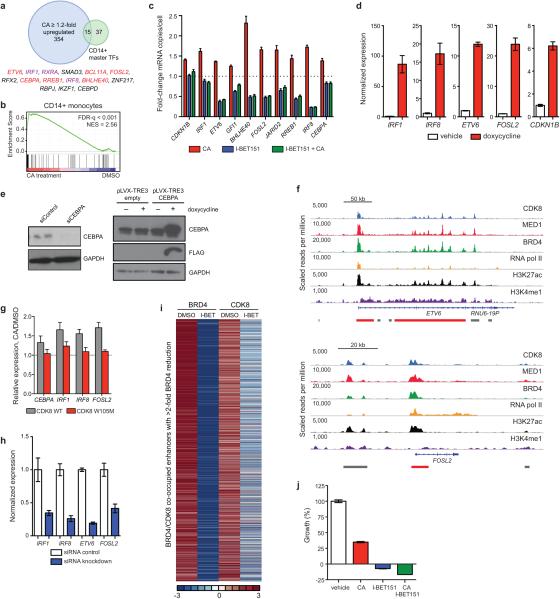
Extended Data Figure 7. Effects of SE-associated gene expression levels on MOLM-14 AML cell proliferation.
(a) Venn diagram showing overlap between CA upregulated genes and CD14+ master TFs. Overlapping genes are listed; SE-associated genes identified by one (purple) or more (red) marks in MOLM-14 are indicated. (b) GSEA plot showing positive enrichment of CD14+ master TFs upon 3 h CA treatment (MOLM-14 differential expression). (c) Fold-change in mRNA copies per cell of selected SE-associated genes upon 3 h treatment with 100 nM CA, 500 nM I-BET151 or 3 h I-BET151 followed by addition of CA for 3 h (mean ± s.e.m., n=3 biological replicates, experiment performed twice). (d,h) mRNA expression levels (d) either 1 day (FLAG-IRF1, FLAG-IRF8) or 3 days (FLAG-CDKN1B, FLAG-FOSL2, FLAG-ETV6) after induction with doxycycline or (h) 2 days after siRNA electroporation (mean, Poisson error, n = 15,000-20,000 technical replicates, experiment performed twice) corresponding to Fig. 3f. (e) Immunoblot showing protein levels of CEBPA 4 days after siRNA electroporation or 1 day after doxycycline-induced expression (experiment performed once) corresponding to Fig. 3f, full scan in Supplementary Figure 1. (f) ChIP-seq binding profiles at the FOSL2 and ETV6 loci. (g) mRNA levels of indicated genes in MOLM-14 cells expressing FLAG-CDK8 (grey) or FLAG-CDK8 W105M (red) after 3 h 25 nM CA treatment (mean ± s.e.m., n=3 biological replicates, one of two experiments shown) (i) Heatmaps showing BRD4 and CDK8 ChIP-seq on regions depleted of BRD4 >2-fold upon I-BET151 treatment for 6 h before and after drug treatment. (j) Effect of 3-day treatment with CA, I-BET151 or the combination of CA and I-BET151 on proliferation of MOLM-14 (mean ± s.e.m., n=6 biological replicates, one of two experiments shown).