INTRODUCTION
This protocol describes imaging of the living mouse brain through a thinned skull using two-photon microscopy. This transcranial two-photon laser-scanning microscope (TPLSM) imaging method allows high-resolution imaging of fluorescently labeled neurons, microglia, astrocytes, and blood vessels, as well as subcellular structures such as dendritic spines and axonal varicosities. The surgical procedure that is required to allow imaging thins the cranium so that it becomes optically transparent. Once learned, the surgery can be performed in ~30 min, and imaging can follow immediately. The procedure can be repeated multiple times, allowing brain cells and tissues to be studied in the same animals over short or long time intervals, depending on the design of the experiment. Two-photon imaging through a thinned and intact skull avoids side effects caused by skull removal and is a minimally invasive method for studying the living mouse brain under physiological and pathological conditions.
MATERIALS.
RECIPE
Please see the end of this article for recipes indicated by <R>.
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
Reagents
Artificial cerebrospinal fluid (ACSF) <R>
Cyanoacrylate glue (Super Glue; Fisher Scientific 11-999-24)
Dextran, rhodamine B, 70,000 MW (Molecular Probes/Invitrogen D1841), 10 mg/mL in phosphate-buffered saline (PBS) (optional; see Step 13)
- Ketamine/xylazine (KX) mixtureMix 20 mg/mL ketamine (Fort Dodge Animal Health) and 3 mg/mL xylazine (Lloyd, Inc.) in saline. Store at room temperature (20°C). Properly stored KX solution is stable for up to 1 yr.
Lubricant eye ointment, sterile (Del Pharmaceuticals, Inc.)
Mice, transgenic, expressing fluorescent proteins within a variety of cell types, such as layer 5 cortical neurons (e.g., thy1-YFP line) or microglia (e.g., CX3CR1-eGFP line) (Feng et al. 2000; Jung et al. 2000)
Equipment
Alcohol preparation pads, sterile (e.g., Fisher Scientific)
Cotton pads
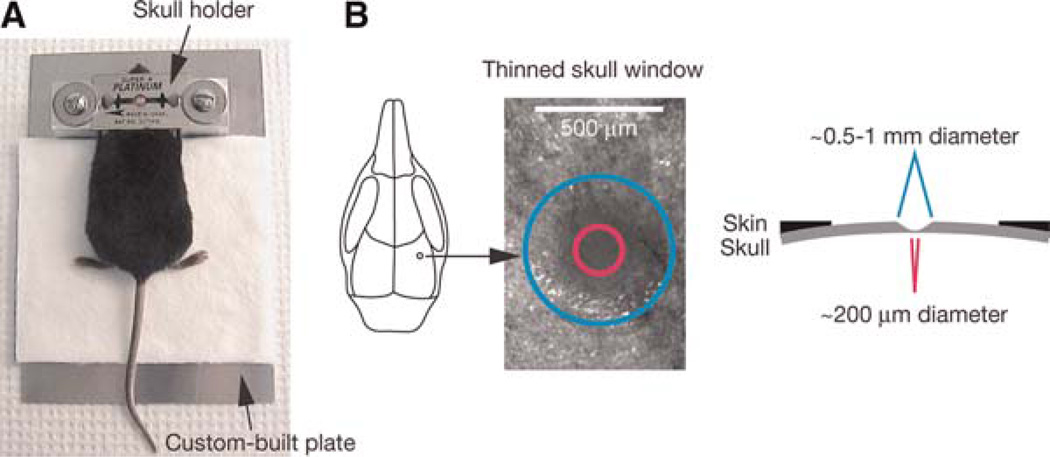
- Custom-built plateAttach two 18 × 18 × 18-mm aluminum blocks to a 14 × 10 × 0.1-cm aluminum plate. Place the blocks 3 cm from each other and ~2 cm from one of the short sides of the plate. Drill a hole with an internal thread on each block to accommodate a ¼ in. screw (Fig. 1A).
Dissecting instruments
Dissecting stereomicroscope (Olympus BX50WI)
Double-edge razor blades (CAMB Machine Knives International LLC CMK169S)
Fluorescence microscope
- High-speed microdrill (Fine Science Tools 18000-17)Make sure that the drill is adequately charged and in good working order, because low drill speeds may result in nicking of the skull or uneven thinning.
- Imaging setupWe use either a custom-built multiphoton microscope equipped with a mode-locked laser system or a Bio-Rad 2001 multiphoton microscope. For both systems, the laser system (Tsunami and Millennia Xs, Spectra-Physics) is tunable from a 690- to 1000-nm wavelength with an 80-MHz pulse repeat and a <100-fsec pulse width. The laser-scanning system is coupled to an upright fluorescence microscope. For the custom-built unit, an Olympus laser-scanning system is used, and detectors (photomultiplier tubes) are placed close to the objective to facilitate the signal detection. We generally use an Olympus 60×, numerical aperture (NA) 1.1 objective for adequate resolution of small structures, such as dendritic spines. A dissecting microscope coupled to a charge-coupled device (CCD) camera is used before two-photon microscope (TPM) imaging for obtaining images of the superficial meningeal vasculature, which are then used as maps for future relocation of the cortical region of interest.
Marker pen
- Microdrill steel burrs (Fine Science Tools 19007-05 or 19007-07)The drill burr should be replaced frequently to assure that it performs properly.
- Microsurgical blades (Surgistar, Inc. 6900)The size, dimensions, and quality of the microsurgical blades are important for achieving proper thinning. For best results, use a new blade with each mouse.
Silk suture, 6-0 (LOOK M552830)
- Skull holderCut the sharp edges of three double-sided razor blades, and then glue the blades to each other with cyanoacrylate to create a thick metal plate. The razor blades have circular holes in the middle for access to the skull. Use two spacers for stabilizing the skull holder (Fig. 1A).
Temperature regulator for mice (optional; see Step 1)
FIGURE 1.
Schematic of thinned skull preparation. A head immobilization device including a custom-built plate and a skull holder is used for reducing movement artifacts during imaging (A). The skull holder/razor blades are glued onto the mouse skull such that the region of interest is exposed in the center of the circular hole of the razor blades. The skull holder/razor blades are then tightened on the aluminum blocks of the custom-built plate. A circular area of skull (typically ~0.5–1mm in diameter, marked with a blue circle) over the region of interest is shaved (B). The thinnest region (marked with a pink circle) for TPLSM imaging is ~20 µm in thickness and ~200 µm in diameter.
FIGURE 1.
Schematic of thinned skull preparation. A head immobilization device including a custom-built plate and a skull holder is used for reducing movement artifacts during imaging (A). The skull holder/razor blades are glued onto the mouse skull such that the region of interest is exposed in the center of the circular hole of the razor blades. The skull holder/razor blades are then tightened on the aluminum blocks of the custom-built plate. A circular area of skull (typically ~0.5–1mm in diameter, marked with a blue circle) over the region of interest is shaved (B). The thinnest region (marked with a pink circle) for TPLSM imaging is ~20 µm in thickness and ~200 µm in diameter.
METHOD
Thinned Skull Preparation
-
1Anesthetize mice by intraperitoneal injection (5–6 µL/g body weight) of KX mixture. Allow sufficient time (5–10 min) for a surgical level of anesthesia to be achieved. Place the mouse on top of a cotton pad. For experiments involving long imaging periods, use a mouse temperature regulator.Experiments should comply with institutional and national animal care guidelines. Continuously monitor the depth of anesthesia by testing the animal’s reflexes during the surgery (e.g., pinching the animal’s foot with a blunt pair of forceps and checking for the lack of withdrawal reflex). Inject more KX as necessary.
-
2
Lubricate both eyes with eye ointment.
-
3
Thoroughly shave the head over most of the scalp with a double-edge razor blade. Clean the scalp with a sterile alcohol preparation pad, and then make a midline scalp incision extending from the neck region between the ears to the frontal area between the eyes. Carefully disrupt the connective tissue located between the scalp and the underlying skull with a pair of spring scissors.
-
4
Localize the brain area to be imaged based on stereotactic coordinates, and label it with a marker pen.
-
5
Remove the connective tissue attached to the skull over the area to be imaged by gently scraping the skull with a blunt microsurgical blade.
-
6Place a small amount of cyanoacrylate glue around the edges of the internal opening of the skull holder, and press the holder against the skull. Make sure that the area to be imaged is exposed in the center of the internal opening (Fig. 1A).Skull immobilization, which is in part determined by the quality of contact and adhesion between the skull holder and the skull, is critical to avoid movement artifacts during imaging.
-
7Draw the loose skin gently up to the edges of the internal opening of the skull holder and apply a small amount of glue to the perimeter of the skin to fix it to the skull.Gluing the skin around the internal opening of the skull holder and the skull aids in holding the ACSF in place during imaging under a water-immersion objective.See Troubleshooting.
-
8
Wait several minutes until the skull holder is firmly glued to the skull. Attach the skull holder to the plate by inserting the lateral edges of the skull holder between the aluminum blocks and spacers/screws. Tighten the screws to completely immobilize the skull holder (Fig. 1A). Wash the opening of the skull holder several times with ACSF to remove remnants of nonpolymerized glue. This helps to prevent the glue from contaminating the microscope objectives during imaging.
-
9Under a dissecting microscope, use a high-speed microdrill to thin a circular area (typically ~0.5–1mmin diameter) (Fig. 1B) over the skull region of interest. Keep the following points in mind:
- Apply a few drops of ACSF to the skull surface during drilling to reduce overheating.
- Perform drilling intermittently to avoid friction-induced overheating.
- Replace the ACSF periodically to wash away the bone debris.The mouse skull consists of two thin layers of compact bone (periosteum and endosteum) sandwiching a thick layer of spongy bone, which contains small cavities and canaliculi that carry blood vessels. Remove the external layer of compact bone and most of the spongy bone with the drill. Bleeding may occur during drilling of the spongy bone portion, but usually stops spontaneously within a few minutes.Do not thin a large region (>1.5 mm) to a thin layer (<50 µm), because it is difficult to exercise fine control of the drilling process and may cause damage to the underlying cortex.
-
10After removing the majority of the spongy bone, remaining concentric structures within the bone can usually be seen under the dissecting microscope, indicating that drilling is approaching the internal compact bone layer. At this stage, skull thickness is still >50 µm. Continue the skull-thinning procedure with a microsurgical blade to obtain a very thin (~20 µm) and smooth preparation (~200 µm in diameter) (Fig. 1B). During the thinning process, repeatedly examine the preparation with a conventional fluorescence microscope until fluorescently labeled structures, such as the dendrites and spines in the area of interest, can clearly be visualized.Hold the microsurgical blade at a ~45° angle during thinning, and take great care not to push the skull downward against the brain surface or to break through the bone, as minor brain trauma or bleeding may induce inflammation and disruption of neuronal structures. In our experience, the best way to know when to stop thinning is by practice and by directly measuring the thickness of the skull from imaging skull autofluorescence with the TPM. For beginners, periodic measurement of skull thickness during thinning greatly helps in preventing skull overthinning.It is critical not to thin a large surface area (>300 µm in diameter) to <15 µm in thickness, as it may cause disruption of neuronal structures and activation of immune cells as evidenced by neurite blebbing, microglia process retraction, and thickening of epidural connective tissues. These events will reduce the quality of subsequent images and may confound experimental results.
Mapping the Imaging Area for Future Relocation
-
11
To identify the same imaged area at a later time point, take a high-quality picture of the brain vasculature with a CCD camera attached to a stereo dissecting microscope (Fig. 2A) or directly to the TPLSM setup.
FIGURE 2.
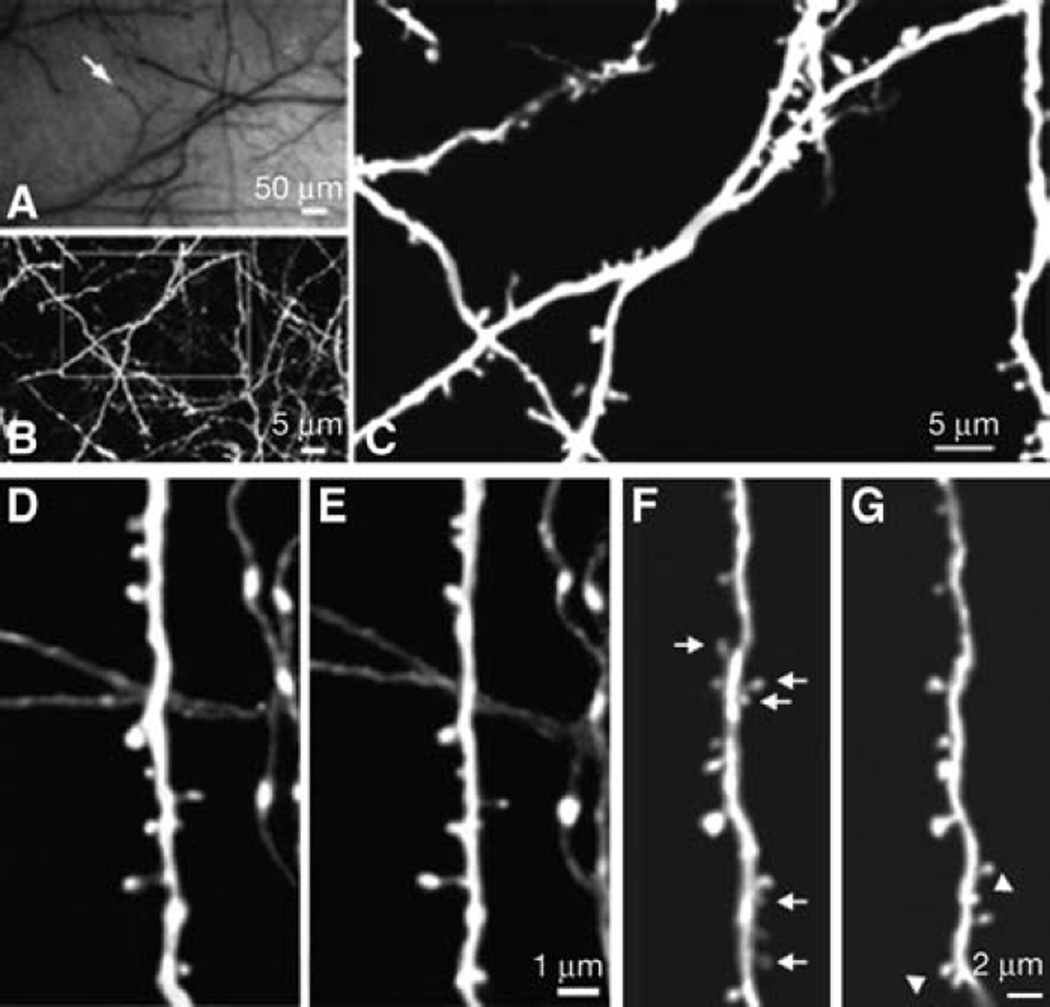
Long-term transcranial TPLSM imaging of fine neuronal structures. (A) CCD camera view of brain vasculature under the thinned skull. The cortical vasculature can clearly be seen through the thinned skull. The vasculature pattern remains stable over months to years and can be used as a landmark to relocate the imaged region at subsequent time points. Arrow indicates the region in which subsequent TPLSM images were obtained. (B) Two-dimensional (2D) projection of a three-dimensional (3D) stack of dendritic branches and axons in the primary visual cortex (60×, ~0.39 µm per pixel). The stack was 50-µm deep (2-µm step size). The boxed region was then imaged at higher magnification. (C) High-power 2D projection of a 3D stack (60×, ~0.13 µm per pixel, 10-µm reconstructed, 0.70-µm step size) reveals clear neuronal structures, including axonal varicosities, dendritic shafts, and dendritic protrusions. (D,E) Axonal and dendritic branches from two animals imaged 3 d apart show the same spines and boutons at the same locations. (F,G) Dendritic branches imaged over 19 mo apart. The arrows indicate spines that are eliminated in the second view. The arrowheads indicate spines that are formed in the second view. Note that most spines in F persist in G. Two-dimensional projections of 3D image stacks containing dendritic segments of interest were used for D–G.
TPLSM Imaging of Neuronal or Glial Structures
-
12
Carefully move the mouse mounted on the custom-built plate to the TPM stage. Select an adequately thinned and uniform area for imaging under a fluorescence microscope, and carefully denote the selected area on the CCD brain vasculature map by observing the pattern of blood vessels adjacent to it (Fig. 2A).
-
13
If performing microglial imaging, inject 100 µL of rhodamine-dextran solution retro-orbitally for vessel labeling before application of eye ointment; alternatively, inject it through the tail vein.
-
14Tune the TPM to the appropriate wavelength (e.g., 890 nm for green fluorescent protein [GFP] and 920 nm for yellow fluorescent protein [YFP]). Although we have used 40× (0.8 NA) and 60× (0.9 NA) in the past, we currently use higher-NA water-immersion objectives (e.g., 60×, 1.1 NA) for imaging small structures, such as dendritic spines.The objective lens should remain immersed in mouse ACSF at all times during imaging. If there is a sudden deterioration of imaging quality, check that the lens remains fully immersed in ACSF, and add more if necessary. Gradual leakage of ACSF is usually caused by inadequate gluing of the scalp around the skull holder openings (Step 7).For neuronal imaging, obtain a low-magnification stack of fluorescently labeled neuronal processes (e.g., 60× objective; 200 µm × 200 µm; 512 × 512 pixel; 2-µm step), which serves as a higher-resolution map for accurate relocation of the same region at later time points in conjunction with the CCD brain vasculature map (Fig. 2B). When performing microglial imaging, take a low-magnification (10× air objective) stack of the rhodamine dextran-labeled vasculature, and mark the selected area before switching to a 40×/60× objective, making sure that objectives (10× and 40×/60×) are cocentered.
-
15Without changing the position of the stage, take high-magnification images (e.g., 66.7 × 66.7 µm; 512 × 512 pixel; 0.75-µm step) (Fig. 2C) from the same area with the TPM. The image stack is typically taken within ~100 µm below the pial surface for spine imaging and within ~200 µm for microglia imaging (Figs. 2, 3).We typically use laser intensities in the range of 10 to 30 mW (measured at the sample) to minimize phototoxicity.See Troubleshooting.
FIGURE 3.
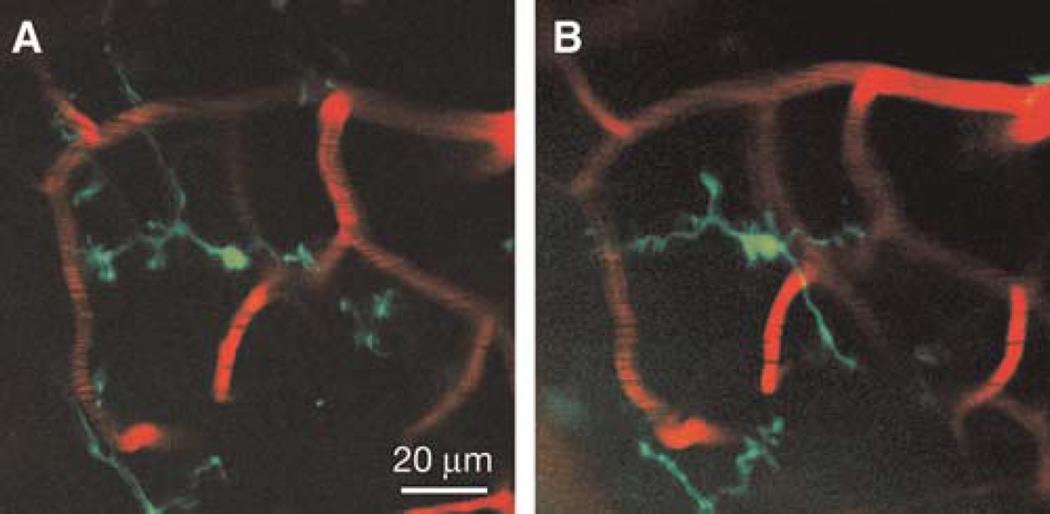
Transcranial TPLSM imaging of enhanced GFP (eGFP)-labeled microglia vasculature within the mouse cerebellum. Two-dimensional projections of a 3D z-stack from the cerebellum (40×, digital zoom=1) of a mouse harboring a single copy of the CX3CR1-eGFP allele driving eGFP expression in a subset of myeloid cells, including central nervous system resident parenchymal microglia imaged 24 h apart (A,B). The cerebellar vasculature has been labeled in red by intravenous injection of a rhodamine-dextran conjugate solution. The stacks are 6 µm in depth (2-µm step size) and are representative of a section of cortex spanning 40–46 µm below the dural surface. eGFP-labeled microglia retain their characteristic highly branched morphology, indicative of a resting state.
Recovery
-
16
When imaging is complete, detach the skull holder from the skull by gently pushing the skull away from the skull holder. When removed, the holder should take with it any remaining glue and be separated from the skin. If glue remains adherent to the skull or skin, gently remove any remaining dried glue with a pair of forceps. Suture the scalp with 6-0 silk, and leave the mouse in a separate cage until it is fully mobile. After recovery, return the mouse to its original housing cage until the next viewing.
Reimaging
-
17Begin by repeating Steps 1–8. Locate the previously thinned region based on the brain vasculature map (CCD for dendrite imaging, 10× rhodamine-dextran map for microglial imaging). Depending on the time of the second view, proceed as follows:
- If the second view is within 1 wk after the first view, carefully remove the connective tissue that has regrown on top of the thinned region using a microsurgical blade, and check the image quality with the TPM. If the image quality is poor (blurry features, high-background fluorescence, or significantly reduced depth of penetration), use a microsurgical blade to carefully shave the skull (as in Step 10) until a clear image can be obtained.
- If the second view is >1 wk after the first view, repeat Steps 9–10.The bone in the thinned area can regenerate substantially if the time interval between imaging sessions is >1 wk. The newly grown bone layer is optically less transparent for TPM imaging, so it is generally necessary to shave the bone so that it is slightly thinner than the previous imaging session.See Troubleshooting.
-
18Roughly locate the previously imaged region under the fluorescence microscope. More precisely, align the region using the TPM in accordance with the low-magnification map, and then zoom in to high magnification to further align it. After the region is precisely aligned with the first view, take images as in Step 15.See Troubleshooting.
-
19For multiple imaging sessions, repeat Steps 16–18.Although we have successfully imaged the same region with high optical clarity up to five times, the number and quality of imaging sessions is dependent on a variety of factors, including the quality of the initial surgery, the intrasession interval, and the experience of the operator. Although extreme thinning of the skull (~15 µm) during the initial surgery may provide excellent clarity during the first imaging session, it will complicate future imaging sessions because of an increased propensity for compensatory bone regrowth. We recommend limiting the number of imaging sessions and skull-thinning procedures to preserve image quality and reduce the perturbation to the brain microenvironment.
TROUBLESHOOTING
Problem (Step 7): The skull holder does not hold ACSF during imaging.
Solution: It is important to seal the internal opening of the skull holder with the surrounding skin. First dry the skin by blotting off excess liquid, and then by gently drying with compressed air. Follow by pulling the skin around the skull holder and by fixing the skin in place with glue.
Problem (Step 15): There are excessive movements of fluorescently labeled structures during imaging.
Solution: Consider the following:
Begin by checking the depth of anesthesia. If the animal begins to wake up, inject more KX, and wait for a few minutes for anesthesia to deepen.
If the animal is under deep anesthesia, check the attachment between the skull and the skull holder. The skull holder must be tightly glued to the skull. Detach the skull holder, and reglue it to the skull if the attachment is loose.
Problem (Steps 15 and 17): The fluorescent image is not clear under the fluorescence microscope/TPM.
Solution: Consider the following:
Check the skull thickness with the TPM. If the skull is too thick, carefully use either the drill or a microsurgical blade to thin the skull, and check again.
The skull may be thin, but uneven or marred by the drill bit or blunt microsurgical blades. An unevenly thinned or marred skull is a result of an improper thinning technique. It may be difficult to correct once part of the bone is already exceedingly thin. Use another animal for the experiment.
Red blood cells that have accumulated anywhere between the objective and the fluorescent structures can substantially degrade the image quality. If the bleeding is from the skull bone, wait until it stops spontaneously, and wash with several changes of ACSF to remove any coagulated blood. Bleeding underneath the bone indicates that the brain has been pushed or has been compromised during the thinning procedure. If this occurs, use another animal for the experiment.
Problem (Steps 15 and 17): Artifacts, such as neurite blebbing or microglial process retraction, appear.
Solution: Use another animal for the experiment. Neurite blebbing is an indicator that neuronal structures have been disrupted and is concurrent with high spine turnover. Microglial process retraction is indicative of an inflammatory response. One or both of these may appear if the skull is pushed downward against the brain surface during thinning, the skull has been overthinned, or the skull has been fractured.
Problem (Step 18): The previously imaged region cannot be located.
Solution: Check the CCD or 10× image of blood vasculature, and try to relocate to the previously imaged region with obvious patterns of blood vasculature and dendritic branches. Try to glue the skull holder such that the imaged region is in the center of the holder opening and parallel to the holder.
DISCUSSION
Studies Using Transcranial Two-Photon Imaging
In vivo imaging of the mouse brain using a TPM (Denk et al. 1990) through a thinned skull is a minimally invasive method that allows for longitudinal studies of cortical structures at high optical resolution over intervals ranging from minutes to years (Grutzendler et al. 2002; Yoder and Kleinfeld 2002; Stosiek et al. 2003; Tsai et al. 2004; Davalos et al. 2005; Nimmerjahn et al. 2005; Zhang et al. 2005; Zuo et al. 2005a, b; Haynes et al. 2006; Majewska et al. 2006; Nishiyama et al. 2007; Xu et al. 2007; Kim et al. 2009; Wake et al. 2009; Wu et al. 2009; Yan et al. 2009; Yang et al. 2009). By creating a thinned skull (thickness ~20 µm) cranial window, it has been possible to image fluorescently labeled structures located up to ~400-µm deep within the cortex (Grutzendler et al. 2002; Tsai et al. 2004; Davalos et al. 2005; Zuo et al. 2005a; Yang et al. 2009). In recent years, the transcranial two-photon imaging approach has been used to study the development and plasticity of synaptic connections (Grutzendler et al. 2002; Tsai et al. 2004; Zuo et al. 2005a,b; Majewska et al. 2006; Nishiyama et al. 2007; Xu et al. 2007; Wu et al. 2009; Yang et al. 2009), neuronal network activity (Stosiek et al. 2003), cerebral microvascular occlusion (Yoder and Kleinfeld 2002), amyloid plaque deposition (Christie et al. 2001; Tsai et al. 2004; Yan et al. 2009), and microglial dynamics and function (Davalos et al. 2005; Nimmerjahn et al. 2005; Haynes et al. 2006; Wake et al. 2009).
Transcranial two-photon imaging has been used successfully to image individual dendritic spines and axonal varicosities in multiple cortical regions over intervals of minutes up to 19 mo (Grutzendler et al. 2002; Zuo et al. 2005a). Figure 2D and E, show examples of images obtained over a 3-d interval in mice at 4 mo of age. Note the remarkable stability of the number and location of adult spines and axonal varicosities between the two views. Most adult spines persist even over a 19-mo interval (Fig. 2F, G). The noninvasive nature of the thinned skull technique also allows microglia to be imaged, which are highly sensitive to their environment and typically respond immediately after various perturbations (Davalos et al. 2005; Xu et al. 2007). Figure 3 shows images of GFP-expressing microglia cells in the cerebellum 24 h apart with minimal changes of their ramified shape under a normal physiological state.
Comparison of Thinned Skull Imaging and Imaging through a Cranial Window
Two-photon imaging through a thinned and intact skull is a relatively straightforward and minimally invasive method for high-resolution study of structural and functional changes in the living cortex. In addition to thinned skull preparations, the cortex can also be visualized through open-skull windows after performing a craniotomy and replacing the skull with a cover glass and/or a layer of agarose on top of the dural layer (see Svoboda et al. 1997; Trachtenberg et al. 2002; Holtmaat et al. 2005, 2006, 2009; De Paola et al. 2006; Lee et al. 2006; Keck et al. 2008; Hofer et al. 2009; Rose 2011). It is important to point out that skull removal can induce a significant inflammatory reaction, and thus could confound efforts to elucidate changes of neurons and glia in the intact brain (Xu et al. 2007; Holtmaat et al. 2009; Yan et al. 2009). The thinned skull window approach not only causes minimal perturbation, but also allows imaging of the cortex immediately after surgery, as opposed to the open-skull method, in which optimal imaging quality is achieved many days after craniotomy (Grutzendler et al. 2002; Trachtenberg et al. 2002; Holtmaat et al. 2005; Zuo et al. 2005a,b; Yang et al. 2009). Therefore, we suggest that two-photon imaging through a thinned skull window, rather than through an open-skull window, should be the method of choice for studying structure and function of the living cortex. In experiments that cannot easily be performed without removing the skull, it is important to interpret the data carefully in the context of the confounding factors associated with open-skull preparations.
Challenges of Preparing an Animal for Transcranial TPM Imaging
The main limitations of the thinned skull technique are related to the fact that skull thickness is critical for image quality and that the optimal thickness exists within narrow limits. For example, a nonuniform skull thickness may cause significant spherical aberrations, resulting in decreased two-photon excitation and distortion of fluorescent structures located deep in the cortex (Helm et al. 2009). We have found that the skull thickness should ideally be ~20 µm to obtain high-resolution images of synapses (Grutzendler et al. 2002; Zuo et al. 2005a; Xu et al. 2007). Overthinning the skull to <15 µm carries the risk of mild cortical trauma resulting from pushing the skull downward with the drill bit or microsurgical blade. In such cases, the thinning process generally leads to activation of microglia as manifested by their amoeboid morphology and is often associated with axonal and dendritic blebbing. Injury of this type can be prevented by carefully using the microsurgical blade at an angle during the thinning procedure and by avoiding pushing the skull against the cortical surface. Because of this issue, achieving an optimal thickness in a consistent way requires significant surgical practice. Furthermore, although multiple imaging sessions within the first few days after the initial surgery can be easily performed and require only minimal removal of debris over the thinned cranium, repeated imaging over intervals >2–3 d requires skull rethinning, which may present a challenge for a beginner. Even with sufficient surgical skill, it is often difficult to perform more than five imaging sessions on the same animal, as the optical properties of the skull may gradually deteriorate with repeated thinning.
TPM imaging through a thinned skull window has emerged in recent years as an important tool for studying structural and functional changes in the living cortex in a minimally invasive manner (Grutzendler et al. 2002; Yoder and Kleinfeld 2002; Stosiek et al. 2003; Tsai et al. 2004; Davalos et al. 2005; Zhang et al. 2005; Zuo et al. 2005a,b; Haynes et al. 2006; Majewska et al. 2006; Nishiyama et al. 2007; Xu et al. 2007; Kim et al. 2009; Wake et al. 2009). With the growing availability of fluorescent reporters to probe the structure and function of the brain (Giepmans et al. 2006; Livet et al. 2007), we envision that transcranial TPM imaging will greatly expand our current understanding of how neural circuits are assembled and are modified throughout life and how cells function in the living brain.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health to W.B.G. and J.G. G.Y. was supported by a postdoctoral fellowship from the NYS Spinal Cord Injury Research Program.
RECIPE
Recipes for items marked with <R> are provided here. Additional recipes can be found online at http://www.cshprotocols.org/recipes.
Artificial cerebrospinal fluid (ACSF)
119 mm NaCl
26.2 mm NaHCO3
2.5 mm KCl
1 mm NaH2PO4
1.3 mm MgCl2
10 mm glucose
Gas with 5% CO2/95% O2 for 10–15 min, then add 2.5-mM CaCl2. Filter sterilize with a 0.22-µm filter apparatus, and store at 4°C. ACSF is stable for 3–4 wk. If overt contamination (solution becomes cloudy) or precipitation is apparent, discard, and make fresh ACSF.
REFERENCES
- Christie RH, Bacskai BJ, Zipfel WR, Williams RM, Kajdasz ST, Webb WW, Hyman BT. Growth arrest of individual senile plaques in a model of Alzheimer’s disease observed by in vivo multiphoton microscopy. J Neurosci. 2001;21:858–864. doi: 10.1523/JNEUROSCI.21-03-00858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- De Paola V, Holtmaat A, Knott G, Song S, Wilbrecht L, Caroni P, Svoboda K. Cell type–specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, Julius D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- Helm PJ, Ottersen OP, Nase G. Analysis of optical properties of the mouse cranium—Implications for in vivo multi photon laser scanning microscopy. J Neurosci Methods. 2009;178:316–322. doi: 10.1016/j.jneumeth.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hubener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–317. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Wilbrecht L, Knott GW, Welker E, Svoboda K. Experience-dependent and cell-type-specific spine growth in the neocortex. Nature. 2006;441:979–983. doi: 10.1038/nature04783. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hubener M, Keck T, Knott G, Lee WC, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck T, Mrsic-Flogel TD, Vaz Afonso M, Eysel UT, Bonhoeffer T, Hubener M. Massive restructuring of neuronal circuits during functional reorganization of adult visual cortex. Nat Neurosci. 2008;11:1162–1167. doi: 10.1038/nn.2181. [DOI] [PubMed] [Google Scholar]
- Kim JV, Kang SS, Dustin ML, McGavern DB. Myelomonocytic cell recruitment causes fatal CNS vascular injury during acute viral meningitis. Nature. 2009;457:191–195. doi: 10.1038/nature07591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Huang H, Feng G, Sanes JR, Brown EN, So PT, Nedivi E. Dynamic remodeling of dendritic arbors in GABAergic interneurons of adult visual cortex. PLoS Biol. 2006;4:e29. doi: 10.1371/journal.pbio.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Majewska AK, Newton JR, Sur M. Remodeling of synaptic structure in sensory cortical areas in vivo. J Neurosci. 2006;26:3021–3029. doi: 10.1523/JNEUROSCI.4454-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Fukaya M, Watanabe M, Linden DJ. Axonal motility and its modulation by activity are branch-type specific in the intact adult cerebellum. Neuron. 2007;56:472–487. doi: 10.1016/j.neuron.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose CR. Two-photon sodium imaging in dendritic spines. In: Helmchen F, Konnerth A, editors. Imaging in neuroscience: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2011. pp. 297–304. [DOI] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Tsai J, Grutzendler J, Duff K, Gan WB. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat Neurosci. 2004;7:1181–1183. doi: 10.1038/nn1335. [DOI] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, Nabekura J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci. 2009;29:3974–3980. doi: 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SH, Arevalo JC, Sarti F, Tessarollo L, Gan WB, Chao MV. Ankyrin repeat-rich membrane spanning/Kidins220 protein regulates dendritic branching and spine stability in vivo. Dev Neurobiol. 2009;69:547–557. doi: 10.1002/dneu.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–551. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- Yan P, Bero AW, Cirrito JR, Xiao Q, Hu X, Wang Y, Gonzales E, Holtzman DM, Lee JM. Characterizing the appearance and growth of amyloid plaques in APP/PS1 mice. J Neurosci. 2009;29:10706–10714. doi: 10.1523/JNEUROSCI.2637-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder EJ, Kleinfeld D. Cortical imaging through the intact mouse skull using two-photon excitation laser scanning microscopy. Microsc Res Tech. 2002;56:304–305. doi: 10.1002/jemt.10002. [DOI] [PubMed] [Google Scholar]
- Zhang ZG, Zhang L, Ding G, Jiang Q, Zhang RL, Zhang X, Gan WB, Chopp M. A model of mini-embolic stroke offers measurements of the neurovascular unit response in the living mouse. Stroke. 2005;36:2701–2704. doi: 10.1161/01.STR.0000190007.18897.e3. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Lin A, Chang P, Gan WB. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005a;46:181–189. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan WB. Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature. 2005b;436:261–265. doi: 10.1038/nature03715. [DOI] [PubMed] [Google Scholar]