in liquid nitrogen and dried by centrifugal evaporation under vacuum. If not analyzed immediately, the intracellular metabolite pellet was stored at −80 °C.
The metabolite pellet was resuspended in 40 mM sodium phosphate buffer (pH 7.4; 40 μL per million cells), and filtered through an Amicon® Ultra centrifugal filter unit (Millipore, 10,000 MWCO). Filtrates and standards were analyzed by high performance anion exchange chromatography (HPAEC; ICS-3000 system, Dionex) using a CarboPac™PA1 column (Dionex) with a pulsed amperometry detector (PAD) and UV-detector (ex: 260 nm).[23, 24] Typically, 20 μL of metabolite was injected into the sample loading loop before entering a guard column (Dionex, 4 × 50 mm) and then an analytical column (Dionex, 4 × 250 mm). The eluents used were 1.0 mM NaOH (C) and 1.0 M NaOAc and 1.0 mM NaOH (D). Low-carbonate NaOH (50% in water) was obtained from Fisher Scientific (SS254-1) and NaOAc was from Sigma (71183). Separation was performed with a flow rate of 1 mL/min and the following gradient elution: T0 min = 95 % C, T30 = 76 % C, T45 = 0 % C, T55 = 0 % C, T60 = 95 % C, T70 = 95 % C. Integrated peaks were compared to commercial standards of CMP-Neu5Ac and synthesized CMP-SiaDAz. Data were normalized to cell number. Experiments were performed in biological duplicate.
For NeuAc-9-P detection, 8 million SW48 cells were cultured with 100 μM Ac4ManNAc for 24 h and intracellular metabolites were harvested as described above. The intracellular metabolite pellet was dissolved in 40 mM sodium phosphate (pH 7.4) buffer and filtered through an Amicon® Ultra centrifugal filter unit (Millipore, 10,000 MWCO). 60 μL of intracellular metabolites was injected into the HPAEC and using the elution gradient with flow rate of 1 mL/min: T0 min = 95 % C, T40 = 70 % C, T45 = 50 % C, T60 = 45 % C, T65 = 0 % C, T75 = 0 % C, T80 = 95 % C, T90 = 95 % C. The peak at 42.5 min was collected. Meanwhile, a Dowex X-50 W-8 cation ion exchange resin (Sigma; 69011-20-7) was equilibrated with 1 M NH4OAc and washed twice with 2 column volumes ddH2O. The collected sample was applied to the column and eluted with ddH2O. Eluent was collected in a Pyrex glass tube (18 × 150 mm) and lyophilized on a Thermoscientific freeze dryer. Samples were submitted for LC-MS analysis at the Shimadzu Center for Advanced Analytical Chemistry, Arlington Texas. Details and mass spectrometry data are presented in Online Resource 1.
Multiple reaction monitoring (MRM) with LC-MS/MS with a triple quadrupole mass spectrometer (3200 QTRAP, ABSCIEX) was used to analyze intracellular metabolites as previously described.[25, 26] The intracellular metabolite pellet was dissolved in solvent A and incubated at 4 °C for 30 min. Insoluble material was removed by centrifugation at 16,000g for 5 minutes at 4° C. Supernatant was filtered twice through 0.2 μm PVDF micro spin filter (Grace Davison Discovery Science, 8604742) at 9000g for 5 min at 4 °C. Intracellular metabolites and standards were directly injected into a Synergi 4u Fusion-RP 80A reverse phase column (Phenomenex) and chromatographed at a flow rate of 0.5 mL/min. The solvent system consisted of 5 mM ammonium acetate in water (solvent A) and 5 mM ammonium acetate in methanol (solvent B). Injected samples were eluted with the following gradient of solvent B: 0 % – 0.2 %/5 min, 0.2 % – 1 %/1 min, 1 % – 3 %/1 min, 3 % – 5%/1 min, 5 % – 25 %/6 min, 25 % – 50 %/4 min, 50 % – 100 %/2 min. The retention time (RT) and mass-to-charge ratio (m/z; positive mode MW+1) of the parent and product ion for Ac5-1-OMe-Neu5Ac were RT: 12–13 min, m/z: 534/414 & 534/129. The parent and product ion pairs were determined empirically by injecting pure standard into the mass spectrometer. Typically, both MRMs for a given metabolite displayed high correlation. The retention time for each MRM peak was matched with the metabolite standard. The area under each peak was quantified using Analyst software, with manual inspection for accuracy.
RESULTS
Metabolism of Ac4ManNDAz to SiaDAz-containing glycoconjugates is cell line-dependent
We reported previously that culturing BJAB K20, BJAB K88, or Jurkat cells with peracetylated, diazirine-modified ManNAc (Ac4ManNDAz) results in production of glycoconjugates containing diazirine-modified sialic acid (SiaDAz; Figure 1).[8, 19, 21, 27] However, attempts to produce SiaDAz-containing glycoconjugates in other cell lines resulted in variable outcomes. To assess the cell line-dependence of SiaDAz incorporation, we cultured fifteen cell lines with 100 μM Ac4ManNDAz for 48–72 h. We isolated the membrane fractions from these cells, released the glycoconjugate-bound sialic acids, and quantified the amounts of Neu5Ac and SiaDAz. Table 1 reports the percent of membrane sialic acid that is SiaDAz. Both Jurkat and BJAB K20 cells displayed high levels of SiaDAz incorporation, with SiaDAz comprising about half of the membrane sialic acid. Since BJAB K20 cells lack the ability to biosynthesize Neu5Ac,[28] membrane Neu5Ac in these cells must derive from the serum used to supplement the media. Indeed, culturing BJAB K20 cells in serum-free media resulted in 100% SiaDAz in the membrane fraction. Other cell lines displayed a variety of lower levels of SiaDAz incorporation; in four cell lines (HeLa, K562, SH-SY, and PC-3) we were unable to detect significant amounts of membrane-associated SiaDAz. Differences were apparent even among related cell lines: the SK-N-SH cell line displays 15% membrane SiaDAz, while a subclone derived from this cell line (SH-SY) does not incorporate SiaDAz into the membrane at a detectable level.
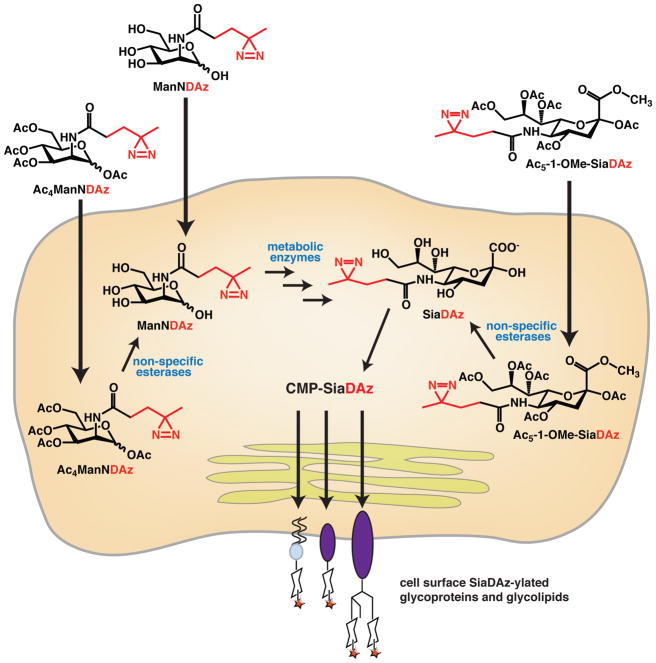
Figure 1. Metabolic oligosaccharide engineering (MOE) to produce glycoconjugates containing a photocrosslinking sialic acid analog, SiaDAz.
A mannosamine analog, ManNDAz, can be provided to the cell in free form or peracetylated form. Peracetylation makes the compound more hydrophobic so that it crosses the plasma membrane more efficiently, allowing a lower concentration of compound to be used. Alternatively, SiaDAz can be added to cell culture media. Esterification of all the free alcohols and the carboxylic acid, forming Ac5-1-OMe-SiaDAz, makes the compound more hydrophobic and allows it to traverse the plasma membrane efficiently. Cellular enzymes convert the analogs to the activated nucleotide sugar form (CMP-SiaDAz), followed by sialyltransferase-catalyzed transfer to glycoproteins and glycolipids.
Metabolism of Ac4ManNDAz and ManNDAz by MDA-MB-231, SW48, and HeLa cells
Based on the diverse levels of membrane SiaDAz we measured in different cell types, we decided to investigate variables that might affect the incorporation of SiaDAz into membrane glycoconjugates. We set out to assess whether there are cell line-specific differences in the ability of cells to metabolize diazirine-containing ManNAc to SiaDAz-containing glycoconjugates, focusing specifically on the abilities of different cell lines to hydrolyze acetoxy groups of protected sugars and to convert ManNDAz to SiaDAz. We focused our initial experiments on three cell lines that exhibited a range of cell surface SiaDAz incorporation in response to addition of Ac4ManNDAz: high incorporation MDA-MB-231 cells (42% membrane SiaDAz), moderate incorporation SW48 cells (10% membrane SiaDAz), and low incorporation HeLa cells (membrane SiaDAz near detection limit).
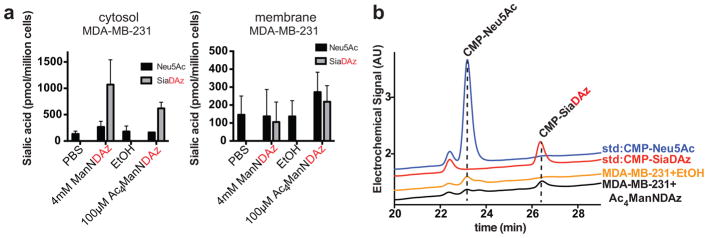
MDA-MB-231 cells, a breast carcinoma cell line, were cultured with 100 μM Ac4ManNDAz for 48 h, after which membrane and cytosolic SiaDAz levels were measured. While SiaDAz comprised only about 44% of membrane sialic acid, about 79% of cytosolic sialic acid was in the form of SiaDAz (Figure 2a). We also cultured the cells with free ManNDAz. Because of the predicted poor membrane permeability of this polar molecule, free ManNDAz was used at a concentration 40 times higher than Ac4ManNDAz. At this high concentration, free ManNDAz was readily metabolized, with SiaDAz detected in both the membrane and cytosol. We also observed production of CMP-SiaDAz in cells cultured with Ac4ManNDAz (Figure 2b).
Figure 2. Metabolism of ManNDAz and Ac4ManNDAz by MDA-MB-231 cells.
MDA-MB-231 cells were cultured with 4 mM ManNDAz or 100 μM Ac4ManNDAz for 48 h. (a) Intracellular and membrane sialic acids were measured by fractionation, DMB derivatization, and HPLC analysis with fluorescence detection. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean. (b) Intracellular CMP-sialic acids were measured by HPAEC with UV detection at 260 nm.
Our results suggest that MDA-MB-231 cells metabolize Ac4ManNDAz to CMP-SiaDAz efficiently and, further, that deacetylation of the protected sugar does not appear to be significant factor limiting its incorporation. For MDA-MB-231 cells cultured with either Ac4ManNDAz or free ManNDAz, SiaDAz is the major sialic acid in the cytosol, but represents only about 40% of the membrane sialic acid. The limited membrane incorporation of SiaDAz suggests impaired transport of CMP-SiaDAz into the Golgi or utilization of CMP-SiaDAz by sialyltransferases as possible limitations in SiaDAz metabolism.
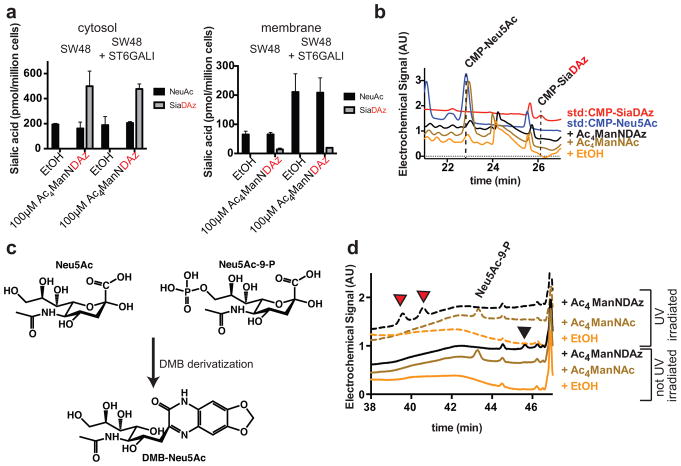
SW48 cells, a colon carcinoma cell line, produce a moderate amount of membrane SiaDAz in response to Ac4ManNDAz supplementation (Table 1). However, SW48 cells also display a very low level of total membrane sialic acid due to low expression of sialyltransferases.[29] To test whether low sialyltransferase expression is the reason for poor SiaDAz incorporation, we prepared SW48 cells stably expressing human ST6GAL1. We chose ST6GALI because it is the dominant sialyltransferase that acts on N-linked glycans and has been shown to be promiscuous in terms of accepting sialic acid analogs.[30, 31] We found that overexpression of ST6GAL1 did not affect the fraction of SiaDAz in the membrane (9%) even though the total amount of membrane sialic acid increased (Figure 3a). SW48 cells were capable of producing SiaDAz from Ac4ManNDAz: about 70% of cytosolic sialic acid was SiaDAz, a result that was relatively unaffected by ST6GAL1 expression (Figure 3a). However, we were unable to detect CMP-SiaDAz in cells supplemented with Ac4ManNDAz, while cells supplemented with the corresponding protected form of the natural sialic acid precursor (Ac4ManNAc) produced high levels of CMP-Neu5Ac (Figure 3b).
Figure 3. Metabolism of peracetylated mannosamines by SW48 cells.
SW48 cells were cultured with 100 μM Ac4ManNAc or 100 μM Ac4ManNDAz for 48 h. (a) Intracellular and membrane sialic acids were measured by fractionation, DMB derivatization, and HPLC analysis with fluorescence. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean. (b) Analysis of SW48 intracellular metabolites by HPAEC-PAD. SW48 cells cultured with Ac4ManNDAz do not produce CMP-SiaDAz, but SW48 cells cultured with Ac4ManNAc increase production of CMP-Neu5Ac. Data presented are a single trial representative of replicate experiments. (c) DMB derivatization of either Neu5Ac or Neu5Ac-9-P produces the same product: DMB-Neu5Ac. (d) Analysis of SW48 intracellular metabolites by HPAEC-PAD. SW48 cells cultured with 100 μM Ac4ManNAc accumulate Neu5Ac-9-P, whose identity was confirmed by mass spectrometry. SW48 cells cultured with 100 μM Ac4ManNDAz accumulate a novel peak (black arrowhead). Upon UV irradiation of metabolites, the novel peak disappeared and was replaced by two faster-migrating peaks (red arrowheads). This is a characteristic finding for diazirine-containing compounds, suggesting SiaDAz-9-P as the identity of the novel peak. Data presented are a single trial representative of triplicate experiments.
Since we were unable to detect CMP-SiaDAz in SW48 cells, we considered whether metabolism might be blocked prior to CMP-sialic acid production. Because the classical DMB labeling protocol that we applied includes an acid hydrolysis step,[32] our measurements of cytosolic sialic acids include free sialic acids, CMP-sialic acids, and also sialic acid-9-Ps (Figure 3c). To gain more insight into why CMP-SiaDAz was not produced, we investigated whether SiaDAz-9-P accumulates in SW48 cells by analyzing intracellular metabolites by HPAEC-PAD. Relative to the control sample, novel peaks were detected in metabolites derived from cells supplemented with either Ac4ManNAc or Ac4ManNDAz (Figure 3d). The novel metabolite from the Ac4ManNAc-supplemented cells was identified as Neu5Ac-9-P by LC-MS IT-TOF analysis (Online Resource 1). The novel metabolite from Ac4ManNDAz-supplemented cells (black arrowhead) eluted with a time similar to Neu5Ac-9-P and degraded to two faster migrating species in response to UV irradiation (red arrowheads), a pattern characteristic of diazirine-containing molecules.[24] While we were unable to obtain sufficient material for unambiguous identification, the most likely assignment for this novel peak is SiaDAz-9-P. Accumulation of SiaDAz-9-P in SW48 cells could reflect inadequate phosphatase activity in these cells, which would limit production of membrane SiaDAz.
When supplemented with Ac4ManNDAz, HeLa cells, a cervical carcinoma cell line, produce levels of membrane SiaDAz that are near our detection limit (Table 1). However, HeLa cells produce substantially more intracellular SiaDAz in response to ManNDAz treatment than in response to Ac4ManNDAz treatment (Figure 4a). A possible explanation for this result is that HeLa cells do not efficiently remove the acetyl groups from Ac4ManNDAz to release free ManNDAz. Acyl protecting groups are typically removed rapidly from peracylated ManNAc analogs through the action of intracellular esterases.[33] However, esterases have varying substrate specificities and different cell types express different esterases.[34, 35] Indeed, esterase expression patterns are known to affect deacylation of pro-drugs.[35] The inability of HeLa cells to deacetylate Ac4ManNDAz does not reflect a general esterase defect because cells cultured with Ac4ManNAc readily increase CMP-Neu5Ac production (Figure 4b). Thus, we hypothesized that the diazirine substituent may interfere with esterase-dependent deprotection in these cells. We hoped to bypass this esterase barrier by culturing cells with free ManNDAz, but unfortunately, HeLa cells cultured with free ManNDAz produced only about 11% membrane SiaDAz (Figure 4a). Thus, HeLa cells may also have a second metabolic barrier, in one of the two enzymatic steps involved in converting SiaDAz to CMP-SiaDAz. The same enzyme produces CMP-Neu5Ac in all cell types, suggesting that the barrier does not occur at this step. Dephosphorylation of Neu5Ac-9-P, on the other hand, could be performed by opportunistic phosphatases if Neu5Ac-9-P phosphatase (NANP) is not expressed. Thus, we speculate that the phosphatase(s) that dephosphorylate Neu5Ac-9-P in HeLa cells may have reduced activity toward SiaDAz-9-P.
Ac5-1-OMe-SiaDAz is poorly metabolized
Our results from SW48 and HeLa cells suggested that dephosphorylation of SiaDAz-9-P might represent a barrier to production of membrane SiaDAz. We hypothesized that direct delivery of a diazirine-modified sialic acid analog would circumvent this metabolic blockade. To investigate this idea, we studied metabolism of a cell-permeable SiaDAz analog in which free hydroxyl groups are protected with acetyl groups and the carboxylate is protected as a methyl ester (Ac5-1-OMe-SiaDAz).[8] We used Jurkat and BJAB K20 cells for these experiments because they readily produce membrane SiaDAz from Ac4ManNDAz, indicating that they do not harbor significant barriers limiting metabolism of cytosolic SiaDAz to membrane SiaDAz.
We found that Jurkat cells produce SiaDAz from Ac5-1-OMe-SiaDAz inefficiently: relatively little free SiaDAz was detected in the cytosolic fraction of Jurkat cell cultured with Ac5-1-OMe-SiaDAz compared to cells cultured with Ac4ManNDAz (Figure 5a). Further, conversion of Ac5-1-OMe-SiaDAz to membrane SiaDAz was also low, with SiaDAz comprising only 35% of membrane sialic acid (compared with about 73% when cells are cultured with Ac4ManNDAz) (Figure 5b). Next, we examined BJAB K20 cells, where SiaDAz metabolism might be expected to be more efficient, since there is little competing endogenous Neu5Ac.[28] While culturing BJAB K20 cells with free ManNDAz or Ac4ManNDAz yielded CMP-SiaDAz, very little CMP-SiaDAz was produced through supplementation with Ac5-1-OMe-SiaDAz (Figure 6a).
Figure 6. BJAB K20 cells poorly metabolize protected sialic acids.
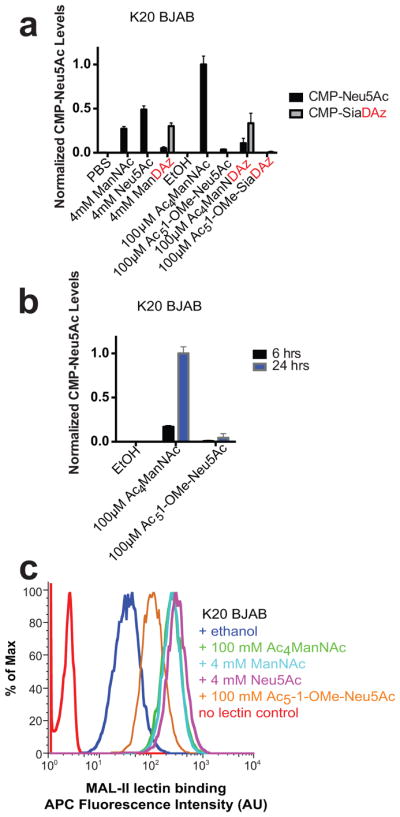
(a) BJAB K20 cells were cultured with indicated molecules for 24 h and intracellular levels of CMP-sialic acids were quantified by HPAEC-UV. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean. (b) BJAB K20 cells were cultured with 100 μM Ac4ManNAc or 100 μM Ac5-1-OMe-Neu5Ac for 6 or 24 h. Intracellular CMP-Neu5Ac was quantified by HPAEC-UV. Analysis was performed in biological duplicate with error bars showing standard deviation of the mean. (c) BJAB K20 cells were cultured with indicated molecules for 48 h. Cell surface α2–3-linked sialic acid was measured by flow cytometry using biotinylated MAL II lectin and APC-streptavidin. Data presented are a single trial representative of replicate experiments.
Protected Neu5Ac is poorly metabolized
Surprised by the poor metabolism of Ac5-1-OMe-SiaDAz, we next assessed metabolism of fully protected Neu5Ac (Ac5-1-OMe-Neu5Ac). Culturing BJAB K20 cells with Ac4ManNAc yields high levels of CMP-Neu5Ac, while addition of free ManNAc or free Neu5Ac leads to lower but significant CMP-Neu5Ac accumulation. But very little CMP-Neu5Ac was detected when BJAB K20 cells were cultured with Ac5-1-OMe-Neu5Ac (Figure 6a). This result does not appear to be due to rapid turnover of CMP-Neu5Ac, since it was also undetectable at an earlier time (Figure 6b). Furthermore, BJAB K20 cells cultured with Ac5-1-OMe-Neu5Ac had lower levels of cell surface sialic acid than cells cultured with Ac4ManNAc, free ManNAc, or free Neu5Ac (Figure 6c). Overall, these results suggest that BJAB K20 cells poorly metabolize fully protected sialic acids, both natural (Ac5-1-OMe-Neu5Ac) and unnatural (Ac5-1-OMe-SiaDAz).
Protected sialic acids accumulate in cells
To understand why protected sialic acids are metabolized poorly, we used liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to detect Ac5-1-OMe-Neu5Ac remaining in the intracellular environment (Figure 7a). BJAB K88 (Figure 7c and d) or Jurkat (Figure 7b and d) cells were cultured with 100 μM Ac5-1-OMe-Neu5Ac for 24 (Figure 7b and c) or 48 h (Figure 7d) and the intracellular metabolites were examined by LC-MS/MS. We detected intracellular Ac5-1-OMe-Neu5Ac in both BJAB K88 cells (28 ± 3 pmol per million cells at 24 h; 39 ± 8 pmol per million cells at 48 h) and Jurkat cells (16 ± 5 pmol per million cells at 24 h; 10 ± 2 pmol per million cells at 48 h) (Figure 7). Assuming a cell volume of 1 pL, these amounts equate to about 10–40 μM intracellular concentration of Ac5-1-OMe-Neu5Ac. Thus, both acetyl and O-methyl protecting groups on Neu5Ac persist in the intracellular environment.
DISCUSSION
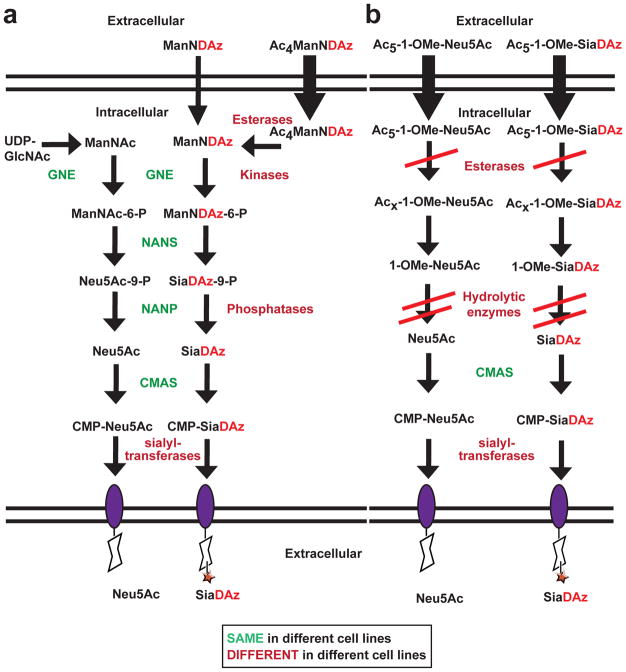
Introduction of unnatural sugar analogs into cellular glycoconjugates through MOE is a powerful tool for efforts to understand glycoconjugate function. However, the utility of this approach is limited by the ability of different cell lines to metabolize the precursor monosaccharides to glycoconjugates. Here we focused on one photocrosslinking sialic acid, SiaDAz, and its cell-permeable mannosamine precursor (Ac4ManNDAz). We discovered cell line-dependent barriers to metabolic production of SiaDAz-containing glycoconjugates (Figure 8). Metabolism of other mannosamine analogs has been reported to be cell line-dependent, [15–17] but additional work will be needed to assess whether the structures of different mannosamine analogs affect the cell type-specific barriers that they experience.
Figure 8. Pathways for metabolizing sialic acid analogs and precursors.
(a) Diazirine-modified ManNAc analogs and their intermediate metabolites compete with endogenous sialic acid precursors for common enzymes. Cell-specific esterases, kinases, and phosphatases aid in the processing of Ac4ManNDAz to ManNDAz and, further, to SiaDAz. (b) Protected sialic acids enter cells. Inefficient deprotection results in their accumulation.
Cell-specific enzyme expression may modulate Ac4ManNDAz metabolism
MOE takes advantage of the promiscuity of cellular enzymes to process analogs of naturally occurring monosaccharides (Figure 8). Specifically, metabolism of peracylated ManNAc analogs requires removal of acyl protecting groups by cellular esterases. Metabolism of ManNAc analogs to CMP-sialic acid analogs requires four additional enzymatic transformations: (1) phosphorylation of O6 position of the ManNAc analog, (2) coupling to pyruvate to yield the O9 phosphorylated sialic acid analog, (3) dephosphosphorylation of the O9 position of the sialic acid analog, and (4) activation to the CMP-sialic acid analog. While annotated sialic acid biosynthetic enzymes (GNE, NANS, NANP, and CMAS) are typically implicated in the metabolism of ManNAc analogs, enzymes outside of this pathway may also play a role.
Effects of cell-specific esterases on Ac4ManNDAz deprotection
Human carboxyesterases have variable substrate specificities and variable expression levels in different tissues.[35] These variations may affect metabolism of peracylated ManNAc analogs. Cell type-specific differences in small molecule deacetylation are precedented: for example, HEK293 cells cannot deacetylate the pro-drug aspirin to form active salicylic acid.[36] Our data suggest that HeLa cells may not effectively deacetylate Ac4ManNDAz, but can deacetylate Ac4ManNAc. In contrast, Yarema and co-workers reported that HeLa cells readily metabolize Ac4ManLev.[12] One possible explanation is that endogenous HeLa esterases may distinguish among structurally similarly molecules, deacetylating Ac4ManNAc and Ac4ManLev, but not Ac4ManNDAz. Yarema et al. have also postulated the existence of a storage “reservoir” for peracylated ManNAc analogs.[12] Our results are also consistent with the idea that Ac4ManNDAz is stored more efficiently than Ac4ManNAc, although the possible molecular basis for such a difference is unknown.
Cell-specific kinases can affect ManNDAz phosphorylation
Specific genes are annotated to function as kinases for individual monosaccharides, but more promiscuous phosphorylation likely occurs. Indeed, BJAB K20 cells do not express GNE (UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase), a bifunctional protein that contains the ManNAc kinase domain.[37] Nonetheless, BJAB K20 cells metabolize ManNDAz to SiaDAz, suggesting that other kinases are able to phosphorylate ManNDAz. Similarly, Jacobs et al. reported that the GNE ManNAc kinase domain has reduced activity toward ManNAc analogs with large N-acyl sidechains.[38] Thus, even in the presence of functional GNE, cells may rely on other kinases to metabolize ManNDAz. Thus, the efficiency of SiaDAz production may depend on which kinases are expressed and the degree to which they phosphorylate ManNDAz.
Cell-specific phosphatases can affect SiaDAz production
Production of sialic acid analogs from ManNAc analogs requires dephosphorylation of the O9 position of the sialic acid. Once again, a single enzyme is annotated as the Neu5Ac-9-P phosphatase (NANP), but other promiscuous phosphatases may also contribute to this activity. We found that SW48 cells cultured with Ac4ManNDAz produce high levels of intracellular SiaDAz but little CMP-SiaDAz or membrane SiaDAz, implying a deficiency in either dephosphorylation of SiaDAz-9-P or activation of SiaDAz to CMP-SiaDAz. We also observed accumulation of Neu5Ac-9-P in SW48 cells cultured with Ac4ManNAc, suggesting that the dephosphorylation step is likely to be the bottleneck. When cultured with Ac4ManNDAz, the SW48 cells accumulated a metabolite that we suspect to be SiaDAz-9-P although we were unable to perform conclusive identification. Further experiments are needed to determine which phosphatase dephosphorylates Neu5Ac-9-P in SW48 cells and whether it is active toward SiaDAz-9-P.
Metabolism of Ac5-1-OMe sialic acids
While MOE of sialic acid is often conducted with ManNAc analogs, an alternative strategy is to culture cells with sialic acid analogs. Use of sialic acid analogs is attractive because many of the metabolic enzymes are bypassed. Unprotected sialic acid analogs can be used, but must be supplied at high (millimolar) concentration. While peracetylation of ManNAc analogs has been pursued as an alternative, several reports indicate that protection of sialic acids does not substantially improve metabolic incorporation.[6, 9] Here we assessed experimentally whether deprotection of sialic acid analogs represents a barrier to metabolism. We found that both Ac5-1-OMe-SiaDAz and Ac5-1-OMe-Neu5Ac are poorly metabolized to membrane sialosides; moreover, we were able to detect intracellular accumulation of fully protected Neu5Ac in both K88 BJAB and Jurkat cells. This finding suggests esterases are slow to remove both the acetyl groups protecting the hydroxyls and the methyl ester protecting the carboxylic acid. Deprotection of the carboxylic acid may be particularly problematic, as Tian et al. demonstrated that bulky substitutions on the acyl chain of ester protecting groups are poorly tolerated by common human esterases.[39]
Effects of ManNAc analogs on Neu5Ac production
In one case, we observed that addition of the unnatural precursor ManNDAz resulted in increased cytosolic Neu5Ac (Figure 4a). Although we did not conduct any experiments to further investigate this surprising observation, one possible explanation involves the feedback regulation of GNE. GNE is strongly and allosterically inhibited by its product, CMP-Neu5Ac. [40, 41] CMP-SiaDAz might bind GNE competitively with CMP-Neu5Ac, but fail to trigger the same allosteric change, resulting a disruption of feedback inhibition. In this way, an unnatural sialic acid precursor might potentiate production of natural sialic acid.
Other factors affecting the efficiency of MOE
We have focused exclusively on identifying metabolic differences among cell lines; other factors not addressed here also play roles. For example, the rates of cell growth and division and the rate at which cell surface glycoconjugates are replaced could affect the rate at which sialic acid analogs are incorporated into glycoconjugates. Since sialic acid analogs must compete with Neu5Ac for incorporation, the levels of Neu5Ac production and Neu5Ac scavenging are also important factors. Cell-specific expression of transmembrane efflux pumps may cause monosaccharide analogs to be removed from cells and not available for metabolism. Monosaccharide analogs can also be degraded by endogenous enzymes,[42] whose expression patterns may vary among cell types. Finally, sialic acid analogs must be added to glycoconjugates by one of the twenty sialyltransferases encoded in the human genome. If sialyltransferases exhibit different abilities to transfer sialic acid analogs, then differential expression of different sialyltransferase family members could underlie some of the cell line-specific differences in sialic acid analog incorporation. Additional experiments will be needed to assess the relative roles of these different factors.
Future directions for MOE
Our results suggest that cell type-specific expression of enzymes including esterases, kinases, and phosphatases may underlie cell type-dependent differences in unnatural monosaccharide metabolism. Identification of the specific promiscuous enzymes that perform these transformations would allow confirmation of this hypothesis. Identification of these enzymes would also offer a strategy to expand the scope of MOE. For example, if the esterase that deprotects Ac4ManNDAz in MDA-MB-231 cells could be identified, it could be overexpressed in HeLa cells cultured with Ac4ManNDAz to test if it improves SiaDAz production. Alternatively, site-directed mutagenesis of enzymes that metabolize naturally-occurring monosaccharides could improve metabolism of diazirine-containing analogs; indeed this has already been accomplished for two enzymes involved in GlcNAc metabolism.[24, 43] Finally, alterations in protecting group chemistry may yield useful compounds for MOE. In particular, use of protected sialic acid analogs is very attractive, since they bypass several metabolic bottlenecks, but deprotection is a limitation. Future sialic acid analogs may include an alternative carboxylate protecting group that is removed by endogenous esterases, or an exogenously introduced deprotecting enzyme. Indeed, specialized pairs of protecting groups and deprotecting enzymes can be used to facilitate cell-type specific experiments. [39, 44]
Supplementary Material
I. Synthesis and characterization of ManNDAz.
II. Mass Spectrometry detection of Neu5Ac-9-P.
Acknowledgments
We thank Sunil Laxman and Benjamin Tu for guidance on LC-MS/MS analysis. We thank Maciej Kukula at the Shimadzu Center for Advanced Analytical Chemistry (SCAAC) at the University of Texas at Arlington for aiding us with mass spec identification of NeuAc-9-P. We thank Yibing Wang, Randy Parker, Amberlyn Wands, Akiko Fujita, and Fan Yang for experimental assistance and thank Akiko Fujita and Amberlyn Wands for comments on the manuscript. We acknowledge support from the National Institutes of Health (NIH R01GM090271), the Cancer Prevention and Research Institute of Texas (CPRIT RP110080), and the Welch Foundation (I-1686). NDP was supported by a predoctoral fellowship from the NIH (F30AG040909) and ACR received support from a training grant from the NIH (T32GM007062).
References
- 1.Varki A. Sialic acids in human health and disease. Trends in Molecular Medicine. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 3.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VDP, Yarema KJ. Metabolic glycoengineering: sialic acid and beyond. Glycobiology. 2009;19:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dube D, Bertozzi C. Metabolic oligosaccharide engineering as a tool for glycobiology. Current Opinion in Chemical Biology. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. Biosynthesis of a nonphysiological sialic acid in different rat organs, using N-propanoyl-D-hexosamines as precursors. J Biol Chem. 1992;267:16934–16938. [PubMed] [Google Scholar]
- 6.Luchansky SJ, Goon S, Bertozzi CR. Expanding the diversity of unnatural cell-surface sialic acids. Chembiochem. 2004;5:371–374. doi: 10.1002/cbic.200300789. [DOI] [PubMed] [Google Scholar]
- 7.Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka Y, Kohler JJ. Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J Am Chem Soc. 2008;130:3278–3279. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- 9.Feng L, Hong S, Rong J, You Q, Dai P, Huang R, Tan Y, Hong W, Xie C, Zhao J, Chen X. Bifunctional unnatural sialic acids for dual metabolic labeling of cell-surface sialylated glycans. J Am Chem Soc. 2013;135:9244–9247. doi: 10.1021/ja402326z. [DOI] [PubMed] [Google Scholar]
- 10.Pham ND, Parker RB, Kohler JJ. Photocrosslinking approaches to interactome mapping. Current Opinion in Chemical Biology. 2013;17:90–101. doi: 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramya TNC, Weerapana E, Liao L, Zeng Y, Tateno H, Liao L, Yates JR, Cravatt BF, Paulson JC. In situ trans ligands of CD22 identified by glycan-protein photocross-linking-enabled proteomics. Molecular & Cellular Proteomics. 2010;9:1339–1351. doi: 10.1074/mcp.M900461-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones MB, Teng H, Rhee JK, Lahar N, Baskaran G, Yarema KJ. Characterization of the cellular uptake and metabolic conversion of acetylated N-acetylmannosamine (ManNAc) analogues to sialic acids. Biotechnology and Bioengineering. 2004;85:394–405. doi: 10.1002/bit.10901. [DOI] [PubMed] [Google Scholar]
- 13.Sarkar AK, Fritz TA, Taylor WH, Esko JD. Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Gal beta 1-->4GlcNAc beta-O-naphthalenemethanol. Proc Natl Acad Sci U S A. 1995;92:3323–3327. doi: 10.1073/pnas.92.8.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemieux GA, Yarema KJ, Jacobs CL, Bertozzi CR. Exploiting differences in sialoside expression for selective targeting of MRI contrast reagents. J Am Chem Soc. 1999;121:4278–4279. [Google Scholar]
- 15.Jacobs CL, Yarema KJ, Mahal LK, Nauman DA, Charters NW, Bertozzi CR. Metabolic labeling of glycoproteins with chemical tags through unnatural sialic acid biosynthesis. Meth Enzymol. 2000;327:260–275. doi: 10.1016/s0076-6879(00)27282-0. [DOI] [PubMed] [Google Scholar]
- 16.Sampathkumar SG, Jones MB, Yarema KJ. Metabolic expression of thiol-derivatized sialic acids on the cell surface and their quantitative estimation by flow cytometry. Nat Protoc. 2006;1:1840–1851. doi: 10.1038/nprot.2006.252. [DOI] [PubMed] [Google Scholar]
- 17.Almaraz RT, Aich U, Khanna HS, Tan E, Bhattacharya R, Shah S, Yarema KJ. Metabolic oligosaccharide engineering with N-acyl functionalized ManNAc analogs: cytotoxicity, metabolic flux, and glycan-display considerations. Biotechnology and Bioengineering. 2012;109:992–1006. doi: 10.1002/bit.24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantey LR, Keppler OT, Pawlita M, Reutter W, Hinderlich S. Efficient biochemical engineering of cellular sialic acids using an unphysiological sialic acid precursor in cells lacking UDP-N-acetylglucosamine 2-epimerase. FEBS Lett. 2001;503:80–84. doi: 10.1016/s0014-5793(01)02701-6. [DOI] [PubMed] [Google Scholar]
- 19.Bond MR, Zhang H, Vu PD, Kohler JJ. Photocrosslinking of glycoconjugates using metabolically incorporated diazirine-containing sugars. Nat Protoc. 2009;4:1044–1063. doi: 10.1038/nprot.2009.85. [DOI] [PubMed] [Google Scholar]
- 20.Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci U S A. 2011;108:12758–12763. doi: 10.1073/pnas.1018510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bond MR, Zhang H, Kim J, Yu SH, Yang F, Patrie SM, Kohler JJ. Metabolism of diazirine-modified N-acetylmannosamine analogues to photo-cross-linking sialosides. Bioconjugate Chem. 2011;22:1811–1823. doi: 10.1021/bc2002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dettmer K, Nürnberger N, Kaspar H, Gruber MA, Almstetter MF, Oefner PJ. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal Bioanal Chem. 2011;399:1127–1139. doi: 10.1007/s00216-010-4425-x. [DOI] [PubMed] [Google Scholar]
- 23.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: Sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 24.Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc Natl Acad Sci U S A. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell. 2013;154:416–429. doi: 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bond MR, Whitman CM, Kohler JJ. Metabolically incorporated photocrosslinking sialic acid covalently captures a ganglioside-protein complex. Mol Biosyst. 2010;6:1796–1799. doi: 10.1039/c0mb00069h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science. 1999;284:1372–1376. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 29.Dallolio F, Chiricolo M, Lollini P, Lau JTY. Human colon cancer cell lines permanently expressing α2,6-sialylated sugar chains by transfection with rat β-galactoside α2,6 sialyltransferase cDNA. Biochem Biophys Res Comm. 1995;211:554–561. doi: 10.1006/bbrc.1995.1849. [DOI] [PubMed] [Google Scholar]
- 30.Gross HJ, Brossmer R. Enzymatic transfer of sialic acids modified at C-5 employing four different sialyltransferases. Glycoconj J. 1995;12:739–746. doi: 10.1007/BF00731233. [DOI] [PubMed] [Google Scholar]
- 31.Gross HJ, Rose U, Krause JM, Paulson JC, Schmid K, Feeney RE, Brossmer R. Transfer of synthetic sialic acid analogues to N- and O-linked glycoprotein glycans using four different mammalian sialyltransferases. Biochemistry. 1989;28:7386–7392. doi: 10.1021/bi00444a036. [DOI] [PubMed] [Google Scholar]
- 32.Galuska SP, Geyer H, Weinhold B, Kontou M, Röhrich RC, Bernard U, Gerardy-Schahn R, Reutter W, Münster-Kühnel A, Geyer R. Quantification of nucleotide-activated sialic acids by a combination of reduction and fluorescent labeling. Anal Chem. 2010;82:4591–4598. doi: 10.1021/ac100627e. [DOI] [PubMed] [Google Scholar]
- 33.Mathew MP, Tan E, Shah S, Bhattacharya R, Adam Meledeo M, Huang J, Espinoza FA, Yarema KJ. Extracellular and intracellular esterase processing of SCFA–hexosamine analogs: Implications for metabolic glycoengineering and drug delivery. Bioorg Med Chem Lett. 2012;22:6929–6933. doi: 10.1016/j.bmcl.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tallman KR, Beatty KE. Far-red fluorogenic probes for esterase and lipase detection. Chembiochem. 2015;16:70–75. doi: 10.1002/cbic.201402548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staudinger JL, Xu C, Cui YJ, Klaassen CD. Nuclear receptor-mediated regulation of carboxylesterase expression and activity. Expert Opin Drug Metab Toxicol. 2010;6:261–271. doi: 10.1517/17425250903483215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawley SA, Fullerton MD, Ross FA, Schertzer JD, Chevtzoff C, Walker KJ, Peggie MW, Zibrova D, Green KA, Mustard KJ, Kemp BE, Sakamoto K, Steinberg GR, Hardie DG. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oetke C, Hinderlich S, Reutter W, Pawlita M. Epigenetically mediated loss of UDP-GlcNAc 2-epimerase/ManNAc kinase expression in hyposialylated cell lines. Biochem Biophys Res Comm. 2003;308:892–898. doi: 10.1016/s0006-291x(03)01471-2. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs CL, Goon S, Yarema KJ, Hinderlich S, Hang HC, Chai DH, Bertozzi CR. Substrate specificity of the sialic acid biosynthetic pathway. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 39.Tian L, Yang Y, Wysocki LM, Arnold AC, Hu A, Ravichandran B, Sternson SM, Looger LL, Lavis LD. Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc Natl Acad Sci U S A. 2012;109:4756–4761. doi: 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornfeld S, Kornfeld R, Neufeld EF, O-Brien PJ. The feedback control of sugar nucleotide biosynthesis in liver. Proc Natl Acad Sci U S A. 1964;52:371–379. doi: 10.1073/pnas.52.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seppala R, Lehto VP, Gahl WA. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am J Hum Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaro BW, Chuh KN, Pratt MR. Chemical reporter for visualizing metabolic crosstalk between carbohydrate metabolism and protein modification. ACS Chemical Biology. 2014;9:1991–1996. doi: 10.1021/cb5005564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez AC, Kohler JJ. Recognition of diazirine-modified O-GlcNAc by human O-GlcNAcase. Medchemcomm. 2014;5:1227–1234. doi: 10.1039/C4MD00164H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang PV, Dube DH, Sletten EM, Bertozzi CR. A strategy for the selective imaging of glycans using caged metabolic precursors. J Am Chem Soc. 2010;132:9516–9518. doi: 10.1021/ja101080y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
I. Synthesis and characterization of ManNDAz.
II. Mass Spectrometry detection of Neu5Ac-9-P.