SUMMARY
Histone acetyltransferase (HAT) complexes are coactivators that are important for transcriptional activation by modifying chromatin. Metazoan SAGA and ATAC are distinct multisubunits complexes that share the same catalytic HAT subunit (GCN5 or PCAF). Here, we show that these human HAT complexes are targeted to different genomic loci representing functionally distinct regulatory elements both at broadly expressed and tissue-specific genes. While SAGA can principally be found at promoters, ATAC is recruited to promoters and enhancers, yet only its enhancer binding is cell-type specific. Furthermore, we show that ATAC functions at a set of enhancers that are not bound by p300, revealing a class of enhancers not yet identified. These findings demonstrate important functional differences between SAGA and ATAC coactivator complexes at the level of the genome and define a role for the ATAC complex in the regulation of a set of enhancers.
INTRODUCTION
Initiation of transcription by RNA Polymerase II (Pol II) is a tightly controlled process (for review, see Venters and Pugh, 2009b). One way of viewing transcription activation is to model this process as a sequential series of events that leads to recruitment of Pol II onto the DNA and allow the production of a RNA molecule. In recent textbook models, the binding of transcription factors specifically recognizing DNA is followed by the recruitment of coactivators that regulate transcription though a variety of enzymatic and nonenzymatic activities. Coactivators act on one side by modulating the chromatin states to modify the accessibility of the DNA and on the other side by favoring the binding of general transcription factors (GTFs) that permit recruitment and loading of Pol II on the DNA to allow proper transcription initiation and elongation.
The metazoan ATAC (Ada-Two-A-Containing) and SAGA (Spt-Ada-Gcn5-Acetyl-transferase) complexes are distinct multisubunit coactivators that share several subunits including the histone acetyltransferase (HAT) enzyme GCN5 (also named KAT2A) or its closely related paralog PCAF (KAT2B). The subunit composition of SAGA and ATAC types of complexes, containing either GCN5 or PCAF, has been described in detail (Gamper et al., 2009; Guelman et al., 2009; Nagy et al., 2010; Suganuma et al., 2008; Wang et al., 2008; Zhao et al., 2008). These complexes are organized in functionally specialized modules. Both the SAGA and ATAC complexes contain a HAT module composed of the HAT enzyme GCN5/PCAF as well as distinct ADA family proteins that have been suggested to modulate the catalytic activity of the HAT enzyme (Gamper et al., 2009). Moreover, in the ATAC complex an additional subunit, ATAC2, has been shown to contain a HAT domain (Guelman et al., 2009; Suganuma et al., 2008). In addition, SAGA was shown to contain a module able to deubiquitinate monoubiqutinated histone H2A and H2B (Zhang et al., 2008a; Zhao et al., 2008). Thus, while these complexes share similarities in protein composition and functional organization, they are functionally distinct due to several specific subunits and functional modules (Nagy et al., 2009; Wang et al., 2008).
Studies analyzing the genome-wide recruitment of yeast GCN5 reported its general binding at promoters and open reading frames of active genes suggesting that its binding correlates with gene activity in yeast (Johnsson et al., 2009; Robert et al., 2004) and that it regulates both initiation and elongation (Govind et al., 2007). Similarly, in human cells, the recruitment of different families of HATs including GCN5 was suggested to be highly redundant at active promoters (Anamika et al., 2010; Zhao et al., 2008). Taken together, these results would suggest that HATs are generally recruited to active genes. In contrast, several studies in yeast and mammalian systems described gene specific HAT requirement (Anamika et al., 2010; Jin et al., 2011; Nagy et al., 2009, 2010; Zanton and Pugh, 2004). At the same time, the general transcriptional coactivator HAT, p300, was not only shown to be associated with promoters, but also with a wide spectrum of active tissue specific enhancers (Heintzman et al., 2007). Thus, at present it is not yet well understood whether genome-wide HATs carry out specific or very broad functions.
In this study, we have determined the genome-wide binding map of the two GCN5/PCAF-containing HAT complexes, SAGA and ATAC, in two human cell lines. We show that the recruitment of the complexes is highly specific as their binding sites show little overlap and they bind to distinct functional elements. ATAC is recruited to enhancers and promoters, while SAGA is mainly recruited to promoters. SAGA and ATAC bound genes are mostly active yet they represent only a subset of active genes suggesting that they function as coactivators for specific expression programs. Finally, we identify a set of functional enhancers, which are ATAC dependent, but p300 independent.
RESULTS
ATAC and SAGA Are Recruited to Distinct Sets of Loci Genome-wide
While SAGA and ATAC subunit composition is well established in several systems, very little is known about where these complexes act in the genome. To gain further insight on the transcription regulation mechanisms by ATAC and SAGA complexes, we performed chromatin immunoprecipitation (ChIP)-coupled high-throughput sequencing (ChIP-seq) against two subunits that based on our work and that of others are complex specific (Krebs et al., 2010; Nagy et al., 2009, 2010; Suganuma et al., 2008, 2010; Wang et al., 2008). We used antibodies against ZZZ3 and SPT20, ATAC- and SAGA-specific subunits, respectively (Figure S1 available online). ChIP-seq was performed in two human cell lines (GM12878 B lymphoblasts [GM] and HeLa cells). Peaks of local enrichment were determined after sequence alignment and normalization to input DNA. In total we identified 480 ATAC and 533 SAGA binding sites in GM cells. The established lists were validated by ChIP-qPCR on randomly selected loci (Figure S2) and a set of high-confidence sites was isolated (372 ATAC and 380 SAGA binding sites) (Figure S2). These high-confidence sites were further confirmed by correlating ChIP-seq enrichments in a second independent biological replicate (Figure S2). Moreover, selected high-confidence ZZZ3- and SPT20-binding sites were further tested by ChIP-qPCR with additional antibodies against ATAC (ATAC2), SAGA (TRRAP), and shared (GCN5) subunits of the complexes. These ChIP experiments further confirmed the specific binding of ATAC and SAGA complexes to the identified sites (Figure S2). Thus, these sets of binding sites were used in all analyses hereafter.
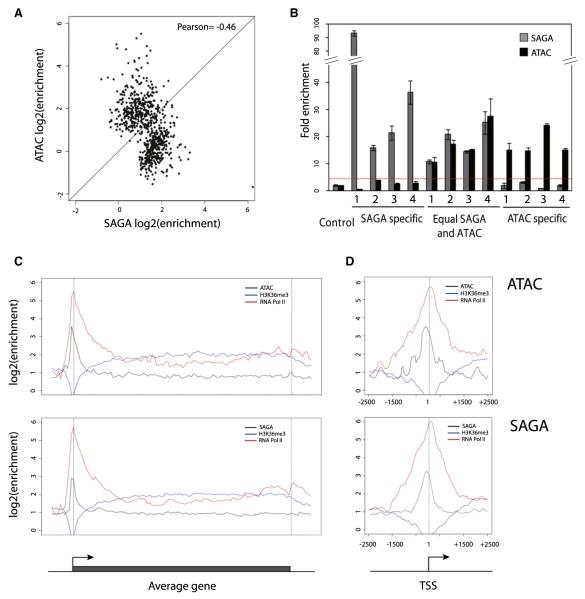
We first asked whether the two complexes bind to common or different loci. A pairwise comparison of the enrichment of ATAC or SAGA of all high confidence binding loci (Figure 1A) reveals that the bindings of both complexes show anticorrelation (Pearson correlation coefficient = −0.46). Thus, we created three groups of loci: bound by ATAC, SAGA, or both. A subset form each group was chosen randomly and successfully validated by ChIP-qPCR (Figure 1B). We conclude that the complexes are in most cases recruited to independent loci with anticor-related intensities, suggesting that their recruitment occurs through functionally distinct mechanisms.
Figure 1. The Function of ATAC and SAGA Is Mutually Exclusive on Most of Their Binding Loci.
(A) Dot plot representing ATAC versus SAGA enrichment over their high-confidence binding sites. High-confidence binding site lists of both complexes were pooled, correcting for sites being present in both lists (within 500 bp) to establish a combined list of binding sites. Then enrichments over input tracks were collected and plotted for SAGA and ATAC.
(B) Randomly chosen loci, belonging to the isolated subsets (High SAGA, Equal SAGA and ATAC, High ATAC), were validated by ChIP-qPCR. Sequence information for each locus (labeled 1 to 4) in each subset can be found in Table S3. Enrichments over background are plotted for SAGA (gray bar) and ATAC (black bar). The horizontal red dotted line indicates the 2-fold enrichments over the control genomic region. Error bars represent the standard deviation for three technical replicates.
(C) Average gene profiles of ATAC (upper panel) and SAGA (lower panel). Enrichments over input were plotted around an average gene structure for ATAC (black) or SAGA (black), as well as for RNA Pol II (red) and H3K36me3 (blue).
(D) Average binding profile of ATAC (upper panel) and SAGA (lower panel) around the TSS. Same color code as in (C) was used.
Since GCN5 has already been reported to have a role at promoters and within coding regions, we analyzed the general binding profile of SAGA and ATAC over the identified bound genes. We created an average profile of ATAC and SAGA binding density along an averaged gene structure (Figure 1C) and also located them around the TSS of bound promoters (Figure 1D). In order to better contrast promoter and coding regions, we compared the obtained profiles with those of RNA Pol II and H3K36me3, a modification present in the body of active genes. We found for both SAGA and ATAC a peak of enrichment 200 bp upstream of TSSs, with no significant enrichment over the coding region (Figure 1C). While SAGA binding appears to be more sharply positioned, ATAC binding around TSSs is less precisely defined (Figure 1D). Human SAGA binding at TSSs is in good accordance with previous reports of SAGA positioning in yeast (Venters and Pugh, 2009a). We do not observe ATAC or SAGA binding in coding regions, however we cannot exclude a role of these complexes in elongation as their binding in these regions could be very dynamic and thus, difficult to capture by ChIP.
ATAC and SAGA Are Binding Distinct Types of Genomic Elements
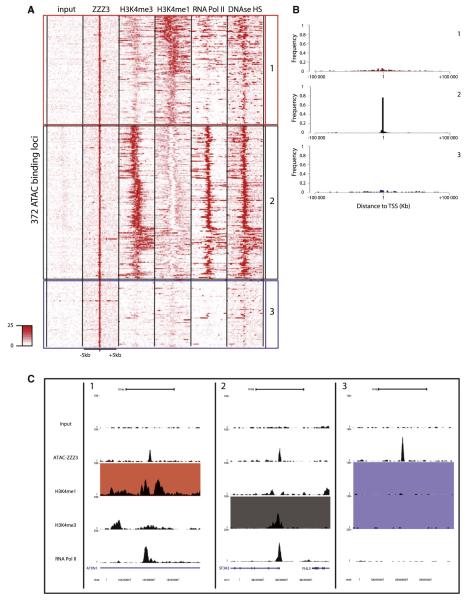
In order to ask whether the distinct binding sites reflect different modes of recruitment and or function, we contrasted the identified ATAC or SAGA binding sites with existing maps of chromatin modifications generated by the ENCODE project (Birney et al., 2007). First, we collected signal densities from raw ChIP-seq data sets for different chromatin marks known to be markers of promoters (H3K4me3) or enhancers (H3K4me1) (Heintzman et al., 2009). Additionally, we interrogated the transcription state using Pol II data as well as the compaction state of the chromatin using DNAase I hypersensitivity sites (DHS) data. These densities were subjected to k means clustering in order to group similar loci in distinct categories (as described in Ye et al., 2011). The resulting heat maps of all the analyzed features for the 372 high-confidence ATAC binding sites are shown in Figure 2A. This analysis identifies three major groups of ATAC targets loci: (1) loci enriched in H3K4me1, with low but detectable H3K4me3 and significant DHSs (red squared and labeled 1), (2) loci enriched in H3K4me3 with high Pol II binding and strong DHSs (black squared and labeled 2), and (3) a group of loci that did not display enrichment for any of the tested features (blue squared and labeled 3). The features of group 2 loci correspond to promoters, while group 1 loci have features of putative enhancers following previous descriptions (Heintzman et al., 2009; Kim et al., 2010; Rada-Iglesias et al., 2011). Note that, similar results were obtained by using the list containing the medium-confidence sites (Figure S3C).
Figure 2. ATAC Associates with Both Promoter- and Enhancer-Type Elements.
(A) Heat map of the signal densities observed on regions surrounding the 372 high-confidence ZZZ3 (ATAC) binding sites (±5 kb) for different genomic features (as indicated). The density map was subjected to clustering in order to create groups of loci sharing the same genomic profile. Loci were classified as putative enhancers (red box and labeled 1) when enriched for H3K4me1 and harboring reduced H3K4me3 signal; promoters (black box and labeled 2) when enriched for H3K4me3 and Pol II, and unassigned (blue box and labeled 3) when none of the tested features were enriched significantly (see also Figure S3).
(B) Frequency plot representing the distance of the ATAC binding sites to the transcription start site (TSS) of the closest gene in the genome in each category previously isolated. Binding sites in both the enhancer (1, red) and unassigned (3, blue) categories do not show any preferential location close to TSSs, while ATAC-binding sites are preferentially found next to TSSs in the promoter category (2, black).
(C) UCSC genome browser tracks of representative examples of loci from each category previously isolated (1, enhancer; 2, promoter; 3, unassigned). Colors of the filled boxes highlight the features used to define the category of the loci (red, enhancer; black, promoter; blue, unassigned).
See also Figures S2 and S3 and Table S2.
An additional distinctive feature between promoters and enhancers is their genomic location. We calculated the distances of ATAC-bound sites to transcriptional start sites for all three categories. The resulting frequency plots show that most of the binding sites of group 2 are located close to transcription start sites (TSSs), while categories 1 and 3 show no preference for TSS (Figure 2B), further suggesting that these are not promoter elements and that category 1-bound ATAC loci may correspond to enhancers based on H3K4me1/3 ratios. Taken together, these results demonstrate that ATAC binds both promoter elements and distal enhancer-type elements. In addition, a third set of ATAC targets (category 3) was identified of yet unknown function. Representative examples of genome browser tracks centered on an ATAC binding site for each category is shown in Figure 2C.
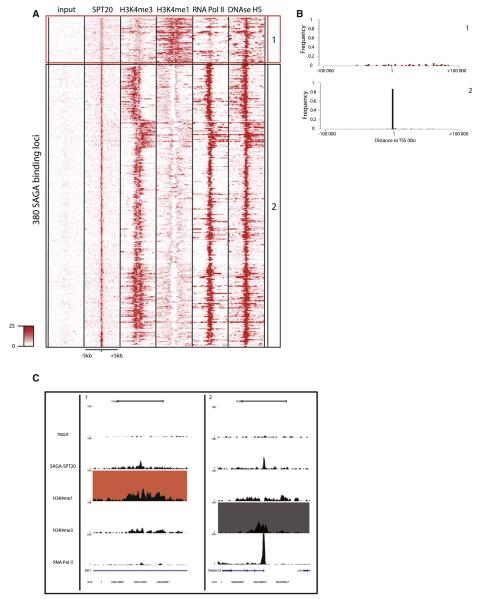
A similar analysis was applied to 380 high-confidence SAGA-bound loci (Figure 3). Based on the different genomic features used, a large majority of SAGA binding sites were located at promoter-type loci (black squared, labeled 2 in Figures 3A–3C). A smaller fraction of SAGA-bound loci appears to be located at putative enhancers (or colocalizing with H3K4me1). Moreover, the intensity of SAGA binding at these H3K4me1 bound loci is significantly lower, when compared to the intensities observed at promoters (Figure 3A) or ATAC binding intensities at the defined enhancers (Figure 2A). This pattern is very similar to those already observed for general transcription factors (TFIIDTAF1) (Heintzman et al., 2007). The number of weakly bound putative enhancers is increased when similar analysis is performed on the list including medium-confidence sites, reinforcing the idea that SAGA is binding with low intensity or more dynamically to these elements (Figures S3D and S3E).
Figure 3. SAGA Associates Preferentially with Promoters.
(A) Heat map of the signal densities observed on regions surrounding the 380 high-confidence SPT20 (SAGA) binding sites (±5 kb) for different genomic features (as indicated). The density map was subjected to clustering in order to create groups of loci sharing the same genomic profile. Loci were classified similarly as in Figure 2 (see also Figure S3).
(B) Frequency plot representing the distance of the SAGA binding sites to the transcription start site (TSS) of the closest gene in the genome in each category previously isolated. Binding sites in the enhancer (1, red) category does not show any preferential location next to TSS, while SAGA-binding sites in the promoter category (2, black) are preferentially found next to TSSs.
(C) UCSC genome browser tracks of representative examples of loci from each category previously isolated (1, enhancer; 2, promoter).
See also Figures S2 and S3 and Table S2.
Interestingly, recent studies have further classified enhancers into active or poised categories based on the modification of H3K27 by acetylation or methylation, respectively (Creyghton et al., 2010; Rada-Iglesias et al., 2011). When analyzed over the ATAC and SAGA-bound loci, we observe acetylation, but not methylation of H3K27 (Figures S3A and S3B), suggesting that ATAC- and SAGA-bound putative enhancers would be in an active state.
These analyses show that SAGA is predominantly recruited to promoters, while ATAC is equally recruited to putative active enhancers, promoters and a set of functionally unassigned loci. These in turn suggest that ATAC and SAGA bind and probably regulate functionally distinct genomic elements.
ATAC or SAGA Regulate a Limited Subset of Active Pol II Genes
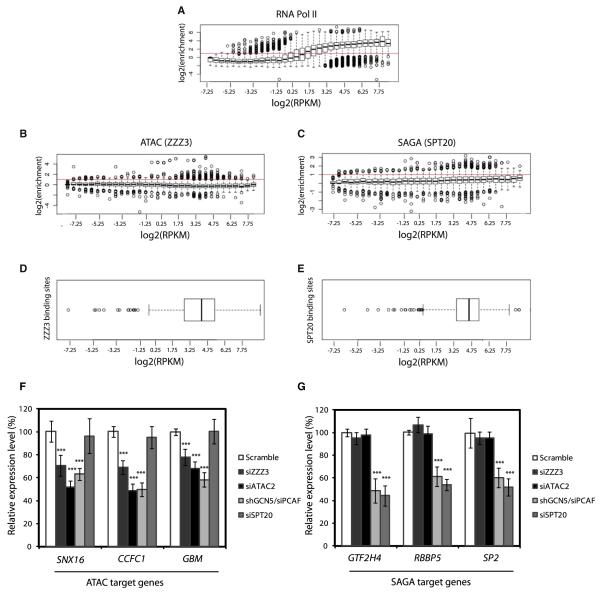
Next, we asked how ATAC or SAGA binding relates to gene activity. We systematically calculated the enrichment level of ATAC, SAGA, and Pol II at promoters of all genes and compared it to the relative expression level of genes as defined by RNA-seq (Birney et al., 2007). As expected, we observed a good correlation between Pol II occupancy and the expression status of the downstream gene (Figure 4A). On the contrary, we did not observe correlation between ATAC or SAGA occupancy and the level of expression of the nearby genes (Figures 4B and 4C). However, for both ATAC and SAGA, a number of high binding intensity values were observed at genes with relative high expression levels [outlier points for log2 (Reads/Kilobase/Million reads-RPKM) > −1], suggesting that the complexes bind in the vicinity of active genes. In order to confirm that these relatively highly expressed genes, bound either by ATAC or SAGA, correspond to the previously detected high confidence binding loci (Figure 1), we identified the gene promoters in the vicinity of these loci. We then analyzed the expression of the identified genes (Figures 4D and 4E). The expression values of these genes distribute in the range of high expression values [log2 (RPKM) > −1], also corresponding to loci where relevant Pol II levels are observed (Figure 4A). These results together demonstrate that genes bound by ATAC and SAGA are indeed active and that only a small subset of active genes is bound by either of the two complexes.
Figure 4. ATAC and SAGA Are Binding a Restricted Set of Active Genes.
(A) Gene expression correlates genome-wide with the RNA Pol II enrichment levels at promoters. Whisker plots representing the enrichment of RNA Pol II at the promoters of genes sharing similar levels of expression. Genes were categorized in increasing expression level (based on RPKM), enrichments over the control were collected at corresponding promoters and Whisker plots were plotted for each category.
(B and C) ATAC (B) and SAGA (C) bind a limited set of active genes. Similar methodology as described above for Pol II was applied to ZZZ3 or SPT20 data set, revealing an absence of global correlation between the recruitment of ATAC, or SAGA, and expression.
(D and E) The distribution of the expression levels of the genes, where ATAC and SAGA were detected at promoters, is displayed by Whisker plot. It shows that ATAC and SAGA identified binding sites are associated with promoters of active genes. Moreover, ATAC or SAGA enrichment over promoters does not correlate genome-wide with the corresponding gene expression.
(F) Depletion of ATAC subunits (ZZZ3, ATAC2, GCN5/PCAF) in HeLa cells decreases the expression of three randomly selected ATAC-bound genes by 30%–50% as compared to the scrambled siRNA control.
(G) Depletion of SAGA subunits (SPT20, GCN5/PCAF) in HeLa cells decreases the expression of three randomly selected SAGA-bound genes by 50%–60% as compared to the scrambled siRNA control. Average results of three biologically independent experiments are shown with standard deviation as error bars. p values marked above the bars with asterisks correspond to an unpaired t test for triplicates (p < 0.01).
To ask whether ATAC and SAGA serve positive regulatory function at bound promoters, we carried out knockdown experiments using small interfering RNA (siRNA) and/or short hairpin RNA (shRNA) knockdown of ATAC- (ZZZ3 and ATAC2) or SAGA-specific (SPT20) subunits or the shared catalytic subunits of the two complexes (GCN5 and PCAF) in HeLa cells (Figure S4). We randomly selected three ATAC- or SAGA-bound genes and assayed their expression level by RT-qPCR after siRNA and/or shRNA knockdown (Figures 4F and 4G). We observed that ATAC subunit knockdowns lead to a significant reduction (30%–50%) in expression of ATAC-bound genes (Figure 4F). SAGA subunit knockdowns lead to a marked decrease (50%–60%) in the expression of SAGA-bound genes (Figure 4G). Consistently, the knockdown of the shared HAT subunits (GCN5 and PCAF) leads to a reduced expression (40%–60%) on both SAGA and ATAC target genes. This in turn suggests that the expression of the regulated genes is dependent on the HAT activity of the complexes. Importantly, on the selected genes the down-regulation of ATAC function has no effect on SAGA-bound genes and vice versa (Figures 4F and 4G). Thus, decrease in the expression levels of ATAC- or SAGA-bound genes by the knockdown of specific subunits demonstrate that the individual coactivator HAT complexes positively regulate the identified genes. Moreover, the fact that knockdown of a SAGA subunits have no effect on the expression of ATAC-bound genes and vice versa, demonstrates that these two complexes function independently from each other.
Taken together, these results show that ATAC and SAGA are not required for active transcription in general but are needed to achieve the proper expression of a subset of Pol II target genes. Thus, our data argue in favor of a model where ATAC and SAGA act as specific coactivators recruited by specific transcription factors.
SAGA and ATAC Contribute to the Regulation of Ubiquitous and Tissue-Specific Transcription Programs
Next, in order to get information on the recruitment specificity of ATAC and SAGA in different cell types, we compared the binding signal of the two HAT complexes between GM and HeLa cells. To this end, we collected binding densities of the two HAT complexes in HeLa over the bound loci identified in GM cells (Figure 5A). The distributions of ChIP-seq signal intensities for both complexes in GM and HeLa cells are comparable (Figures S5I–S5K), confirming that differences in SAGA or ATAC binding in GM and HeLa cells are not because of technical limitations. We observed that, while ATAC or SAGA binding is lost on a majority of the putative enhancer loci (red squared labeled 1), their binding is mostly invariant at the promoter bound loci across the two cell lines (black squared labeled 2). This observation is supported by the low correlations observed in the binding of ATAC or SAGA across cell types at putative enhancers (Figure 5B). Note that a substantial set of HeLa specific binding sites were also identified for both ATAC and SAGA (Figure S5). However, these sites cannot be further categorized as H3K4me1/3 data are not yet available for HeLa cells from the ENCODE project. We validated these categories by ChIP-qPCR, which corroborate the fact that promoter binding is widely conserved across cell types, while putative enhancer binding is not (Figure S5). These results are in agreement with previous reports describing that enhancers are the most variable class of transcriptional regulatory elements between cell types (Heintzman et al., 2009). Indeed, our results suggest that ATAC, and to a lesser extent SAGA, by binding to enhancers will contribute to regulation of cell specific transcription programs.
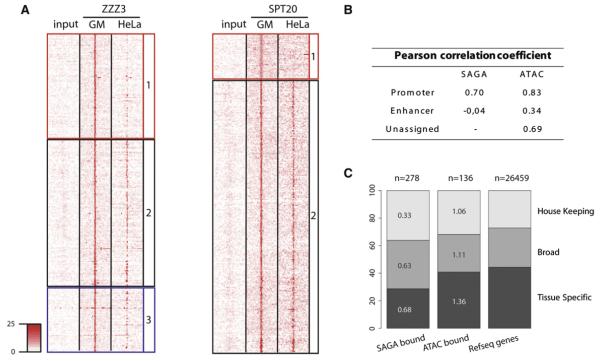
Figure 5. ATAC and SAGA Binding Exert Higher Tissue Specificity on Enhancers Compared to Promoters.
(A) Left: Comparative heatmap of the ATAC (ZZZ3) ChIP-seq signal density in GM12878 and HeLa cells around the high confidence ZZZ3 binding sites. Similar loci organization as in Figure 2A was conserved in order to illustrate differences in cell-type binding specificity between enhancer (red), promoter (black), and undefined (blue) type of loci. Right: Similar analysis has been carried out for SAGA-bound loci.
(B) Table presenting Pearson correlation coefficients calculated by comparing ChIP-seq signal in GM12878 and HeLa cells for each of the described categories (as indicated).
(C) Piled bar diagram representing the type of genes, bound by ATAC or SAGA compared to the distribution of these genes over the reference gene set (RefSeq transcripts). It reveals that neither SAGA nor ATAC show a preferential association with any of the tested categories. The proportion of genes regulated by ATAC and SAGA from the total set in each category is depicted in the corresponding subbars (in percentage).
In order to further dissect the function of the two HAT complexes, we analyzed the expression status of ATAC and SAGA-bound genes across 81 tissues and classified them according to the wideness of their expression spectra. Our analysis indicates that ATAC- or SAGA-bound promoters show a similar distribution to the total reference gene set (Figure 5C). They contain a comparable proportion of widely expressed genes and genes expressed only in a given tissue. Further Gene Ontology (GO) classification of ATAC- or SAGA-bound genes (Table S1) did not reveal any pathways specifically enriched by the studied HAT complexes, suggesting that they are involved in a variety of processes rather than specific pathways. Taken together, these observations indicate that ATAC and SAGA contribute to the regulation of both ubiquitous and tissue specific transcription programs.
Identification of Transcription Factors as Candidates for Recruiting ATAC or SAGA to Pol II Promoters
In order to get more insights in the recruitment of the studied HAT complexes at different loci, we screened ATAC- or SAGA-bound DNA sequences for known transcription factor (TF) binding motifs (JASPAR) (Portales-Casamar et al., 2010) (Figure 6 and Figure S6). In the ATAC- and/or SAGA-bound promoter subsets, several TF binding motifs were found enriched (Figure 6A). The binding site of the B cell-specific factor, SPIB, was found enriched at ATAC-bound promoters, and the binding site for the widely expressed factor CREB1 was enriched at SAGA-bound promoter loci in GM cells. In addition, within both ATAC- and SAGA-bound promoter-type sites, several factors belonging to the ETS family factors, ELK1, ELK4, and GABPa, were found to be highly enriched (Figure 6A). These factors have almost similar recognition motifs, and are widely expressed (Figure 6A). We failed to detect enriched motifs at distal loci (Figure S6). This result could be explained by observations from Heintzman et al. (2009) suggesting that enhancers are regulated by specific combination of TFs, for which motifs are not yet well defined. Thus, recruitment by SBIP or CREB could explain the specificity differences observed between ATAC and SAGA binding and function.
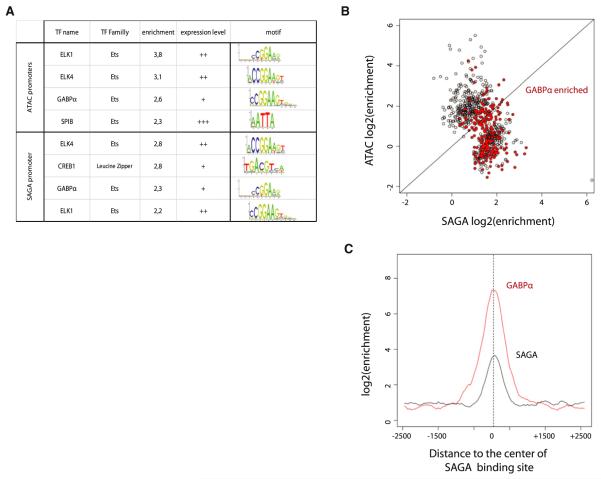
Figure 6. A Screen of Transcription Factor Motifs Identifies Candidate Transcription Factors for SAGA and ATAC Recruitment.
(A) Table summarizing the JASPAR motifs identified as enriched in the sequence of each category of ATAC or SAGA binding sites compared to the corresponding control set (see the complete analysis in the Supplemental Information). The motifs are ranked by enrichment over the control set in each category. The expression level category of the significant matches (enrichment > 2) is represented based on RNA-seq data in GM cells (RPKM categories: − [−1 < ]; + [−1:1]; ++ [1:2]; +++ [2 < ]).
(B) Dot plot highlighting the significant enrichments of GABPα ChIP-seq signal (fold enrichment > 2 in red) over the ATAC and SAGA bound sites.
(C) Average profile of enrichment of GABPα (red) and SAGA (compared to input) over a 5 kb window centered on the SAGA sites, where co-occupancy was observed.
Next, to confirm these TF screen results, we used available GABPα ChIP-seq data to determine whether ATAC and/or SAGA binding sites are co-occupied by GABPα (Figure 6B). We observe a significant GABPα binding at a large proportion of the SAGA bound sites. GABPa is also found at a limited number of ATAC binding loci, but most of these (82%) are also co-occupied by SAGA, with very few sites being ATAC specific (Figure 6B and Figures S6B and S6C). This suggests that GABPα enrichment over the ATAC binding loci is unlikely to reflect a direct TF/coactivator functional relationship. However, the high frequency of GABPa occupancy at SAGA-specific sites, clearly suggest a direct functional TF-coactivator recruitment. Moreover, the observation that GABPα average enrichment over the SAGA binding loci (Figure 6C) precisely overlaps with the center of the SAGA peak further validates the potential recruitment of SAGA by GABPα. Taken together, these results suggest that several factors, or combination of TFs, are likely to participate in ATAC or SAGA recruitment. Moreover, it reveals GABPα as a strong candidate for SAGA recruitment at several gene promoters. As GABPα is a critical factor for B cell differentiation (Xue et al., 2007), it suggests that SAGA is a crucial cofactor in regulating transcriptional pathways necessary for B cell differentiation.
ATAC Is Binding and Regulating a Set of Enhancers that Are Independent of p300
p300 was shown to be widely recruited to active enhancer loci and thus, was suggested to play a preponderant role in enhancer function (Heintzman et al., 2007, 2007; Kim et al., 2010). Since ATAC and to a much lesser extent SAGA are recruited to a set of enhancer-type elements (Figures 2 and 3), we analyzed the overlap between ATAC, SAGA, and p300 at these sites. To this end, we first investigated the p300 enrichment levels (Figure 7A, blue bars) and compared them with the ATAC or SAGA enrichments (red bars) at the previously isolated putative enhancer- or promoter-type loci for both complexes (Figures 7A and 7B). Interestingly, the distribution of the enrichments reveals that p300 and ATAC, as well as SAGA and p300, are equally distributed at promoters (Figures 7A and 7B, promoter panel), with distributions showing no significant statistical differences (Wilcoxon > 0.05). These results suggest a strong functional overlap between either ATAC and p300, or SAGA and p300, at the regulated promoters. Interestingly, when compared across cell types ATAC or SAGA with p300 they are codetected at promoters, suggesting a coordinated action of these HAT coactivators (Figures S7B and S7C).
Figure 7. ATAC Is Defining a Class of Enhancers that Are Independent of p300.
(A and B) Analysis of the distribution of p300 enrichment over enhancers and promoters of ATAC and SAGA-bound loci. For each category, the enrichment for p300 were calculated and compared to ATAC (A) or SAGA (B) enrichment, respectively. For each category a fixed step distribution histogram was plotted representing back-to-back the enrichments (log2) of ATAC or SAGA (left, red) and p300 (right, blue). The p value was calculated using a Wilcoxon test.
(C) UCSC Genome browser tracks of representative examples of ATAC specific putative enhancers (left), or enhancers bound by both ATAC and p300 (right). The color of the filled boxes highlights the features used to define the category of the loci (red, enhancer).
(D) Validation of ZZZ3- and/or p300-bound enhancers by ChIP-qPCR quantification. Sequence information for each enhancer in each subset can be found in Table S3. Enrichment of ATAC and p300 over loci, where no binding is expected (control), is calculated for ZZZ3 (black bar) and p300 (white bar). The red line indicates the 2-fold enrichments over the background.
(E) Enhancer reporter (luciferase) assay was carried out to measure the activity of ATAC (ZZZ3)-bound enhancers (E1–E4) and randomly selected genomic regions (N1–N2) compared to empty vector in GM (white bars) and HeLa cells (black bars). Known p300 bound HeLa cell-specific enhancers (H4 and H9) (Heintzman et al., 2009) were used as positive controls. ATAC bound enhancers identified in GM cells are not active in HeLa cells. The red line indicates the highest activity obtained with negative controls. Error bars represent the standard deviation for three biological replicates.
In contrast, when we looked at ATAC-bound enhancer-type loci in GM cells the large majority of these sites had no, or only weak, p300 binding (Figure 7A, enhancer panel; Wilcoxon < 0.05). ATAC-bound enhancers can be clearly separated in two groups: p300-independent (Figure 7C, left) and p300-dependent enhancers (Figure 7C, right). In order to validate these categories, we performed ChIP-qPCR on randomly chosen putative enhancer loci from both categories and known p300-dependent enhancers (Heintzman et al., 2009), further confirming the existence of ATAC-bound putative enhancer loci that are free of p300 (Figures 7D). In contrast, we did not find any SAGA-bound enhancers, which are p300 independent (Figure S7), further demonstrating functional differences between ATAC and SAGA complexes. In conclusion, our analyses strongly suggest the existence of an until now overlooked enhancer-type loci that are bound by the ATAC HAT complex, but are devoid of p300 binding. Furthermore, ATAC-bound enhancers show cell-type specificity (Figures 5A and 5B), similarly to p300-dependent enhancers (Figures S7B and S7C) (Heintzman et al., 2009).
To verify the enhancer function of the identified ATAC-bound p300-independent distal regulatory elements, we cloned four ATAC-bound, H3K4me1- and H3K27ac-enriched regions that lack p300 binding and two randomly selected negative regions devoid of any regulatory mark in a luciferase reporter construct. As controls we have used known p300-bound enhancers from HeLa cells (Heintzman et al., 2009). Interestingly, when transfecting these reporter constructs to GM cells the selected ATAC-bound enhancers show a 2- to 5-fold increase in luciferase activity over the negative genomic regions (Figure 7E), demonstrating that ATAC-bound, but p300-independent, enhancer-type loci are functionally active in GM cells. Interestingly, these ATAC-bound enhancers identified in GM cells are not active in HeLa cells, confirming their cell-type specificity (Figure 7E; and see above). In contrast, the p300-bound enhancers are only active in HeLa cells. Thus, we conclude that ATAC binds to a set of functional distal regulatory regions that share similarities with previously described enhancers, but are not bound by p300. These results together demonstrate the existence of an enhancer category that is bound by the ATAC HAT complex, but is devoid of p300 binding.
DISCUSSION
The HAT Coactivator Complexes, ATAC and SAGA, Do Not Act at Every Expressed Pol II Gene
In this study, we mapped the presence of the GCN5/PCAF-containing HAT coactivator complexes, ATAC and SAGA, genome-wide in two different cell types. Previous genome-wide studies of HAT complexes have suggested that GCN5 is generally recruited to active genes (Robert et al., 2004; Wang et al., 2009). Interestingly, by separating the function of different human GCN5/PCAF-containing complexes, our study demonstrates that ATAC and SAGA are both recruited to specific and limited set of genes. One could argue that this result is a consequence of technical limitations and lack of sensitivity in our experimental set up. While we cannot exclude that an increase in the detection sensitivity could potentially reveal more targets for the two complexes, it is clear from our correlation studies (Figure 4) that this limited number of sites is not a technical issue. Indeed, if the complexes would have been binding to the most active genes, we would observe ATAC and SAGA binding only at the top highly expressed genes. However, we observe ATAC and SAGA binding at genes with highly variable expression levels, showing that we are able to detect the complexes also at weakly expressed genes. Moreover, knockdown of ATAC or SAGA subunits clearly affect specific genes predicted to be bound by the respective complex. Thus, together our results indicate that these distinct human HAT complexes have a narrower spectrum of action than previously estimated.
This observation argues in favor of a model where HAT coactivators are recruited to their particular set of target genes by specific transcription factors. Indeed, we were able to isolate a set of transcription factors for each complex, likely explaining their recruitment specificity. One can further speculate that on the other hand specific subunits of ATAC or SAGA would participate in such a recruitment mechanism. Similarly to TFIID (Cler et al., 2009) and Mediator (Malik and Roeder, 2010), specific subunits would function as interaction partners or binding surfaces for different set of transcription factors to recruit these complexes differentially.
The systematic mapping of several HAT enzymes, suggested a general redundancy in their recruitment to promoters of active genes (Anamika et al., 2010; Wang et al., 2009). Here, we systematically compared the overlap between ATAC or SAGA binding with that of p300 (Figure 7A). These analyses confirmed a strong overlap between the recruitment of ATAC and p300, as well as SAGA and p300, at promoter-type loci and SAGA and p300 at SAGA-bound distal regulatory regions. In contrast, the different ATAC-bound enhancer-type loci are mostly devoid of p300 binding. Note, however, that if we would pool all ATAC and SAGA binding events, the majority of the binding sites would show an overlap with p300. Thus, the current analysis is not contradictory with the previously observed strong genome-wide overlap between PCAF/GCN5 and p300 HATs (Anamika et al., 2010; Wang et al., 2009). However, it emphasizes the importance of detailed genome-wide analyses that can unambiguously separate functionally distinct complexes containing the same HAT enzymes. Our analysis also suggests that at promoters there is a higher redundancy of HAT action, as compared to enhancers, where the action of HATs is more specific.
GCN5/PCAF-Containing HAT Complexes Are Not Restricted to Stress Regulated Pathways
A series of genomic studies in yeast suggested that SAGA function is mainly dedicated to the transcriptional regulation of stress regulated genes (SAGA dependent), while housekeeping genes are regulated mostly by the general transcription factor TFIID (SAGA independent) (Ghosh and Pugh, 2011; Huisinga and Pugh, 2004; Zanton and Pugh, 2004). Since all SAGA subunits and the overall structural organization of the complexes are highly conserved through evolution (Brand et al., 1999; Wu et al., 2004), the functions of the complexes were assumed to be conserved. Moreover, at the single gene scale metazoan SAGA and ATAC have been described to regulate different sets of stress induced genes (Nagy et al., 2009, 2010). Here, we demonstrate that both ATAC and SAGA are also recruited to housekeeping genes under normal conditions in human cells. Thus, our study expands the role of ATAC and SAGA in higher eukaryotes and shows that the role of ATAC and SAGA is not only restricted to stress induced pathways, but that both complexes can regulate housekeeping as well as tissue specific genes.
The Discovery of p300-Free ATAC-Dependent Enhancers Opens Perspectives in the Understanding of Mechanisms Regulating Enhancer Function
Distal enhancer elements are known to play a key role in long-distance transcriptional regulation. However, defining the genomic locations of such regulatory elements and their regulation is still not fully understood. The recent technology advancements and the mapping of numerous biological features have allowed defining a minimal set of rules to identify enhancer loci (Heintzman et al., 2009; Rada-Iglesias et al., 2011). Here, we report the binding of ATAC on a limited set of active enhancer loci that share several common features with previously defined enhancers, but differ in their coactivator requirement. Indeed, we demonstrate that the broadly bound p300 HAT is not a universal marker of all enhancers, since most of the identified ATAC-bound enhancer-type loci, which are enriched in H3K4me1 and H3K27ac, have low Pol II and detectable DHSs, are p300 independent. Our results have three implications. First, the high recruitment specificity of ATAC on a set of enhancer-type loci suggests an until now nondescribed role for this complex at this type of long-distance regulatory elements. Second, it demonstrates that p300 that was previously considered as a “general” marker of enhancer is not the only HAT recruited to these functional elements. Third, it suggests that enhancer regulation complexity was previously underestimated and that enhancers can be regulated through a variety of regulatory signals including the specific recruitment of HAT complexes. Our data raises the possibility that the binding of distinct HAT coactivator complexes likely in combination with other coactivators, define different sets of enhancers. This issue is particularly important since enhancers have been shown to be the most important regulatory elements to control tissue specific transcription (Heintzman et al., 2009). Thus, the existence of complex and probably combinatorial mechanisms specific for each enhancer subsets and dedicated to particular transcriptional pathways can be expected. Interestingly, we also observed the binding of ATAC to a set of distal loci not marked by previously described enhancer features (H3K4me1, DHSs, Pol II). This intriguing observation suggests that ATAC binds to a set of previously uncharacterized genomic elements. Even though the function of these elements remains to be established, one can speculate on their nature. Two major hypotheses can be raised, they could either be long-distance regulatory elements not sharing the canonical enhancer features in chromatin marking, or an independent class of regulatory element for which the genomic features have not yet been defined. Further systematic investigation of the function of these loci should reveal unexpected functions in the regulatory repertoire of the multi faced ATAC complex.
EXPERIMENTAL PROCEDURES
Cell Culture and Antibodies
GM12878 cells were obtained from the Coriell Cell Repositories. GM and HeLa cells were grown according to the instructions of the suppliers. Description and characterization of anti-SPT20 (3006) antibody is provided in the Supplemental Experimental Procedures. The anti-hZZZ3 (2616) polyclonal antibody was described earlier (Nagy et al., 2010). Transient transfections of the cells and luciferase reporter assay are described in the Supplemental Experimental Procedures.
Chromatin Immunoprecipitation-High-Throughput Sequencing
ChIP-seq and RNA-seq data were obtained from the ENCODE project repository (Birney et al., 2007). Details on the data sets used, sequencing statistics, and laboratory contribution can be found in Table S2.
ChIP was carried out as described previously (Nagy et al., 2010). Assessment of the individual enrichment over the control genomic region was performed by ChIP-qPCR in triplicate with primers specific for these regions with SYBR Green master mix (Roche). Information of the primers used for ChIP-qPCR is provided in Table S3. Details of ChIP-seq method are provided in the Supplemental Experimental Procedures.
Bioinformatics Procedures
A full description of Bioinformatics procedures can be found in the Supplemental Experimental Procedures. In brief, ZZZ3 and SPT20 enrichment clusters were detected with MACS (Zhang et al., 2008b). Peaks were ranked base on tag density and a cutoff was determined according to ChIP-qPCR validation results (Figure S2). The organization of distinct loci, density heat maps, and enrichment calculations were done with seqMINER (Ye et al., 2011). Comparative correlation analyses were obtained with ad hoc R scripts.
Supplementary Material
ACKNOWLEDGMENTS
The authors are very grateful to the ENCODE project participants from which released data were used in the analyses. The authors are grateful to T. Ye, L. Burger, C. Keime, and S. Le Gras for bioinformatics support; the Institut de Génétique et de Biologie Moléculaire et Cellulaire Next Generation Sequencing platform for data generation; D. Dembele for statistical support; and G. Duval and P. Eberling for antibody and peptides generation and the IGBMC cell culture service. We thank D. Schübeler and F. Müller for critical reading of the manuscript. A.R.K. was a recipient of a fellowship from Institut National de la Santé et de la Recherche Médicale Région Alsace and the Association pour la Recherche sur le Cancer. This work was funded by grants from the Agence Nationale pour la Recherche (GenomATAC; ANR-09-BLAN-0266), the European Union (EUTRACC and EPIDIACAN), and the Centre National de la Recherche Scientifique (LEA-SkinChroma).
Footnotes
ACCESSION NUMBERS
The sequencing data have been deposited in the GEO database under the accession number GSE31052.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at doi:10.1016/j.molcel.2011.08.037.
REFERENCES
- Anamika K, Krebs AR, Thompson J, Poch O, Devys D, Tora L. Lessons from genome-wide studies: an integrated definition of the coactivator function of histone acetyl transferases. Epigenetics Chromatin. 2010;3:18. doi: 10.1186/1756-8935-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286:2151–2153. doi: 10.1126/science.286.5447.2151. [DOI] [PubMed] [Google Scholar]
- Cler E, Papai G, Schultz P, Davidson I. Recent advances in understanding the structure and function of general transcription factor TFIID. Cell. Mol. Life Sci. 2009;66:2123–2134. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Kim J, Roeder RG. The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol. Cell. Biol. 2009;29:266–280. doi: 10.1128/MCB.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Pugh BF. Sequential recruitment of SAGA and TFIID in a genomic response to DNA damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011;31:190–202. doi: 10.1128/MCB.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, Zha J. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol. Cell. Biol. 2009;29:1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson A, Durand-Dubief M, Xue-Franzén Y, Rönnerblad M, Ekwall K, Wright A. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 2009;10:1009–1014. doi: 10.1038/embor.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Demmers J, Karmodiya K, Chang NC, Chang AC, Tora L. ATAC and Mediator coactivators form a stable complex and regulate a set of non-coding RNA genes. EMBO Rep. 2010;11:541–547. doi: 10.1038/embor.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol. Cell. Biol. 2009;29:1649–1660. doi: 10.1128/MCB.01076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci. 2010;67:611–628. doi: 10.1007/s00018-009-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38(Database issue):D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Mushegian A, Swanson SK, Abmayr SM, Florens L, Washburn MP, Workman JL. The ATAC acetyltransferase complex coordinates MAP kinases to regulate JNK target genes. Cell. 2010;142:726–736. doi: 10.1016/j.cell.2010.07.045. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009a;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 2009b;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher-Reilley J, Wu Z, Spolski R, Jing X, Zhang YC, McCoy JP, Leonard WJ. The transcription factor GABP is a critical regulator of B lymphocyte development. Immunity. 2007;26:421–431. doi: 10.1016/j.immuni.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. USA. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008a;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008b;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.