Abstract
MicroRNAs have emerged as crucial regulators of neuronal function, suggesting that aberrant microRNA expression might contribute to pathologies of the nervous system. In this issue of The EMBO Journal, Emde et al (2015) report a global decrease in microRNAs as common hallmark of different forms of amyotrophic lateral sclerosis (ALS). Strikingly, enhancing microRNA biogenesis has beneficial effects on the neuromuscular function in mouse models of ALS. Thus, the microRNA pathway represents a promising novel target for therapeutic intervention in neurodegeneration.
Subject Categories: Neuroscience, RNA Biology
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease with an incidence of 1–3 in 100,000 per year. The characteristic degeneration of motor neurons results in muscle weakness, muscular atrophy, and premature death usually within 3–5 years after clinical manifestation. This pathology is considered to have a complex etiology involving multiple genes and environmental factors. Despite intense research, there is no effective treatment for ALS available up to now. The only FDA‐approved drug for ALS (Riluzole) modestly slows disease progression. Thus, a better understanding of the molecular mechanisms underlying the pathogenesis of ALS is a prerequisite for the development of innovative therapeutic strategies.
Various pathological pathways including protein aggregation, defects in stress response, and aberrant RNA metabolism have been described for ALS. Importantly, mutations in several genes encoding RNA‐binding proteins (RBPs), such as TAR DNA‐binding protein 43 (TDP‐43) or fused in sarcoma (FUS), have been linked to this pathology (Ling et al, 2013). Noteworthy, both TDP‐43 and FUS were previously reported to localize to stress granules (SG), specialized cytoplasmic foci that harbor translationally stalled mRNA (Buchan & Parker, 2009). SGs form upon different kinds of cellular stress and are observed more frequently in several neurodegenerative diseases.
Precise control of gene expression by microRNAs (miRNAs), a class of small non‐coding RNAs, is important for the physiological function of neurons (Schratt, 2009), and emerging evidence suggests that dysregulations in the miRNA pathway are causative for neurodegenerative diseases (Abe & Bonini, 2013). The biogenesis of miRNAs involves stepwise processing: The long primary miRNA (pri‐miRNA) transcript is first cleaved by Drosha in the nucleus to produce a hairpin‐shaped miRNA precursor (pre‐miRNA). In the cytoplasm, the pre‐miRNA is further processed by Dicer to generate the mature miRNA, which binds to target mRNAs via an imperfect complementarity, thereby inhibiting protein synthesis (Ha & Kim, 2014). Importantly, TDP‐43 was found to promote miRNA biogenesis at the Drosha and Dicer processing step (Kawahara & Mieda‐Sato, 2012) and also FUS enhances miRNA production via interaction with Drosha (Morlando et al, 2012), providing functional links between miRNAs, SG‐related RBPs, and ALS. Furthermore, several studies reported aberrant expression of various miRNAs in the pathology of ALS (Paez‐Colasante et al, 2015). However, the findings of these publications show only a small overlap, which could be explained by the wide range of ALS subtypes and/or the different preparations used for miRNA profiling.
In the present study, Emde et al (2015) investigated a potential role of altered miRNA biogenesis in ALS. To test this hypothesis, the authors took a comprehensive approach by combining the analysis of patient tissue, mouse models, and cell culture experiments. Expression profiling of motor neurons from patients by microarray revealed global downregulation of miRNAs in two distinct forms of ALS. This finding was recapitulated by inducing stress in a cell line using chemical stressors or overexpression of ALS‐causing mutant genes. The authors observed reduction in several candidate miRNAs and concomitant pre‐miRNA accumulation upon stress induction, as measured by qPCR. Together, these results represent a major advance compared to previous studies since they suggest defective miRNA biogenesis due to impaired Dicer processing as common denominator in ALS.
How can the observed impaired miRNA biogenesis be explained? Interestingly, while the expression levels of Dicer did not change, both chemical stress and overexpression of ALS‐causing genes led to reduced processing activity in an in vitro Dicer assay. Furthermore, the authors showed that these stress‐induced changes result from remodeling of the Dicer complex and SG formation. Strikingly, Enoxacin, a molecule known to boost miRNA biogenesis (Shan et al, 2008), partially rescued the impairments in pre‐miRNA processing in vitro and in vivo. Most notably, Enoxacin also improved the neuromuscular function in vivo in two genetically unrelated ALS mouse models. Taken together, the authors provide evidence for an extensive downregulation of miRNAs in motor neurons due to stress‐induced remodeling of the Dicer complex as common hallmark of multiple forms of ALS (Fig 1).
Figure 1. Potential link between dysregulated miRNA biogenesis and ALS .
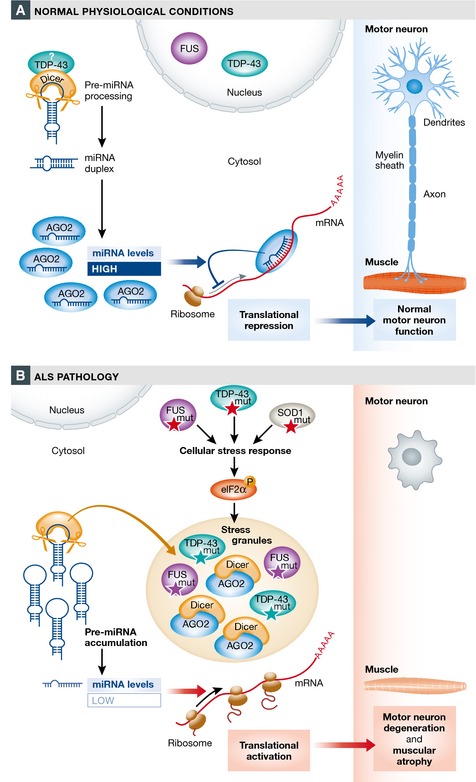
(A) Under normal physiological conditions, pre‐miRNAs are by default processed by the Dicer complex in the cytosol. The resulting mature miRNA binds to specific target mRNAs, thereby blocking protein production. Interactions of Dicer with RBPs (e.g. TDP‐43) can enhance miRNA biogenesis. (B) In ALS, mutations in multiple genes (including TDP‐43, FUS, and SOD1) have been found to activate a stress response involving phosphorylation of eIF2α and SG formation. Remodeling of the Dicer complex leads to reduced pre‐miRNA processing and concomitantly decreased miRNA levels. As a consequence, the translation of target mRNAs is aberrantly relieved from inhibition by miRNAs, ending in degeneration of motor neurons.
While these findings suggest a new mechanism for ALS, they also raise a number of important questions that should be addressed in the future. First, since not all miRNAs are equally affected by ALS mutations, what makes some miRNAs more vulnerable to changes in Dicer activity than others? Such studies could provide new insight into the regulation of pre‐miRNA processing. Second, what are the precise mechanisms whereby different ALS mutations impinge on the stress pathway? Studying molecular links between ALS proteins and SG components, such as eIF2α, would be very informative. Finally, and probably most importantly, what are the miRNA targets that mediate the detrimental effects downstream of impaired miRNA processing? Addressing the biological consequences of the aberrant miRNA levels promises to shed light on the molecular mechanisms that lead to motor neuron degeneration, thereby expanding our understanding of ALS.
Beyond that, the results of this study lay a framework for the development of novel diagnostic tools. A particular problem of neurodegenerative diseases is their late diagnosis due to long asymptomatic phases preceding first clinical manifestations. Circulating miRNAs might serve as valuable non‐invasive biomarkers to diagnose and treat neurodegeneration already at an early stage. Furthermore, uncovering specific miRNA expression signatures would be useful to discriminate distinct forms of ALS and to improve the classification of neurodegenerative diseases.
Finally, the present study not only highlights the importance of an intact miRNA network to confer neuronal integrity upon cellular stress but also points to Dicer as promising target for therapeutic approaches. On a broader scope, finding a pharmacological agent that efficiently targets the miRNA pathway has an exceptional potential for future treatments not only of neurodegeneration but also of other diseases that are accompanied by global changes in miRNA expression, such as cancer.
See also: A Emde et al (November 2015)
References
- Abe M, Bonini NM (2013) MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol 23: 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde A, Eitan C, Liou L‐L, Libby RT, Rivkin N, Magen I, Reichenstein I, Oppenheim H, Eilam R, Silvestroni A, Alajajian B, Ben‐Dov IZ, Aebischer J, Savidor A, Levin Y, Sons R, Hammond SM, Ravits JM, Möller T, Hornstein E (2015) Dysregulated miRNA biogenesis downstream of cellular stress and ALS‐causing mutations: a new mechanism for ALS. EMBO J 34: 2633–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Mieda‐Sato A (2012) TDP‐43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA 109: 3347–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79: 416–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I (2012) FUS stimulates microRNA biogenesis by facilitating co‐transcriptional Drosha recruitment. EMBO J 31: 4502–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez‐Colasante X, Figueroa‐Romero C, Sakowski SA, Goutman SA, Feldman EL (2015) Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat Rev Neurol 11: 266–279 [DOI] [PubMed] [Google Scholar]
- Schratt G (2009) microRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, Chan AW, Shi Z, Liu Q, Wahlestedt C, He C, Jin P (2008) A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol 26: 933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]


