Abstract
The linker histone H1 family members are a key component of chromatin and bind to the nucleosomal core particle around the DNA entry and exit sites. H1 can stabilize both nucleosome structure and higher-order chromatin architecture. In general, H1 molecules consist of a central globular domain with more flexible tail regions at both their N- and C-terminal ends. The existence of multiple H1 subtypes and a large variety of posttranslational modifications brings about a considerable degree of complexity and makes studying this protein family challenging. Here, we review recent progress in understanding the function of linker histones and their subtypes beyond their role as merely structural chromatin components. We summarize current findings on the role of H1 in heterochromatin formation, transcriptional regulation and embryogenesis with a focus on H1 subtypes and their specific modifications.
Keywords: chromatin, epigenetics, linker histone, modifications, subtypes
Introduction
In eukaryotic cells, the genomic DNA needs to be densely packed but still accessible for fundamental processes such as transcription, replication and DNA repair. This is achieved by arranging the DNA in a hierarchical and dynamic manner in a large nucleoprotein complex called chromatin. The fundamental building block of chromatin is the nucleosomal core particle in which approximately 147 bp of DNA are wound around an octamer of the four core histones H2A, H2B, H3 and H4 1. These core particles are connected by a short stretch of linker DNA, forming a structure resembling beads on a string. The linker histone H1 binds to the entry/exit sites of DNA on the surface of the nucleosomal core particle and completes the nucleosome. It influences the nucleosomal repeat length (NRL) 2 and is required to stabilize higher-order chromatin structures such as the so-called 30-nm fibre 3. In vitro polynucleosomes form this 30-nm fibre only at high ionic strength or in the presence of H1 4. For a long time, the linker histones were seen as a rather rigid, merely structural component of chromatin and thus a general repressor of transcription 5. However, for more than a decade, it has been known that linker histones are in fact rather dynamic components of chromatin. FRAP studies with H1-GFP fusion proteins revealed that linker histones have residency times in the range of 3–4 min 6, 7. In contrast, core histones have residency times on a timescale of hours (for a review on H1 mobility see 8).
Another unexpected finding came from numerous knockout studies in different eukaryotes. Assuming a fundamental role in the maintenance of higher-order chromatin structure, depletion of H1 was anticipated to have major effects on nuclear structure and hence also cell viability. Depletion of H1 in Tetrahymena thermophila revealed that H1 is not essential in this organism and that only a specific subset of genes is up- or downregulated 9. Even if full viability without H1 was somewhat surprising, this was the first notion that H1 was in fact not a general repressor but rather a regulator of specific genes. In vertebrates, knockout of H1 is complicated by the presence of multiple subtypes. Whereas knockout of only one H1 subtype in mouse did not cause a pronounced phenotype 10–13, the simultaneous knockout of three H1 subtypes was embryonically lethal, for the first time demonstrating the essential role of linker histones in mammals. Cells obtained from these triple H1-null embryos contained about 50% of the normal H1 amount 14 leading to a global reduction in nucleosomal repeat length and local decompaction of chromatin. Likewise, chicken complete knockout cells displayed decreased global nucleosome spacing and increased nuclear volume 15, but are viable. Remarkably, in all organisms analysed, the reduction in H1 levels did not cause global upregulation of transcription but rather affected a specific set of genes 9,15–19. For a more detailed overview on H1 knockout studies, we would like to refer the reader to the review of Izzo et al, 2008 20.
To increase the puzzling complexity of linker histones, H1 proteins can—like core histones—be modified at multiple sites by different posttranslational modifications (e.g. 21–27), which opens up many possibilities for the regulation of linker histone function. We will discuss these modifications in the second half of this review in more detail.
During the last few years, the development of some highly specific antibodies, advanced knockout techniques and the use of new, genomewide analysis methods have shed new light on the role of H1 in chromatin organization and dynamics. This review aims to point out the most recent findings concerning the functions of H1 subtypes and modifications. For additional information, the reader may be referred to recent reviews on other aspects of the linker histone family (e.g. 3,20,28–31).
Linker histone structure and position in the nucleosome
In metazoans, H1 histones are relatively small (∼200 aa) proteins with a three-domain structure consisting of a short N-terminal tail (∼20–35 aa), a central globular domain (∼70 aa) and a long, extremely basic C-terminal tail (∼100 aa). The central globular domain is highly conserved among all H1 subtypes and contains a winged-helix motif 32. The C-terminal domain (CTD) is largely unstructured in aqueous solution but adopts an extensively folded structure upon interaction with DNA 33.
In lower eukaryotes, linker histone-like proteins can be found. In Tetrahymena, H1 molecules are similar to the CTD of metazoan H1 without a globular domain 34. In Saccharomyces cerevisiae, the linker histone-like protein Hho1p possesses two globular domains 35, whereas S. pombe seems to lack a linker histone 36.
Since there is currently no crystal structure of a nucleosome containing H1 available, many attempts have been made to determine the exact position of H1 (or at least its globular domain) within the nucleosome and its precise interaction with the linker DNA. This issue still remains a matter of debate. Based on data from cryo-electron microscopy, hydroxyl radical footprinting and nanoscale modelling, Syed et al 37 developed a model where the globular domain of histone H1 interacts with the DNA minor groove located at the centre of the nucleosome and symmetrically contacts a 10-bp region of each linker DNA. Analysing in vitro reconstituted mammalian 30-nm fibres with cryo-electron microscopy combined with fitting of the chicken histone H1 globular domain structure, Song et al 38 described a similar model with the H1 globular domain asymmetrically binding to the minor groove and contacting both linker DNA strands. In contrast, Zhou and colleagues analysed the Drosophila melanogaster H1–nucleosome complex by solution NMR spectroscopy. They report that the globular domain of H1 uses two positively charged surfaces to bridge the nucleosome core and the linker DNA asymmetrically and interacts tightly with only one 10-bp stretch of linker DNA 39. This supports previous results obtained by combining FRAP assays for measuring the binding of wild-type or mutant globular domains of histone H1.0 to DNA in vivo 40. All of these models are not necessarily contradicting each other because H1 might have more than one mode of binding. Most of these studies are based on in vitro experiments and isolated or reconstituted chromatin/nucleosomes in the absence of many other chromatin components (such as other chromatin proteins or chaperones) and histone modifications and therefore do not necessarily fully reflect the in vivo situation.
Histone H1 subtypes and their binding affinity to chromatin
The linker histones display much higher sequence variability between different species than the evolutionary extremely conserved core histones. Additionally, higher eukaryotes contain multiple H1 subtypes. For example, 11 H1 genes have been described in mice and humans. The five H1 family members H1.1–H1.5, the so-called somatic linker histone subtypes, are widely expressed in many different cell types in a mainly replication-dependent manner with a peak of expression in S phase 41. These somatic subtypes are encoded together with the core histone genes in the histone gene cluster 42,43. This is remarkable regarding the fact that the core histone genes have their origin in archeabacteria, whereas linker histones have an eubacterial ancestor 44. H1.0 and H1x are expressed independent of the cell cycle, and it has been suggested that H1.0 replaces somatic H1 subtypes in terminally differentiated cells 45,46. Four H1 subtypes are found in germ cells (H1oo in oocytes and H1t, H1T2 and HILS1 in spermatids or spermatocytes) 47–49. For comprehensive reviews on H1 subtypes, see 20,28.
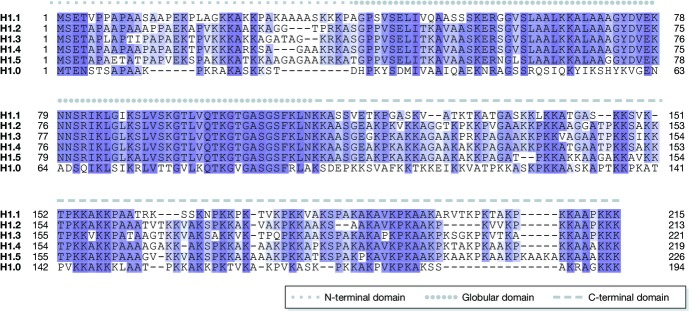
At this point, the question arises why multiple H1 proteins exist and why they are conserved through evolution. The genes of the mammalian H1 subtypes originated from gene duplication events are paralogs, whereas H1 subtypes in two different species are orthologs (such as human H1.4 and mouse H1e). Comparing the paralog linker histone family members within a single species, the highest conservation is found in the globular domain, whereas both tails are more variable. This is especially true for the mammalian somatic linker histone subtypes H1.1–H1.5. They share an almost identical amino acid sequence in the globular domain, as depicted in Fig1 for the human somatic H1 subtypes and H1.0. When comparing different species, it is remarkable that the tail regions of corresponding H1 subtypes are highly conserved. For example, human H1.4 shares 93.5% sequence identity with its mouse ortholog H1e 20. The concept that orthologs such as human H1.4 and mouse H1e share more similarity than paralogs within a species is a good argument in favour of a functional significance for the multiple subtypes. Furthermore, the high conservation of the tails suggests that functional differences are mainly conferred via the more variable tail regions.
Figure 1. Alignment of the somatic human H1 isotypes and H1.0.
The globular domain (solid line) is highly conserved, whereas the N- and C-terminal domains (dotted and dashed lines, respectively) are more variable 130. Conserved residues are highlighted by shades of blue. Darker colour represents higher conservation.
In this context, differences in the most basic functional aspects of H1—namely binding affinity to chromatin and influence on NRL—were investigated by different approaches. In FRAP experiments with human cell lines expressing H1 subtypes with N-terminal eGFP fusions, different recovery times were observed for different subtypes 50,51. Whereas subtypes with shorter C-terminal tails (H1.1 and H1.2) recover within 1–2 min, the subtypes with longer C-terminal tails (H1.4 and H1.5) need up to 15 min to reach equilibrium again. However, in these studies effects of the GFP fusion cannot be excluded.
The effects of different H1 subtypes on NRL have been evaluated both in vitro and in vivo. Chromatin assembled with different human H1 subtypes in vitro in H1-free preblastodermic Drosophila embryo extracts displays distinct nucleosomal repeat length. H1.5 and H1.4 (subtypes with high chromatin affinity in the same study) result in the longest NRL, while H1.2, H1.3 and H1.0 in an intermediate and H1.1 and H1x in the shortest NRL 52. These results are supported by an in vivo study performed by Öberg et al 53 in Xenopus oocytes. However, in this study, H1.5 showed a rather low affinity for chromatin as was observed also in several other in vitro studies 54,55. A likely explanation for these differences might be that H1 affinity to chromatin is also regulated by other factors like chaperones, competing proteins or H1 modifications 8. In these rather artificial systems, cofactors specific for H1.5 might be missing.
This work shows that almost 50 years after Kinkade and Cole described the first biochemical fractionation of histone H1 subtypes 56, the specific functional role of individual H1 subtypes is still not fully understood. However, despite the potential limitations of the different in vitro and in vivo approaches mentioned, they nourish the idea of subtype-specific H1-chromatin binding.
Subnuclear distribution of linker histone subtypes
The finding that H1 is involved in the regulation of specific subsets of genes suggests that H1 may not be uniformly distributed in the genome. To date, only a handful of studies provide detailed genomewide binding studies of histone H1 in diverse organisms using different methodologies. These studies all demonstrate a characteristic binding pattern of H1 with dips of low H1 occupancy at transcription start sites mainly at active genes that seems to be evolutionary conserved 57–62.
Many mapping studies were performed in organisms either with only one H1 subtype, for example Drosophila melanogaster, or with antibodies that cannot discriminate between subtypes. These studies give interesting insight in H1 localization, however not about subtype specificity. Braunschweig et al 58 mapped the single H1 subtype in the embryonic D. melanogaster cell line Kc167 with the so-called DamID technique. This method applies fusion of H1 with a DNA adenine methyltransferase (Dam) that specifically methylates adenine in GATC sequences, which can be detected by DpnI restriction digestion coupled with microarrays or sequencing. The authors report a pervasive binding pattern of H1 with similar levels in euchromatin and heterochromatin. The characteristic H1-depleted regions (dips) are mainly correlated with active transcription start sites or putative regulatory elements in intergenic regions. Furthermore, the histone H3 subtype H3.3 was identified as a negative regulator of H1 binding. Interestingly, this H3 histone has been linked with gene activation via opening of higher-order chromatin 63. A similar depletion pattern for H1 was described in human MCF-7 cells by ChIP combined with microarrays with an H1 antibody that does not distinguish between H1 subtypes 57. In this case, the authors compared the binding of H1 with PARP-1 enzyme occupancy and demonstrated reciprocal binding of the two proteins with a negative correlation of H1 binding and gene expression. A recent study in Drosophila S2 cells uses ChIP-sequencing (ChIP-seq) to localize H1 and HMGD-1, another chromatin architectural protein, genome wide 61. The authors again observe depletion of H1 at transcription start sites of active genes. However, they combine their study with biochemical fractionation of chromatin, and in contrast to the study of Braunschweig et al, they claim a relative enrichment of H1 in heterochromatin whereas HMG-1 preferentially associates with euchromatin.
Studying the differential distribution of the individual linker histone family members can be extremely challenging in a mammalian system since currently no subtype-specific ChIP-grade antibodies are commercially available. The generation of such antibodies is complicated due to the high sequence similarity of H1 family members. Only in the last 2 years, several groups have revisited this question using genomewide methodologies. In mice ESCs, Cao et al 60 knocked in tagged versions of the most abundant H1 subtypes H1d and H1c (corresponding to human H1.3 and H1.2, respectively), and overexpressed a tagged replacement subtype H1.0 to perform ChIP-seq. In this study, H1d and H1c show very similar binding patterns; however, they also possess different binding specificities. Both are enriched in major satellite repeats. These repeats constitute the pericentromeres that form chromocentres with a more stable and condensed chromatin conformation 64. H1.0 often correlates with H1d and H1c; however, it also displays differential binding preferences with enriched binding at minor satellites and LINE-1 elements. In support of this enrichment pattern, triple knockout of H1d, H1c and H1e (corresponding to human H1.2, H1.3 and H1.4) leads to vastly increased transcription from major satellite repeats in parallel with shorter NRL and clustering of chromocentres 60.
Izzo et al, 59 mapped the location of the five somatic H1 subtypes (H1.1–H1.5) in human lung fibroblasts with the above-described DamID technique 65. Millan-Arino et al 62 approached the problem by overexpressing H1.2-H1.5 with a C-terminal HA-tag in a human breast cancer cell line followed by ChIP-seq. Both groups find a non-uniform distribution of H1 molecules over the genome with differences between the H1 subtypes. In general, H1 has a wide occupancy over the genome but is depleted from active transcription start sites. In the study of Izzo et al, H1.2–H1.5 show a rather similar binding profile with depletion at CpG-dense regions and active regulatory elements, whereas H1.1 displays a strikingly different binding pattern with even a slight enrichment at some active chromatin domains. Millan-Arino et al did not address H1.1 distribution, since H1.1 is not expressed in the cell type they analysed. Interestingly, both studies find a connection of H1 subtype occupancy with regions in the genome that are associated with 3D organization of chromatin. H1.2-H1.5 59 or H1.2 62, respectively, were found to be enriched in gene-poor regions, low GC content or lamina-associated domains (LAD). When comparing these two approaches, it is important to consider that the DamID technique does not rely on overexpression with the risk of “atypical” binding and compensatory downregulation of other subtypes; however, it has limited resolution and does not provide full insights into repetitive elements lacking GATC sequences.
A further layer of complexity is added by changes in the distribution of certain H1 subtypes during cellular differentiation. Li et al demonstrated recently that H1.5 shows a different distribution in human ESCs compared with differentiated fibroblasts. Most strikingly, H1.5 associates with transcriptionally silent genes encoding membrane-associated proteins in differentiated cells, but not in undifferentiated ESCs 66. Regarding the replication-independent subtypes H1.0 and H1x, some interesting features were observed. The replacement subtype H1.0, which accumulates in terminally differentiated cells, is enriched in nucleolar-associated DNA regions, for example the repetitive sequences coding for ribosomal DNA. The least characterized subtype H1x also displays a “typical” H1 binding profile with depletion at active transcription start sites, but interestingly is enriched at sites positive for RNAP II and thus ongoing transcription in gene bodies 67.
For the future, it will be very important to develop novel tools to study H1 subtypes such as subtype-specific antibodies that can be used in ChIP, or knock in systems where all expressed endogenous H1 subtypes are tagged. Alternatively, other techniques could be applied such as MALDI imaging, which can be used to measure mass spectra from tissues and thereby protein distribution, such as the distribution of H1 subtypes.
Based on the above-discussed studies, we favour a model in which there are regions in the genome where H1 subtypes might be interchangeable, providing basic H1 functions (e.g. global chromatin organization). However, distinct H1 subtypes might be needed for specific functions at specific regions in the genome, for example the precise regulation of genes. The distribution of H1 subtypes seems to vary between species, cell types and also during cell differentiation, raising the question of the role of H1 in cellular differentiation and development. Thus, we are only beginning to understand how the H1 subtypes help to organize chromatin in time and space in the nucleus.
Potential H1 functions
H1 and heterochromatin formation
A fundamental role for H1 in heterochromatin formation was recently identified in D. melanogaster. When the protein level of the single somatic H1 subtype was reduced to ∼20% of the normal content with an RNAi approach, the larvae depleted of H1 did not develop to adult flies 68. Strikingly, pronounced effects on heterochromatin structure and function were observed, for example H1 was a strong suppressor of position effect variegation (PEV) and essential for the organization of pericentromeric regions in polytene chromosomes. Normal salivary gland polytene chromosomes display a pattern of bands and interbands with prominent chromocentre staining for heterochromatin protein 1 (HP1) and the repressive mark H3K9me2/3. Depletion of H1 resulted in a massively altered chromosome structure with loss of chromosome banding. H3K9me2 staining was undetectable and instead in a single chromocentre several HP1-foci were observed 68. The same group reported very recently that loss of H1 caused derepression of more than 50% of transposable elements (TEs) but only about 10% of protein-coding genes. TEs in D. melanogaster are thought to be mainly located in heterochromatin. The authors demonstrated that H1 plays a critical role in silencing TEs by physically interacting with the histone methyltransferase Su(var)39 and tethering it to heterochromatin, thus facilitating methylation of H3K9 69 and providing a binding platform for HP1. Additionally, H1 can also bind directly to HP1. A second pathway for H1-dependent heterochromatin formation involves the single STAT protein in D. melanogaster STAT29E. Besides its role in JAK-STAT signalling, STAT29E was shown to be a widespread component of chromatin with an important function in formation of pericentric heterochromatin. Interestingly, STAT29E localization to chromatin is dependent on its interaction with H1 70.
Several reports have also described links between H1 and heterochromatin components in mammalian cells. Perhaps the most well-characterized interaction is between H1 and the HP1 protein, which is thought to be subtype specific and dependent on a specific methylation of H1 71–74 (see H1 modifications). Trojer et al 75 described an additional interaction of methylated H1 with L3MBTL1, a transcriptional repressor binding to specific mono- and dimethylated lysine residues on histones and locally compacting chromatin. Findings in ESCs from triple H1 knockout mice also underline the role of H1 in structural maintenance of heterochromatin 60 (see “subnuclear distribution of H1 subtypes”). Furthermore, in mouse mature photoreceptors, H1c promotes heterochromatin condensation 76. Given this seemingly fundamental role of H1 in heterochromatin formation in D. melanogaster, it will be a challenging task to determine the significance of H1 in heterochromatin formation in mammals in more detail.
H1 in specific gene expression
Manipulating H1 levels was shown to lead to both up- and downregulation of specific genes. Here, we want to give a short overview of the many studies that have described this effect in various organisms that express one or multiple H1 subtypes. In early studies, Shen et al 9 demonstrated that H1 regulates specific genes in Tetrahymena. Hashimoto et al 15 generated the first vertebrate total H1 knockout in chicken cells, removing all six H1 subtypes genetically. They found that in histone H1-deficient cells, the expression of multiple genes was affected, mainly by downregulation. In D. melanogaster, RNAi-induced H1 depletion preferentially affected genes in heterochromatic regions, and H1 was required for silencing of transposable elements, as mentioned above 18.
Interestingly, the Skoultchi group found that in mouse models, depletion of individual H1 subtypes can affect PEV and gene expression 77. In their seminal paper describing the H1 triple-knockout ESCs and a reduction of total H1 by 50%, the same group reported that only very few but specific genes are up- or downregulated, demonstrating a clear role of H1 in fine-tuning gene expression 14. In an approach alternative to knockout studies, Sancho et al 19 created stable T47D breast cancer cell lines carrying inducible vectors encoding shRNAs that should target and deplete H1.0, H1.2, H1.3, H1.4 or H1.5 and found that different subsets of genes were affected upon knockdown of specific H1 subtypes. This approach enabled rapid depletion of a single H1 subtype, circumventing some of the problems caused by potential compensatory effects due to upregulation of other H1 subtypes in conventional knockout studies. However, in shRNA approaches low levels of the target protein may remain.
In addition to these studies of the general effects of H1 depletion on gene expression, histone H1 proteins have been implicated in the regulation of specific model genes, by both activation and repression. Some of these studies provide very detailed mechanistic insight into the modes of action of H1, such as hormone-induced transcription at the mouse mammary tumour virus (MMTV) promoter. The earliest work on this topic described H1 displacement upon hormonal induction 78. Later it became clear that the situation was more complex and that the presence of H1 prior to hormone induction facilitates efficient transcriptional activation 79. In fact, it was demonstrated that binding of H1 to the MMTV promoter induces a distinct conformation of chromatin, facilitating the binding of hormone receptor and transcription factors 80,81. The subsequent displacement of H1 from the promoter involves H1 phosphorylation 80,82 (see also the section on H1 modifications).
This direct influence on chromatin architecture is however only one of the means of linker histone-mediated transcriptional regulation. It is more and more apparent that direct interaction of H1 with both transcriptional activators and repressors also plays an important role. In a recent report, Kim et al 83 demonstrated that H1.2 selectively binds the Cul4A ubiquitin ligase and the PAF1 elongation complexes as well as the phosphorylated C-terminal domain of RNAP II. Cul4A ubiquitinates H4K31, which in turn was shown to be necessary for H3K4 and H3K79 methylation, both marks associated with active transcription. Based on these findings, the authors postulated an interesting cooperative model in which H1.2 is tethered to actively transcribed target loci via recognition of RNAP II Ser2 phosphorylation and recruits Cul4A and PAF1 to help maintain active gene transcription.
H1 has also been linked to repression of specific genes. For example, a role of H1 in the regulation of interferon (IFN)-stimulated genes has been shown 84. Multiple transcriptional repressors have been found in a complex with H1. An example of such H1 binding partners linked to repressive chromatin states is Msx1, a negative regulator of muscle cell differentiation or the HP1 protein. Msx1 recruits mouse H1b to a key regulatory element in the MyoD gene, inducing a repressive chromatin state, which resulted in repressed muscle cell differentiation 85. The binding of HP1 to human H1.4 methylated at K26 was demonstrated in both in vitro and in vivo systems 72, but has thus far not been directly linked to the transcription of specific genes. Only recently, Studencka et al 86 provided evidence that the interaction of HP1 with the homologous linker histone of Caenorhabditis elegans HIS24 methylated at K14 is in fact involved in the regulation of innate immune genes and genes regulating stress response. Further examples where H1 acts as a repressor include its interaction with the p53 protein or nuclear ribosomal proteins. In mammals, p53 has been reported to directly interact with an H1.2 complex, where H1.2 acts as a repressor of p53-mediated transcription 87. In D. melanogaster, ribosomal proteins have been found to interact with H1 and could connect H1 and gene repression 88.
H1 also seems to regulate DNA methylation and hence gene expression. Fan et al 16 found that nearly one-third of the genes misregulated in H1 triple-knockout mice were thought to be regulated by DNA methylation. Yang et al 89 recently analysed more closely the role of H1 in silencing of h19 and Gtl2, two genes whose expression is regulated by DNA methylation of their imprinting control regions (IRCs) in mouse ESCs. They demonstrated that some H1 subtypes specifically interact with the DNA methyltransferases DNMT1 and DNMT3B and thus can recruit these enzymes to ICRs. Furthermore, H1 interfered with the setting of an “active” histone mark, namely H3K4 methylation by Set7/9. Interestingly, these effects were H1 subtype specific, providing more evidence for subtype-specific functions of the H1 family members. It is of note that there is now first evidence that H1 can also regulate antisense transcription: H1.3 has been found to inhibit h19 noncoding RNA expression 90.
These studies provide some first insight into the many genes regulated by H1 and support the concept that H1 can not only block the binding of other proteins to chromatin but also could serve as a recruitment platform for transcriptional activators or repressors. Since H1 is often considered a fine-tuner of gene expression, it will be interesting for the future to focus on the global role of H1 subtypes in an unbiased manner in gene induction for example during stress responses and on the consequences on biological fitness, rather than merely on steady state levels of gene expression.
H1 in DNA repair
DNA damage and subsequent DNA repair involve major chromatin reorganization events, including local opening of chromatin. Therefore, it is conceivable that histone H1 might play a role in this context too. Indeed, the S. cerevisae H1 homolog, Hho1p, has a global role in DNA repair by inhibiting homologous recombination, giving rise to hyper-resistance to DNA damage upon loss of Hho1p, a rather unusual phenotype in yeast 91.
Also in higher eukaryotes, H1 has been linked to DNA repair: Hashimoto et al described a unique role for the chicken histone H1 subtype H1R in the DNA damage response. Although the underlying molecular mechanism of the reported altered sensitivity of the H1R−/− mutants to DNA-damaging agents is still unknown 92, some evidence might come from findings in the famous mouse triple-knockout cells. These cells are hyper-resistant to DNA damage, most likely due to a more open chromatin structure that could facilitate genome surveillance mechanisms 93. Recently, the Mailand laboratory identified histone H1 as a key target of ubiquitination by RNF8-Ubc13 in double-stand break (DSB) repair. Ubiquitylated forms of H1 proteins were found to serve as an initial binding platform for the ubiquitin ligase RNF168 that ubiquitinates other targets and to be more loosely associated with chromatin, suggesting that ubiquitylation of H1 may play a role in facilitating chromatin remodelling to allow for efficient repair 94. This, as shown previously for H1.4K34ac, is another example where H1 modifications can act as recruiters of specific factors as well as directly on H1 dynamics. However, in the case of H1 ubiquitination, it is still unclear whether this is an H1 subtype-specific effect. Furthermore, H1 phosphorylation has also been implicated in DNA repair; for details, see the H1 Phosphorylation section.
H1 in early embryogenesis
It seems a common theme in metazoan development that in germ cells, the so-called germ cell-specific H1 subtypes can at least partly replace somatic H1 subtypes. For a long time, D. melanogaster seemed to be an exception, with only one H1 protein characterized. Yet, in 2013, dBigH1, an embryonic linker histone, was identified with an unusually long N-terminal tail enriched in negatively charged amino acids. dBigH1 is present in germ cells and during the first hours of embryogenesis, vanishing upon the onset of cellularization when it is replaced by the somatic dH1. dBigH1 is essential for early embryonic development and prevents premature zygotic genome activation 95.
In Xenopus, maternally expressed B4 is the main linker histone found in eggs and is replaced by somatic H1 after the midblastula transition concomitant with zygotic gene activation 96. It seems that B4 favours a more open chromatin conformation and allows ATP-dependent chromatin remodelling 97. Interestingly, B4 plays a major role in successful reprogramming of somatic mammalian cell nuclei via transplantation into Xenopus oocytes. Following nuclear transfer, the somatic H1 subtypes are rapidly lost from chromatin and exchanged by B4. The incorporation of B4 is required for successful reprogramming, possibly by supporting the reactivation of pluripotency genes 98.
In mammals, oocyte-specific H1oo is expressed until the late two-cell embryonic stage 49,99. Prolonged ectopic expression leads to a prolonged expression of pluripotent marker genes and prevents differentiation 100. However, it is still unclear whether during this period also somatic H1 subtypes are present.
From these studies, it is obvious that germ cell-specific H1 subtypes seem to play an important role in early embryogenesis and reprogramming, even though further work will be required to better understand the function of these more specialized H1 subtypes with very restricted expression patterns. This will require a combination of in vivo and in vitro approaches.
Linker histone modifications
Already in the 1970s, phosphorylation of histone H1 was discovered (see, e.g. 101), but only during the last decade, it has become clear that the linker histones are posttranslationally modified by many more chemical groups on residues in both their tail regions and globular domain, similar to the core histones. In a number of mass spectrometric studies H1 methylation, acetylation, ADP-ribosylation, ubiquitination, formylation and PARylation have been identified 21,23,25–27,102–106. These studies come from diverse organisms such as Drosophila, chicken, mouse and humans. With these findings, the question has emerged whether the so-called histone code hypothesis, suggesting specific functions for distinct modifications, could also be extended to the linker histones. However, due to the lack of site- and modification-specific antibodies, the H1 modification field is currently much less advanced than the core histone modification field. In the following sections, we mainly focus on modifications described in mammalian model systems.
H1 Phosphorylation
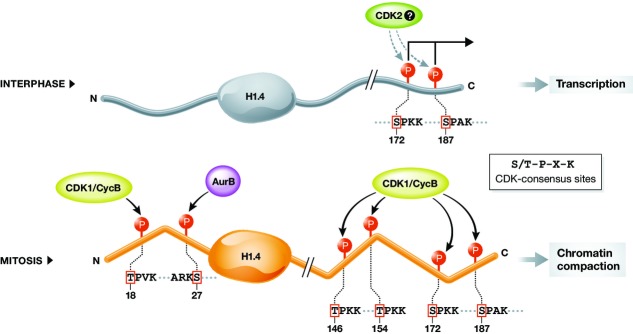
Phosphorylation is so far the most well-characterized H1 modification. Phosphorylation of linker histones occurs mainly in the tail regions, especially the C-terminal tail, where several {(S/T)-P-X-(K/R)} motifs are located that are recognized by cyclin-dependent kinases (CDKs). Phosphorylation levels are lowest during the G1 phase of the cell cycle, rise during S phase and reach maximum levels in mitosis, sharply dropping in telophase. Talasz et al 107 showed for H1.5 that during G1 and S phase, mainly Ser residues are modified and that Thr phosphorylation occurs mainly during mitosis. In mitosis, CDK1/CycB is predominantly responsible for H1 phosphorylation, but involvement of other kinases has also been reported. For instance, H1.4S27, H1.4S35 and H1.5T10 are phosphorylated by Aurora B kinase, protein kinase A and glycogen synthase kinase-3, respectively 108–110. Figure2 depicts cell cycle-dependent phosphorylation of H1.4. Whether H1 phosphorylation is indeed necessary for correct chromosome condensation in mitosis has been debated; however, a study has demonstrated that the linker histone itself is indeed required for metaphase chromosome compaction 111. Furthermore, agents causing H1 dephosphorylation in mitotic cells concomitantly lead to chromosome decondensation 112,113.
Figure 2. Cell cycle-dependent phosphorylation of H1.4.
In interphase (top panel), two phosphorylated serine residues have been detected in the C-terminal tail, S172 and S187 116,131. Both residues are part of a CDK consensus sequence (S/T-P-X-K) and are likely to be phosphorylated by Cdk2 82,115,132. S172p and S187p have been linked to active transcription by RNAP I and II 116. Phosphorylation levels are thought to increase during S phase and are highest in mitosis (bottom panel), where up to six phosphorylation sites have been identified 116 in both the N- and C-terminal tails. Threonine residues are phosphorylated mainly during mitosis 131. Ser/Thr residues located in CDK consensus sites are thought to be targeted by the mitotic kinase CDK1/CycB 133. H1.4S27 is phosphorylated by Aurora B, a member of the chromosomal passenger complex 108. Full phosphorylation during mitosis may allow for a structural rearrangement inducing chromatin compaction 118. p: phosphorylation, S/T-P-X-K: CDK consensus site.
In contrast to the situation in mitosis, in G1 and S phase H1 phosphorylation is involved in processes requiring local opening of chromatin. Alexandrow et al 114 detected histone H1 phosphorylation at replication sites in S phase and hypothesized that phosphorylation of H1 favours chromatin decondensation and thus facilitates fork progression.
Several publications confirm a link between H1 phosphorylation and active transcription. According to the current model, phosphorylation of H1 could weaken its binding affinity to chromatin and favours removal of H1 from active promoter regions. This has been studied in detail for hormone-induced MMTV promoter activation 80,81,115. Vicent et al 82 provided a detailed study of the events taking place during the first minute after progesterone induction of the MMTV promoter. The chromatin remodelling complexes NURF and ASCOM are recruited to the promoter by the activated progesterone receptor. The ASCOM complex contains methyltransferase subunits that specifically trimethylate H3K4. This mark then stabilizes NURF binding. NURF remodelling in turn facilitates recruitment of the Cdk2/cyclin A kinase, which phosphorylates H1, leading to its removal and thus facilitating access of, for example, transcription factors to the MMTV promoter region.
Zheng et al 116 identified additional interphase phosphorylation sites (H1.2S173p, H1.4S172p and H1.4S187p) in human HeLaS3 cells and showed that these phosphorylations are enriched in nucleoli. With ChIP experiments, they could further demonstrate that H1.4S187p is associated with active rRNA promoters and is induced at hormone response elements supporting a role of specific H1 phosphorylations in both RNAP I- and RNAP II-mediated transcription.
It is logical to assume that histone H1 phosphorylation and its effects on chromatin binding could also be implicated in DNA damage repair. Indeed, it has been proposed that the phosphorylation status of H1 could indicate the degree of DNA damage to the cell 117. At low levels of DNA damage, only few H1 molecules would become phosphorylated and would be released from chromatin, allowing chromatin decondensation and the access of repair proteins. However, if the DNA damage is very strong, much more H1 would be phosphorylated and released from chromatin, signalling that the DNA damage is beyond repair.
Is seems counterintuitive that H1 phosphorylation would mediate both chromatin condensation and decondensation. A model to solve this conundrum based on structural data comes from Roque et al who analysed the effect of partial and full phosphorylation of the H1 C-terminal domain on its secondary structure when bound to DNA. Depending on the phosphorylation level of the CTD different proportions of α-helixes, β-structures and also unstructured regions were observed, suggesting phosphorylation-dependent structural rearrangements 118. Moreover, partial phosphorylation of full-length H1 impaired its capacity to aggregate chromatin 119. Taken together, this model suggests that site-specific H1 phosphorylations could bring about distinct structural changes influencing chromatin architecture in different ways, revealing that this modification can have complex effects.
Methylation
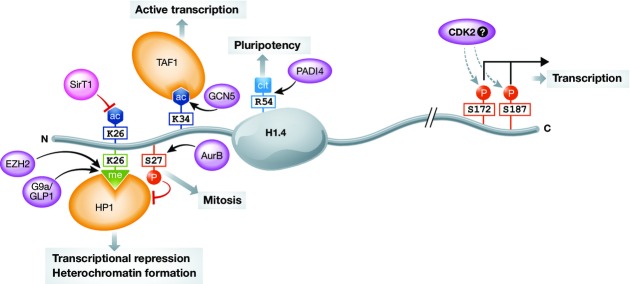
The most prominent methylation sites in linker histones have been found within the N-terminal tail of H1 (Fig3). Methylation of lysine K26 in the N-terminal tail of H1.4 is the most abundant methylation occurring on a human linker histone 23. Interestingly, K26 methylation is conserved in vertebrates and was, for example, also found among the H1 modifications in D. melanogaster (K27me2), suggesting a conserved function for this modification 26. In mammalian cells, it has been described that H1.4K26me is catalysed by the Ezh2 methyltransferase as part of the PRC2 complex and by the methyltransferases G9a (together with its interaction partner Glp-1), whereas it is removed by the lysine demethylase JMJD2/KDM4 74,120,121. Methylated H1.4K26 provides a binding platform for HP1 and L3MBTL1, both proteins with prominent roles in heterochromatin. Interestingly, H1.4K26 is part of an “ARKS” motif, and concomitant phosphorylation of the adjacent Ser27 inhibits HP1 binding, thus providing an example for crosstalk between modifications on H1 72.
Figure 3. H1.4 modifications and their cellular functions.
Among the various modifications of linker histones, only few were characterized with site-specific antibodies. Mostly modifications on the subtype H1.4 have been characterized. H1.4 is methylated at K26, which is catalysed by G9a/GLP1 and potentially also by EZH2 120,134. This methylation provides a binding platform for HP1 and is thus linked to transcriptional repression and heterochromatin formation. Phosphorylation of S27 inhibits binding of HP1 to K26me 72. The C-terminal phosphorylations S172p and S187p are present on H1.4 also in interphase and have been linked to active transcription 116. H1.4K34ac is set by the acetyltransferase GCN5 and is enriched at active transcription start sites. It can positively regulate transcription by (i) recruiting the bromodomain-containing TAF1 subunit of the TFIID transcription complex and (ii) increasing H1 mobility 122. The conversion of R54 to citrulline by PADI4 has been shown to occur on several mouse H1 subtypes, among them H1.4. This modification in a DNA-binding site results in eviction of H1 and global chromatin decondensation in pluripotent cells. p: phosphorylation, ac: acetylation, me: methylation, cit: citrullination, violet: enzymes, orange: readers.
Due to the lack of H1 methylation site-specific modification antibodies, there is currently very little information about the function of distinct methylation sites. However, Weiss et al 120 demonstrated that G9a/Glp1 methylate different H1 isotypes at different sites (namely H1.4 on K26, H1.2 on K187, but not on K26/27) and that different methylation sites have different readers, supporting the idea of specific functions for specific methylations.
Acetylation
Acetylation of H1 occurs in both the tail regions and the globular domain. Interestingly, the acetylation sites in the globular domain are mostly residues that are thought to be directly involved in DNA binding 21. For core histones, acetylation has been generally linked to opening of chromatin structure and active transcription. It is likely that acetylation of H1 at these sites directly affects DNA binding and may contribute to H1 displacement. However, it is often difficult to distinguish effects of linker histone acetylation from core histone acetylation, especially in studies that rely on inhibitors of histone deacetylases.
A role of H1 acetylation in transcription has been demonstrated for an N-terminal H1 acetylation site. A thorough analysis of H1.4K34 acetylation provided evidence that the histone acetyltransferase GCN5 acetylates H1.4K34 (Fig3). This acetylation mark can activate transcription by recruiting TAF1, a subunit of the transcription factor TFIID, and also directly by reducing H1 affinity to chromatin 122. Acetylation of H1.4K26 has also been reported; however, the corresponding acetyltransferase has not yet been identified. SirT1 can remove K26 acetylation, thus providing the possibility for methylation and subsequent heterochromatin formation 123. T47D cells expressing a H1.4K26A mutant, but no endogenous H1.4, display defects in cell proliferation and misregulation of specific genes, underlining the importance of modifications of H1.4K26 124. Since the same enzymes can acetylate/deacetylate core histones and H1, this of course raises the question of functional links between H1 and core histone acetylation.
Other H1 modifications
In 2000, Pham and Sauer described potential H1 monoubiquitination in Drosophila by TAFII250, a subunit of the TFIID, and herewith linked it to transcription 125. Only recently, specific ubiquitination sites were identified in D. melanogaster H1 on K23, K27 and K165 26. Ubiquitination occurs also on mammalian linker histones 21, but so far its function has not been analysed.
Most recently, H1 citrullination was described by Christophorou et al 126. The conversion of an arginine residue to citrulline by peptidylarginine deiminases (PADIs) results in the loss of a positive charge. PADI4 citrullinates several H1 subtypes at R54 in mouse ESCs. Surprisingly, citrullination of a single arginine residue, H1K54, was reported to evict H1 from chromatin and to result in global chromatin decondensation (Fig3).
Overall, during the last few years, multiple laboratories have managed to gain striking new insights in some histone H1 modifications; however, the H1 modification field is still lagging far behind the core histone modification field. One reason for this is the surprisingly low success rate when attempting to raise H1 modification- and subtype-specific antibodies. In our experience, the success rate to get such antibodies—after purification and stringent quality controls—is < 1 in 100, independent of the species we tried for the immunization. Novel approaches are urgently needed to solve this issue.
Conclusion
The view on the linker histone family has changed dramatically from a rather static chromatin component stabilizing higher-order structures to a versatile protein that can regulate multiple DNA-dependent processes. In many cases, H1 subtypes may be redundant, explaining overlapping binding profiles in genomewide localization studies and compensation effects upon single H1 knockouts. However, there is now increasing evidence that there are also subtype-specific functions, where H1 subtypes act as binding platforms for interacting proteins, or selectively regulate chromatin organization. Table1 provides an overview over the H1 subtype-specific functions discussed in this manuscript. It has also become clear that additionally, H1 subtype functions can be regulated by subtype-specific modifications (see Fig4 for a model of the multiple H1 functions). Further interest in histone H1 and its subtypes will come from the finding that subtype-specific recurrent histone H1 loss-of-function mutations could be drivers in cancer 127.
Table 1.
Examples of H1 subtypes and their specific functions
| H1 subtypea | Functions | References |
|---|---|---|
| Saccharomyces cerevisiae (1 subtype) | ||
| Hho1p | DNA repair, inversely linked with gene expression | 91 |
| Tetrahymena thermophila (1 subtype) | ||
| H1 | Depletion results in up- and downregulation of specific genes, not essential | 9 |
| Caenorhabditis elegans (8 subtypes) | ||
| HIS24me | Regulation of innate immune genes and stress response genes | 86 |
| Drosophila melanogaster (2 subtypes) | ||
| Somatic H1 | Heterochromatin formation, silencing of TEs (involved in Su(var)39/HP1 pathway, JAK/STAT29E pathway) essential | 69,70 |
| dBigH1 | Prevents premature zygotic genome activation | 95 |
| Xenopus laevis (5 subtypes) | ||
| B4 (germ cell-specific) | Open chromatin conformation, ATP-dependent chromatin remodelling required for successful reprogramming of somatic mammalian cell nuclei in transplantation experiments | 97,98 |
| Chicken (7 subtypes) | ||
| H1R | DNA damage response | 92 |
| Mouse (11 subtypes) | ||
| H1a-H1e | Somatic, replication-dependent subtypes, both general and specific functions | 41 |
| H1c, H1d, H1e (H1.2, H1.3, H1.4) | Triple knockout is embryonic lethal | 14 |
| H1b (H1.5) | Interacts with Msx1, repression of muscle cell differentiation | 85 |
| H1c (H1.c) | Heterochromatin condensation in photoreceptors | 76 |
| H1(0) (H1.0) | Replacement subtype in differentiated cells | 45 |
| H1x (H1.10) | Function not clarified, replacement subtype | |
| H1oo (H1.8) | Expression of pluripotency genes in early embryos | 49,100 |
| H1t2 (H1.7) | Sperm cell differentiation | 48 |
| Human (11 subtypes) | ||
| H1.1-H1.5 | Somatic, replication-dependent subtypes, both general and specific functions | 41 |
| H1.2 | Maintenance of active gene transcription, recruitment of Cul4A ubiquitin ligase/PAF1 | 83 |
| H1.2S173p: active transcription | 116 | |
| Repressor of p53-mediated transcription | 87 | |
| H1.3 | Inhibition of h19 noncoding RNA transcription | [90,90] |
| H1.4 | H1.4K26me: heterochromatin formation via binding to HP1 and L3MBTL1 | 72,75 |
| H1.4K34ac: recruitment of TAF, active transcription | 122 | |
| H1.4S172p, H1.4S187p: active transcription | 116 | |
| H1.0 | Replacement subtype in differentiated cells | 45 |
| H1x (H1.10) | Mitotic progression | 128 |
| H1oo, H1t, H1T2, HILS1 (H1.8, H1.6, H1.7, H1.9) | Germ cell-specific subtypes, function not clarified | 47,48,129 |
For further description, see also 20.
For mouse and human subtypes/variants, the systematic names are in parentheses.
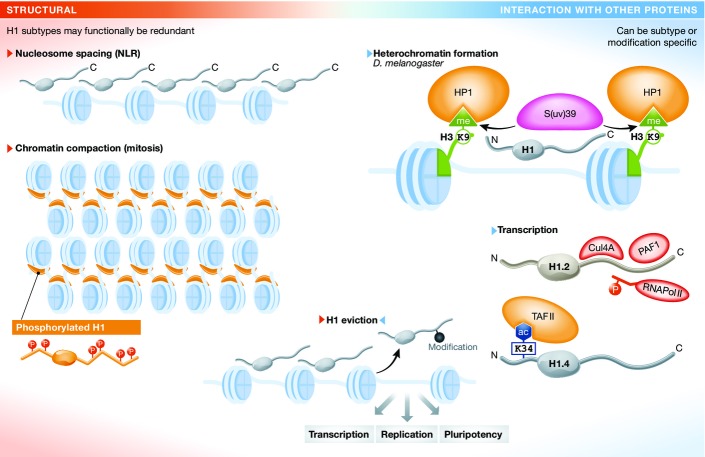
Figure 4. Model of H1 modes of action.
Based on current knowledge, we propose a dual model of H1 function: (i) H1 is a key structural component of chromatin. It can stabilize nucleosome structure and influence nucleosome spacing, and it is required for chromatin compaction (left panel). These functions often seem to be redundant between different subtypes. (ii) H1 also functions through interaction with other proteins that will in turn modify chromatin or take part in DNA-based processes (right panel). In Drosophila melanogaster, H1 recruits S(uv)39 to chromatin, which is required for heterochromatin formation 69. These functions are often subtype specific, as, for example, the recruitment of Cul4A and PAF1 by H1.2 to support target gene transcription 83. Often, H1 modifications come into play, as was demonstrated for HP1 binding to H1.4K26me or TAFII binding to H1.4K34ac 72,122. Of course, these two modes of action are not always separable. For example, interaction with other proteins or H1 modifications can be required for H1 eviction from chromatin (f), so that it will no longer exert its stabilizing function, resulting in chromatin opening for DNA-based processes like transcription, replication and repair.
Acknowledgments
We thank all former and current members of the RS group for discussions about H1 and its mysteries. Work in RS laboratory is supported by the Fondation pour la Recherche Médicale (FRM), the Agence Nationale de Recherche (CoreAc) and La Ligue National Contre La Cancer (Equipe Labellise). We apologize that we were not able to cite all H1 literature.
Glossary
- ASCOM
ASC-2 complex, coactivator of nuclear receptors
- AurB
Aurora B
- CDK
cyclin-dependent kinase
- ChIP
chromatin immunoprecipitation
- CTD
C-terminal domain
- Cul4A
Cullin 4A
- CycB
Cyclin B
- DamID
DNA methyltransferase identification
- DNMT
DNA methyltransferase
- DSB
double-strand break
- eGFP
enhanced green fluorescent protein
- ESC
embryonic stem cell
- Ezh2
enhancer of zeste 2
- FRAP
fluorescence recovery after photobleaching
- G9a
G9A histone methyltransferase
- GCN5
general control of amino acid synthesis 5
- GLP1
G9a-like protein 1
- HMGD-1
high mobility group protein D 1
- HP1
heterochromatin protein 1
- ICR
imprinting control region
- IFN
interferon
- JAK
Janus kinase
- JMJD2
Jumonji domain-containing protein 2, histone demethylase
- KDM4
lysine demethylase 4
- L3MBTL1
L(3)mbt-like 1
- LAD
lamina-associated domain
- MALDI
matrix-assisted laser desorption ionization
- MMTV
mouse mammary tumour virus
- Msx1
Msh homeobox 1
- MyoD
myogenic differentiation D
- NMR
nuclear magnetic resonance
- NRL
nucleosomal repeat length
- NURF
nucleosome remodelling factor
- PADI
protein arginine deiminase
- PAF1
RNA polymerase II-associated factor
- PARP-1
poly (ADP-ribose) polymerase 1
- PEV
position effect variegation
- PRC2
polycomb repressive complex 2
- RNAi
RNA interference
- RNAP II
RNA polymerase II
- RNF8
ring finger protein 8, E3 ubiquitin ligase
- Set7/9
SET-domain histone methyltransferase-7/9
- shRNA
short hairpin RNA
- SirT1
sirtuin 1
- STAT
signal transducer and activator of transcription
- Su(var)39
suppressor of variegation 3-9
- TAF
transcription initiation factor TFIID subunit 1
- TE
transposable element
- TFIID
transcription factor IID
- Ubc13
ubiquitin-conjugating enzyme 13, E2 ubiquitin ligase
Conflict of interest
The authors declare that they have no conflict of interest.
Sidebar A: In need of answers.
How exactly does H1 bind to the nucleosomal core particle and how does this mediate higher-order structures?
What is the role of H1 subtypes in 3D organization of chromatin in the nucleus?
Which H1 functions are redundant and which are subtype specific?
How does H1 subtype distribution vary between different cell types and change during development?
How are specific H1 subtypes incorporated at distinct genomic regions?
What are the roles of H1 modifications? Which H1 modifications are still undiscovered?
What are the enzymes setting and removing H1 modifications and what are the readers of these modifications?
References
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Rhodes D. Structure of the “30 nm” chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS, Brown DD. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984;37:903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- Sirotkin AM, Edelmann W, Cheng G, Klein-Szanto A, Kucherlapati R, Skoultchi AI. Mice develop normally without the H1(0) linker histone. Proc Natl Acad Sci USA. 1995;92:6434–6438. doi: 10.1073/pnas.92.14.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabent B, Saftig P, Bode C, Doenecke D. Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem Cell Biol. 2000;113:433–442. doi: 10.1007/s004180000146. [DOI] [PubMed] [Google Scholar]
- Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21:7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabini S, Franke K, Saftig P, Bode C, Doenecke D, Drabent B. Spermatogenesis in mice is not affected by histone H1.1 deficiency. Exp Cell Res. 2000;255:114–124. doi: 10.1006/excr.1999.4767. [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Takami Y, Sonoda E, Iwasaki T, Iwano H, Tachibana M, Takeda S, Nakayama T, Kimura H, Shinkai Y. Histone H1 null vertebrate cells exhibit altered nucleosome architecture. Nucleic Acids Res. 2010;38:3533–3545. doi: 10.1093/nar/gkq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Hellauer K, Sirard E, Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J Biol Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- Vujatovic O, Zaragoza K, Vaquero A, Reina O, Bernues J, Azorin F. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012;40:5402–5414. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho M, Diani E, Beato M, Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4:e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A, Kamieniarz K, Schneider R. The histone H1 family: specific members, specific functions? Biol Chem. 2008;389:333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Krüger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Busby SA, Barber CM, Shabanowitz J, Allis CD, Hunt DF. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J Proteome Res. 2004;3:1219–1227. doi: 10.1021/pr0498887. [DOI] [PubMed] [Google Scholar]
- Lu A, Zougman A, Pudełko M, Bȩbenek M, Ziółkowski P, Mann M, Wiśniewski JR. Mapping of lysine monomethylation of linker histones in human breast and its cancer. J Proteome Res. 2009;8:4207–4215. doi: 10.1021/pr9000652. [DOI] [PubMed] [Google Scholar]
- Deterding LJ, Bunger MK, Banks GC, Tomer KB, Archer TK. Global changes in and characterization of specific sites of phosphorylation in mouse and human histone H1 isoforms upon CDK inhibitor treatment using mass spectrometry. J Proteome Res. 2008;7:2368–2379. doi: 10.1021/pr700790a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Garea A, Imhof A. Fine mapping of posttranslational modifications of the linker histone H1 from Drosophila melanogaster. PLoS ONE. 2008;3:e1553. doi: 10.1371/journal.pone.0001553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet-Costa C, Vilaseca M, Diema C, Vujatovic O, Vaquero A, Omeñaca N, Castejón L, Bernués J, Giralt E, Azorín F. Combined bottom-up and top-down mass spectrometry analyses of the pattern of post-translational modifications of Drosophila melanogaster linker histone H1. J Proteomics. 2012;75:4124–4138. doi: 10.1016/j.jprot.2012.05.034. [DOI] [PubMed] [Google Scholar]
- Sarg B, Lopez R, Lindner H, Ponte I, Suau P, Roque A. Identification of novel post-translational modifications of linker histones in chicken erythrocytes. J Proteomics. 2015;113:162–177. doi: 10.1016/j.jprot.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010;20:519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman SW, Young NL, Parthun MR, Freitas MA. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C, Snijders A, Williamson J, Reynolds C, Baldwin J, Dickman M. Post-translational modifications of the linker histone variants and their association with cell mechanisms. FEBS J. 2009;276:3685–3697. doi: 10.1111/j.1742-4658.2009.07079.x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Roque A, Iloro I, Ponte I, Arrondo JLR, Suau P. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J Biol Chem. 2005;280:32141–32147. doi: 10.1074/jbc.M505636200. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Hayashi H, Iwai K. Tetrahymena histone H1. Isolation and amino acid sequence lacking the central hydrophobic domain conserved in other H1 histones. J Biochem. 1987;102:369–376. doi: 10.1093/oxfordjournals.jbchem.a122063. [DOI] [PubMed] [Google Scholar]
- Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J Biol Chem. 1998;273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- Godde JS, Widom J. Chromatin structure of Schizosaccharomyces pombe. A nucleosome repeat length that is shorter than the chromatosomal DNA length. J Mol Biol. 1992;226:1009–1025. doi: 10.1016/0022-2836(92)91049-u. [DOI] [PubMed] [Google Scholar]
- Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci USA. 2010;107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu R-M, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- Zhou B-R, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y. Structural insights into the histone H1-nucleosome complex. Proc Natl Acad Sci USA. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H10 with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Sirotkin AM, Buchold GM, Skoultchi AI, Marzluff WF. The mouse histone H1 genes: gene organization and differential regulation. J Mol Biol. 1997;271:124–138. doi: 10.1006/jmbi.1997.1166. [DOI] [PubMed] [Google Scholar]
- Kasinsky HE, Lewis JD, Dacks JB, Ausió J. Origin of H1 linker histones. FASEB J. 2001;15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Doenecke D. Histone H1 zero: a major player in cell differentiation? FASEB J. 1994;8:1260–1268. doi: 10.1096/fasebj.8.15.8001738. [DOI] [PubMed] [Google Scholar]
- Happel N, Schulze E, Doenecke D. Characterisation of human histone H1x. Biol Chem. 2005;386:541–551. doi: 10.1515/BC.2005.064. [DOI] [PubMed] [Google Scholar]
- Drabent B, Kardalinou E, Doenecke D. Structure and expression of the human gene encoding testicular H1 histone (H1t) Gene. 1991;103:263–268. doi: 10.1016/0378-1119(91)90284-i. [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci USA. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- Th’ng JPH, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus: control of binding and localization by the c-terminal domain. J Biol Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th’ng JPH. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS ONE. 2009;4:e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öberg C, Izzo A, Schneider R, Wrange Ö, Belikov S. Linker histone subtypes differ in their effect on nucleosomal spacing in vivo. J Mol Biol. 2012;419:183–197. doi: 10.1016/j.jmb.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Orrego M, Ponte I, Roque A, Buschati N, Mora X, Suau P. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 2007;5:22. doi: 10.1186/1741-7007-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasz H, Sapojnikova N, Helliger W, Lindner H, Puschendorf B. In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J Biol Chem. 1998;273:32236–32243. doi: 10.1074/jbc.273.48.32236. [DOI] [PubMed] [Google Scholar]
- Kinkade JM, Cole RD. The resolution of four lysine-rich histones derived from calf thymus. J Biol Chem. 1966;241:5790–5797. [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009;28:3635–3645. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A, Kamieniarz-Gdula K, Ramírez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z, Lee EK, Wu H, Medrzycki M, Pan C, et al. High-resolution mapping of H1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalabothula N, McVicker G, Maiorano J, Martin R, Pritchard JK, Fondufe-Mittendorf YN. The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation. BMC Genom. 2014;15:1–14. doi: 10.1186/1471-2164-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Arino L, Islam ABMMK, Izquierdo-Bouldstridge A, Mayor R, Terme JM, Luque N, Sancho M, Lopez-Bigas N, Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, Huang L, Wen Z, Li W, Li X, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Li J-Y, Patterson M, Mikkola HKA, Lowry WE, Kurdistani SK. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012;8:e1002879. doi: 10.1371/journal.pgen.1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Izquierdo-Bouldstridge A, Millán-Ariño L, Bustillos A, Sampaio C, Luque N, Jordan A. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J Biol Chem. 2015;290:7474–7491. doi: 10.1074/jbc.M114.617324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Kavi H, Kim BJ, Guzzardo PM, Emelyanov AV, Xu N, Hannon GJ, Zavadil J, Fyodorov DV, et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Emelyanov AV, Fyodorov DV, Skoultchi AI. Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenetics Chromatin. 2014;7:16. doi: 10.1186/1756-8935-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- Hale TK, Contreras A, Morrison AJ, Herrera RE. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol Cell. 2006;22:693–699. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. J Biol Chem. 2009;284:8395–8405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, III, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Popova EY, Grigoryev SA, Fan Y, Skoultchi AI, Zhang SS, Barnstable CJ. Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina. J Biol Chem. 2013;288:17895–17907. doi: 10.1074/jbc.M113.452144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci USA. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick EH, Bustin M, Marsaud V, Richard-Foy H, Hager GL. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Brown DT. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res. 1999;27:3355–3363. doi: 10.1093/nar/27.16.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop R, Di Croce L, Beato M. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 2003;22:588–599. doi: 10.1093/emboj/cdg052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S, Astrand C, Wrange O. Mechanism of histone H1-stimulated glucocorticoid receptor DNA binding in vivo. Mol Cell Biol. 2007;27:2398–2410. doi: 10.1128/MCB.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballare C, Beato M. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 2011;25:845–862. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee B, Kim J, Choi J, Kim J-M, Xiong Y, Roeder RG, An W. Linker histone H1.2 cooperates with Cul4A and PAF1 to drive H4K31 ubiquitylation-mediated transactivation. Cell Rep. 2013;5:1690–1703. doi: 10.1016/j.celrep.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota S, Nagata K. Silencing of IFN-stimulated gene transcription is regulated by histone H1 and its chaperone TAF-I. Nucleic Acids Res. 2014;42:7642–7653. doi: 10.1093/nar/gku485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. Msx1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- Studencka M, Konzer A, Moneron G, Wenzel D, Opitz L, Salinas-Riester G, Bedet C, Kruger M, Hell SW, Wisniewski JR, et al. Novel roles of Caenorhabditis elegans heterochromatin protein HP1 and linker histone in the regulation of innate immune gene expression. Mol Cell Biol. 2011;32:251–265. doi: 10.1128/MCB.05229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J-Q, Liu L-P, Hess D, Rietdorf J, Sun F-L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-M, Kim BJ, Norwood Toro L, Skoultchi AI. H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proc Natl Acad Sci USA. 2013;110:1708–1713. doi: 10.1073/pnas.1213266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrzycki M, Zhang Y, Zhang W, Cao K, Pan C, Lailler N, McDonald JF, Bouhassira EE, Fan Y. Histone H1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Sonoda E, Takami Y, Kimura H, Nakayama T, Tachibana M, Takeda S, Shinkai Y. Histone H1 variant, H1R is involved in DNA damage response. DNA Repair. 2007;6:1584–1595. doi: 10.1016/j.dnarep.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang S-M, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, et al. Histone H1 couples initiation and amplification of ubiquitin signaling after DNA damage. Nature. 2015 doi: 10.1038/nature15401. (in press) [DOI] [PubMed] [Google Scholar]
- Pérez-Montero S, Carbonell A, Morán T, Vaquero A, Azorín F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 2013;26:578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Dworkin-Rastl E, Kandolf H, Smith RC. The maternal histone H1 variant, H1M (B4 protein), is the predominant H1 histone in Xenopus pregastrula embryos. Dev Biol. 1994;161:425–439. doi: 10.1006/dbio.1994.1042. [DOI] [PubMed] [Google Scholar]
- Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci USA. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci USA. 2010;107:5483–5488. doi: 10.1073/pnas.1000599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kihara M, Meczekalski B, King GJ, Adashi EY. H1oo: a pre-embryonic H1 linker histone in search of a function. Mol Cell Endocrinol. 2003;202:5–9. doi: 10.1016/s0303-7207(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Ohgane J, Tanaka S, Yagi S, Shiota K. Oocyte-specific linker histone H1foo is an epigenomic modulator that decondenses chromatin and impairs pluripotency. Epigenetics. 2014;7:1029–1036. doi: 10.4161/epi.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R, Chalkley R, Granner D. Lysine-rich histone phosphorylation. A positive correlation with cell replication. Biochemistry. 1972;11:1094–1098. doi: 10.1021/bi00756a023. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Mann M. N(epsilon)-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008;36:570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders APL, Pongdam S, Lambert SJ, Wood CM, Baldwin JP, Dickman MJ. Characterization of post-translational modifications of the linker histones H1 and H5 from chicken erythrocytes using mass spectrometry. J Proteome Res. 2008;7:4326–4335. doi: 10.1021/pr800260a. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: “PAR-laying” NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Poirier GG, Niedergang C, Champagne M, Mazen A, Mandel P. Adenosine diphosphate ribosylation of chicken-erythrocyte histones H1, H5 and high-mobility-group proteins by purified calf-thymus poly(adenosinediphosphate-ribose) polymerase. Eur J Biochem. 1982;127:437–442. doi: 10.1111/j.1432-1033.1982.tb06891.x. [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci USA. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasz H, Sarg B, Lindner HH. Site-specifically phosphorylated forms of H1.5 and H1.2 localized at distinct regions of the nucleus are related to different processes during the cell cycle. Chromosoma. 2009;118:693–709. doi: 10.1007/s00412-009-0228-2. [DOI] [PubMed] [Google Scholar]
- Hergeth SP, Dundr M, Tropberger P, Zee BM, Garcia BA, Daujat S, Schneider R. Isoform-specific phosphorylation of human linker histone H1.4 in mitosis by the kinase Aurora B. J Cell Sci. 2011;124:1623–1628. doi: 10.1242/jcs.084947. [DOI] [PubMed] [Google Scholar]
- Happel N, Stoldt S, Schmidt B, Doenecke D. M phase-specific phosphorylation of histone H1.5 at threonine 10 by GSK-3. J Mol Biol. 2009;386:339–350. doi: 10.1016/j.jmb.2008.12.047. [DOI] [PubMed] [Google Scholar]
- Chu C-S, Hsu P-H, Lo P-W, Scheer E, Tora L, Tsai H-J, Tsai M-D, Juan L-J. Protein kinase A-mediated serine 35 phosphorylation dissociates histone H1.4 from mitotic chromosome. J Biol Chem. 2011;286:35843–35851. doi: 10.1074/jbc.M111.228064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Freedman B, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley LR, Valdez JG, Buchanan JS. Characterization of the mitotic specific phosphorylation site of histone H1. Absence of a consensus sequence for the p34cdc2/cyclin B kinase. J Biol Chem. 1995;270:27653–27660. doi: 10.1074/jbc.270.46.27653. [DOI] [PubMed] [Google Scholar]
- Th’ng JP, Guo XW, Swank RA, Crissman HA, Bradbury EM. Inhibition of histone phosphorylation by staurosporine leads to chromosome decondensation. J Biol Chem. 1994;269:9568–9573. [PubMed] [Google Scholar]
- Alexandrow MG. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J Cell Biol. 2005;168:875–886. doi: 10.1083/jcb.200409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee RN, Banks GC, Trotter KW, Lee HL, Archer TK. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol Cell Biol. 2001;21:5417–5425. doi: 10.1128/MCB.21.16.5417-5425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, Nardulli AM, Hager GL, Kelleher NL, Mizzen CA. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol. 2010;189:407–415. doi: 10.1083/jcb.201001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JE, Rea S. Core and linker histone modifications involved in the DNA damage response. Subcell Biochem. 2010;50:17–42. doi: 10.1007/978-90-481-3471-7_2. [DOI] [PubMed] [Google Scholar]
- Roque A, Ponte I, Arrondo JLR, Suau P. Phosphorylation of the carboxy-terminal domain of histone H1: effects on secondary structure and DNA condensation. Nucleic Acids Res. 2008;36:4719–4726. doi: 10.1093/nar/gkn440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R, Sarg B, Lindner H, Bartolome S, Ponte I, Suau P, Roque A. Linker histone partial phosphorylation: effects on secondary structure and chromatin condensation. Nucleic Acids Res. 2015;43:4463–4476. doi: 10.1093/nar/gkv304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM, Dundr M, Garcia BA, Daujat S, Schneider R. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin. 2010;3:7. doi: 10.1186/1756-8935-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Kamieniarz K, Izzo A, Dundr M, Tropberger P, Ozretic L, Kirfel J, Scheer E, Tropel P, Wisniewski JR, Tora L, et al. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev. 2012;26:797–802. doi: 10.1101/gad.182014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Terme J-M, Millán-Ariño L, Mayor R, Luque N, Izquierdo-Bouldstridge A, Bustillos A, Sampaio C, Canes J, Font I, Sima N, et al. Dynamics and dispensability of variant-specific histone H1 Lys-26/Ser-27 and Thr-165 post-translational modifications. FEBS Lett. 2014;588:2353–2362. doi: 10.1016/j.febslet.2014.05.035. [DOI] [PubMed] [Google Scholar]
- Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JCR, Zernicka-Goetz M, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J, Bödör C, Wang J, Araf S, Yang C-Y, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Matsunaga S, Morimoto A, Ono-Maniwa R, Uchiyama S, Fukui K. H1.X with different properties from other linker histones is required for mitotic progression. FEBS Lett. 2007;581:3783–3788. doi: 10.1016/j.febslet.2007.06.076. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kato S, Tanaka M, Kuji N, Yoshimura Y. Structure and expression of the human oocyte-specific histone H1 gene elucidated by direct RT-nested PCR of a single oocyte. Biochem Biophys Res Commun. 2003;304:351–357. doi: 10.1016/s0006-291x(03)00610-7. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarg B, Helliger W, Talasz H, Förg B, Lindner HH. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J Biol Chem. 2006;281:6573–6580. doi: 10.1074/jbc.M508957200. [DOI] [PubMed] [Google Scholar]
- Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baatout S, Derradji H. About histone H1 phosphorylation during mitosis. Cell Biochem Funct. 2006;24:93–94. doi: 10.1002/cbf.1293. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]