Abstract
Hhex encodes a homeodomain transcription factor that is widely expressed in hematopoietic stem and progenitor cell populations. Its enforced expression induces T-cell leukemia and we have implicated it as an important oncogene in early T-cell precursor leukemias where it is immediately downstream of an LMO2-associated protein complex. Conventional Hhex knockouts cause embryonic lethality precluding analysis of adult hematopoiesis. Thus, we induced highly efficient conditional knockout (cKO) using vav-Cre transgenic mice. Hhex cKO mice were viable and born at normal litter sizes. At steady state, we observed a defect in B-cell development that we localized to the earliest B-cell precursor, the pro-B-cell stage. Most remarkably, bone marrow transplantation using Hhex cKO donor cells revealed a more profound defect in all hematopoietic lineages. In contrast, sublethal irradiation resulted in normal myeloid cell repopulation of the bone marrow but markedly impaired repopulation of T- and B-cell compartments. We noted that Hhex cKO stem and progenitor cell populations were skewed in their distribution and showed enhanced proliferation compared to WT cells. Our results implicate Hhex in the maintenance of LT-HSCs and in lineage allocation from multipotent progenitors especially in stress hematopoiesis.
Keywords: Hematopoiesis, Lymphopoiesis, Stem cells, Homeobox, Homeodomain
Introduction
Hematopoietically expressed homeobox (Hhex, formerly Hex or Prhx) is a highly conserved gene with orthologs in all metazoan species examined to date. Hhex knockout in mice is early embryonic lethal at E10.5 so most investigations have focused on Hhex's role in development and the cause of fetal demise. Similar to other homeobox genes, Hhex is required for embryonic patterning and organogenesis. Hhex−/− die in utero from major developmental malformations, especially in liver and heart primordial structures [1–5]. Hhex was originally cloned from human bone marrow (BM) and peripheral blood leukocytes and was found in diverse hematopoietic cell lines and in embryonic blood islands and endothelial precursors [6–8]. Embryoid bodies derived from Hhex−/− embryonic stem (ES) cells show defects in granulocyte-monocyte colony formation and in the formation of definitive hematopoietic cell colonies [1]. Hhex encodes a 30 kDa transcription factor with repressive activity that may involve oligomerization, binding to Groucho/TLE family of corepressors, and displacement of TATA binding protein although activation of targets has also been described [4, 9–15]. Hhex protein binds DNA via a well-conserved homeodomain that is flanked at the carboxyl terminus by an acidic domain and by an amino-terminal proline-rich domain that has little similarity to other proteins.
Hhex is strongly linked to both murine and human hematologic neoplasms [16–19]. Hhex is the second most frequent integration site in retroviral insertional mutagenesis screens in AKXD mouse models of leukemias and lymphomas [18]. Enforced expression of Hhex in murine BM transduction followed by transplantation induces T-cell acute lymphoblastic leukemia (T-ALL) in recipient mice [16]. In human T-ALL, HHEX is highly expressed in the treatment-resistant subtype, early T-cell precursor-ALL (ETP-ALL), where it is a direct transcriptional target of the LIM domain Only-2 (LMO2) protein complex [20]. HHEX is part of an ETP-ALL gene signature that is also observed in Lmo2 transgenic mouse models, which have T-cell progenitor differentiation arrest, quiescence, and enhanced self-renewal [21]. In thymocyte adoptive transfer experiments, Hhex overexpression confers enhanced self-renewal, in the same manner as Lmo2 [22]; and, deletion of Hhex markedly attenuates Lmo2-induced T-ALL, underscoring Hhex's role as a critical downstream mediator of Lmo2's oncogenic actions [20]. In contrast to these studies supporting Hhex as an oncogene, data from human acute myeloid leukemia (AML) suggests that HHEX is a tumor suppressor through post-transcriptional regulation of mRNA transport with the eukaryotic initiation factor 4E [23]. HHEX is also part of a rare chromosomal translocation, t(10;11) (q23;p15), in human AML creating a NUP98-HHEX fusion protein [24]. Most of HHEX is expendable for AML induction by this fusion protein except for the homeodomain, which contributes to DNA binding, and NUP98's transcriptional activating domains.
Study of Hhex's role in hematopoiesis has been limited to primitive stages due to the early embryonic lethality of knockout mice. In this study, we induced a conditional deletion of Hhex using vav-Cre, which generated viable mice with efficient gene deletion allowing analysis of postnatal hematopoiesis. We found a severe defect in B-cell development at steady state which was observed in Rag1−/− blastocysts complemented by Hhex−/− ES cells [25]. Additionally, we discovered severe phenotypes in the setting of stress hematopoiesis. Hhex conditional knockout (cKO) BM was severely compromised in competitive BM transplantation assays and after sublethal irradiation, Hhex cKO mice could not repopulate lymphoid cells whereas myeloid repopulation was normal. We discovered that Hhex cKO mice had skewed proportion of stem and progenitor cell populations with increased proliferation. Our studies show that Hhex is required at multiple stages of hematopoietic stem and progenitor cell differentiation.
Materials and Methods
Mice
Floxed Hhex mice were created at NCI Frederick as previously described and detailed in Supporting Information Methods [20]. The floxed Hhex mice used for analyses in this article were generated by backcrossing Hhexlox/+ to B6 for 10 generations followed by intercrossing to create homozygous floxed mice, Hhexlox/lox. B6.Vav-Cre transgenic mice [26] (B6.Cg-Tg(Vav1-cre)A2Kio/J) were purchased from Jackson Laboratories (Bar Harbor, ME, http://www.jax.org) and were crossed with the floxed mice to create Hhex cKO mice (Hhex cKO) [27]. Both Hhexlox/lox and cKO mice (i.e., equivalent genetic background) were used for in vitro and in vivo studies with the former referred to as wild type (WT) throughout the manuscript. B6.SJL (CD45.1) mice were host mice for transplantation and purchased from Charles River (Frederick, MD, http://www.criver.com). All mice were housed in specific-pathogen-free facilities at Vanderbilt University with approved protocols from the IACUC.
Genotyping
Genomic DNA was isolated from mouse BM, spleen, and thymus using Qiagen DNeasy Blood and Tissue kit per manufacturer's instructions (cat#69504). Primer sequences for polymerase chain reaction (PCR) amplification of the Hhex floxed and cKO alleles were 5′-GCTCTCCAGCCACTTTGGAG-3′, 5′-GCACACCTGT GGCTAAATGCA-3′, and 5′-CATCAGGGTATGAGGAGAAG-3′.
Peripheral Blood and Hematopoietic Tissue Analyses and Proliferation
For complete blood counts, peripheral blood was collected retro-orbitally and analyzed by Hemavet instrument (Drew Scientific, Dallas, TX, http://www.drew-scientific.com/). Mono-nuclear cells were purified by density centrifugation in lymphocyte separation medium (LSM, Mediatech, http://www.cellgro.com/) after acid-citrate lysis of erythrocytes. For fluorescence activated cell sorting (FACS) analysis and sorting, antibodies were purchased from BD Pharmingen (San Diego, http://www.bdbiosciences.com/index[lowen]us.shtml) and eBiosciences (Supporting Information Table S1 for full list). FACSAria, LSRFortessa, LSRIII, and FACS Canto II flow cytometers were used for sorting and analysis. Additional analyses were performed with Flojo software. For proliferation analyses, mice were intraperitoneally injected with 1 mg of bromodeoxyuridine (BrdU) in 150 μL phosphate buffered saline (PBS) and sacrificed 2 hours later. The stem and progenitor populations were assessed for BrdU incorporation per manufacturer's instructions (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com) and as previously described [21, 28, 29].
Retroviral Transductions
To produce retroviruses, the MSCV-Hhex-iresGFP (MIG-Hhex) or MSCV-iresGFP (empty MIG) and pCL-Eco plasmids were cotransfected into the Phoenix packaging cell line using calcium phosphate precipitation as described and viral supernatant collected 48 hours later; this supernatant was clarified by centrifugation at 1,000g and used for transductions [30]. BM cells from Hhex cKO or WT mice were flushed from two femurs, followed by erythrocyte lysis, and separation by LSM. BM cells were transduced using spinfection with viral supernatant at 2,000 rpm with 8 μg/mL of polybrene (Sigma) for 1 hour at 4°C. Cocultures with OP9 and OP9-DL1 cells were performed as previously described (Supporting Information) [21, 31].
Competitive BM Transplant Assays
Competitive BM transplantation assays were performed by intravenous injection of mixed CD45.2 donor (3 × 106) whole BM cells with CD45.1 competitor (0.5 × 106) BM cells at a 6:1 ratio. WT and Hhex cKO BM are CD45.2+. Recipient CD45.1+ mice were lethally irradiated with 9.5 Gy.
Sublethal Irradiation Assay
WT and Hhex cKO littermates were sublethally irradiated with a dose of 6.5 Gy and were sacrificed at 3, 6, and 9 weeks post-irradiation for fluorescence-activated cell sorting (FACS) analysis of BM, spleens, and thymi.
Gene Expression Analysis
Hhex gene expression was done in accordance with the Immgen data release policy and is available through this resource (www.immgen.org). Total RNA was purified by RliaPrep RNA Cell Mini-Prep system (Promega, Madison, WI, http://www.promega.com) and prepared for Illumina sequencing as previously described [21, 32]. See Supporting Information methods for preparation of cDNA libraries for next generation sequencing (i.e., RNA-seq).
Results
Hhex cKO Mice Show Reduced BM and Splenic Cellularity
To determine the normal expression pattern of Hhex, we analyzed data from the Immunological Genome Project (ImmGen) where the gene expression of sorted hematopoietic cells of various lineages was analyzed by microarray [33]. As shown in Figure 1(A), Hhex expression was observed in multiple hematopoietic lineages; highest levels were seen in developing B cells, hematopoietic stem and progenitor cells, and myeloid cells (also Supporting Information Fig. S1A). Hhex expression was also high in ETP and double negative (CD4−CD8−) cells but was not observed in progenitors of subsequent T-cell differentiation stages (Supporting Information Fig. S2). Hhex clustered in fine-grained module 138, along with Lmo2, Lyl1, and Mef2c, a gene signature also observed in mouse and human Lmo2-induced T-ALLs [20, 34, 35].
Figure 1.
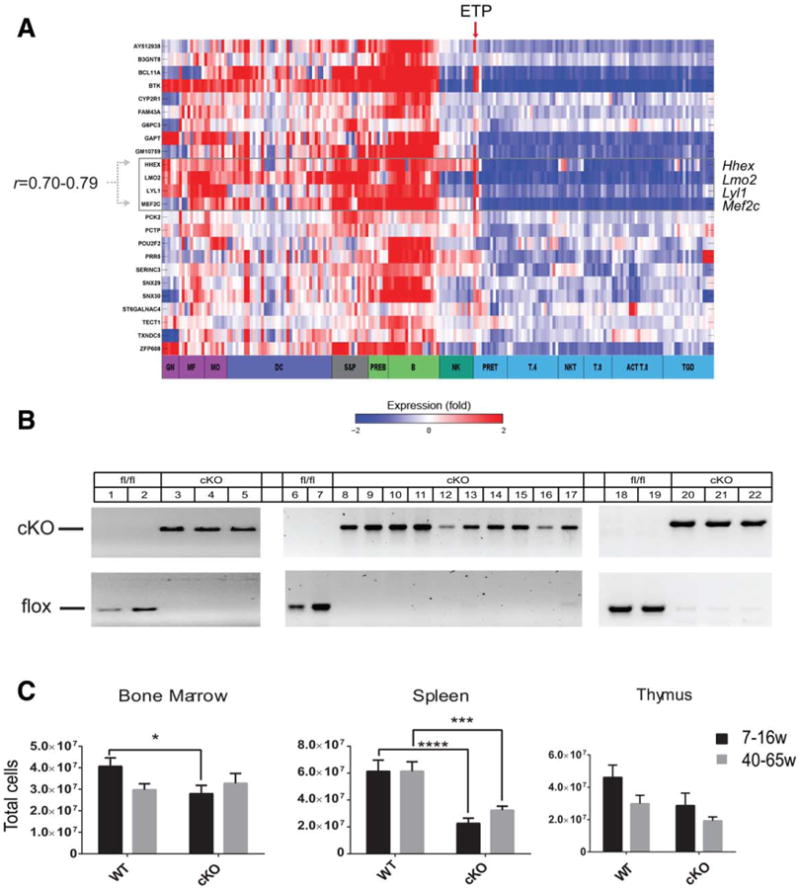
Hhex cKO mice show reduced bone marrow and splenic cellularity. (A): Heat map of 24 significantly enriched genes in hematopoietic cells across ImmGen datasets with red indicating high expression and blue indicating low expression on log2 expression scale. Cell types grouped together are color coded. Column showing expression in early T-cell precursor is indicated by red arrow. Hhex is highly expressed in myeloid and B lineages and is highly correlated (Pearson correlation shown) with the boxed genes. (B): Agarose gel of polymerase chain reaction genotyping of bone marrow, spleen, and thymus cells taken from WT (same as Hhexlox/lox) and Hhex cKO (Hhexlox/lox; Tg[vav-Cre]) mice. Genotyping of splenocytes was done on sorted B220+ (lanes 6, 8, 10, 12, 14, and 16) and B220− (lanes 7, 9, 11, 13, 15, and 17) cells. (C): Bone marrow cellularity (femurs and tibiae) of WT and Hhex cKO mice between 7–16 weeks and 40-65 weeks (WT 7–16 weeks, n = 22; WT 40-65 weeks, n = 12; cKO 7-16 weeks, n = 21; cKO 40–65 weeks, n = 12). Splenic cellularity of WT and Hhex cKO mice is shown for mice ages 7–16 weeks and 40–65 weeks (WT 7–16 weeks, n = 20; WT 40–65 weeks, n = 11; cKO 7–16 weeks, n = 22; cKO 40–65 weeks, n = 11). Thymic cellularity of WT and Hhex cKO mice between 7–16 weeks and 40–65 weeks, n = 11 each cohort. Total cellularity was determined by counting live cells. Bar graphs are means ± SEM; two tailed Student's t test generated p values: *, p ≤.05; **, p ≤ .01; ***, p ≤ .001; ****, p ≤ .0001. If brackets and asterisks are not shown then pairwise comparisons were not significant. Abbreviations: ACTT.8, activated CD8+ T cells; B, B cells; cKO, conditional knockout; DC, dendritic cells; GN, granulocytes; Hhex, hematopoietically expressed homeobox; Lmo2, LIM domain only-2; MF, macrophages; MO, monocytes; NK, NK cells; NKT, NK/T; PREB, pre-B; PRET, pre-T; S&P, stem and progenitor; T.4, CD4+ T cells; T.8, CD8+ T cells; TGD, γδ T cells; WT, wild type.
We created cKO mice to analyze postnatal hematopoiesis. The generation of mice with loxP sites flanking Hhex exons 2– 4 has been described [20]. We bred floxed Hhex mice to vav-Cre transgenic mice to generate Hhexlox/lox; Tg(vav-Cre), hereafter denoted Hhex cKO, which were viable, fertile, and born in normal litter sizes. We observed highly efficient Cre-mediated deletion of floxed Hhex in the BMs, spleens, and thymi of Hhex cKO mice where there were no detectable floxed alleles and no mRNA from exons 2–4 (Fig. 1B; Supporting Information Fig. S1). The peripheral blood counts of Hhex cKO mice were normal (Supporting Information Fig. S3) compared to age-matched littermates with intact Hhex (i.e., Hhexlox/lox denoted WT hereafter) but Hhex cKO mice had reduced total cellularity of the BM and spleen (Fig. 1C); the total BM cellularity of older mice was not significantly different between WT and cKO; and, there was no statistical difference in thymic cellularity in young or aged mice.
Hhex cKO Mice Have Markedly Reduced Numbers of Mature and Developing B Cells
On gross dissection, the lymph nodes and thymi of Hhex cKO mice appeared similar to WT mice, however, the spleens were smaller in size and had distorted architecture with smaller follicles and denser periarteriolar lymphoid sheaths (Fig. 2A). Hhex cKO spleens had reduced numbers of mature CD4+ and CD8+ T cells and granulocytes and macrophages but their progenitor populations were similar to WT thymi and BM, respectively (Supporting Information Fig. S3; Fig. 5 for myeloid progenitor comparison, LK or Lin−Kit+). Immunohistochemical staining for CD3 and B220 showed that Hhex cKO splenic follicles were composed of T cells and B cells were notably absent (Fig. 2A). We analyzed both mature and immature B-cell populations in hematopoietic tissues by FACS. The absolute numbers and proportions of B cells in Hhex cKO BMs and spleens were markedly reduced compared to WT mice (Fig. 2B, 2C). Hhex cKO mice had reduced proportions and absolute numbers of B-cell progenitors at multiple stages of differentiation from pro-B cells to recirculating B cells (Fig. 2E). We sorted the BM and spleen into B220− and B220+ populations and confirmed complete knockout of the floxed Hhex allele (Fig. 1B). Hhex cKO BM had an increased aberrant population of B220−CD19+ cells (Fig. 2C, 2D), which were sorted and analyzed and showed complete deletion of Hhex (data not shown). Hence, some mature B cells developed without Hhex but at markedly reduced number. This was also confirmed by similar V-D-J recombination patterns in genomic DNA isolated from the B cells of WT and Hhex cKO mice (Supporting Information Fig. S7). In summary, our in vivo data showed impaired B-cell differentiation in Hhex cKO BM and markedly reduced absolute numbers of the earliest B-cell progenitors, the pro-B cells, suggesting a defect in B-cell commitment.
Figure 2.
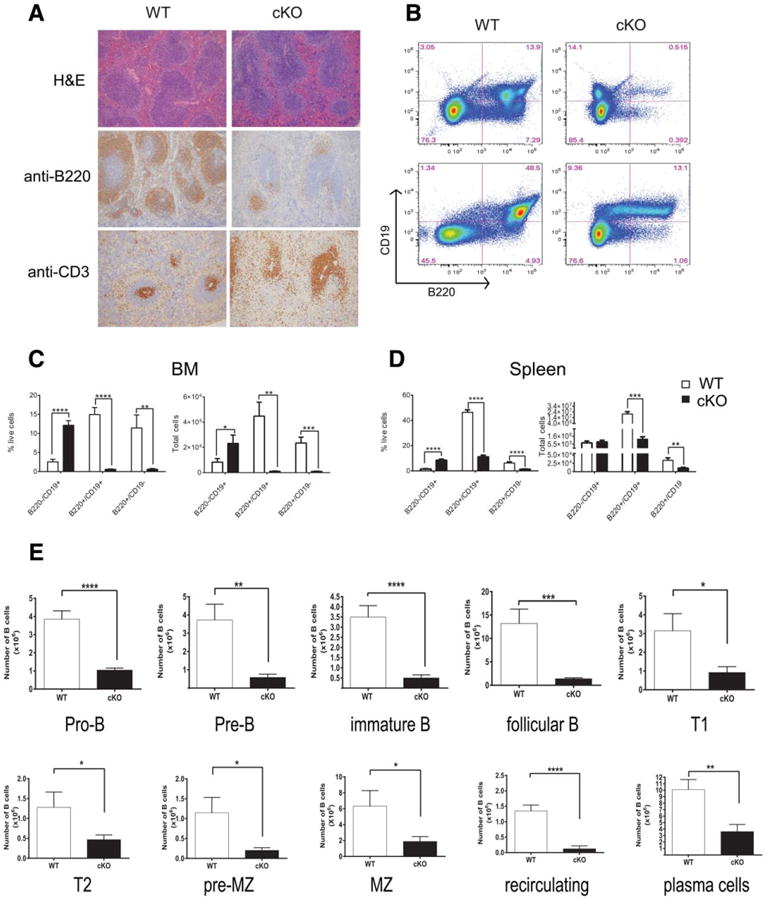
Hhex cKO mice show a defect in B-cell development. (A): Histological evaluation of spleens from WT (aka Hhexlox/lox) and hematopoietically expressed homeobox (Hhex) cKO mice. Sections were stained with H&E (hematoxylin and eosin), anti-B220, or anti-CD3 antibodies. (B): Representative fluorescence activated cell sorted plots for B220 and CD19 for BM and spleen cells isolated from WT and Hhex cKO mice. (C): Bar graphs show mean (± SEM, n = 9) proportions (% live cells) and absolute numbers of B cells in the BM. Absolute numbers were calculated based on the total number of cells multiplied by the percentage of each population. (D): Bar graphs show mean (± SEM, n = 8) proportions (% live cells) and absolute numbers of B cells in the BM. Calculation was same as for BM. (E): Bar graphs show the mean numbers (± SEM, WT: n = 10 and Hhex cKO: n = 9) of B-cell subsets in the BM and spleen (Supporting Information Table S2) of WT and Hhex cKO mice. WT values are shown in white and Hhex cKO values are in black. Two-tailed Student's t tests were applied: *, p ≤ .05; **, p ≤ .01; ***, p ≤.001; ****, p ≤ .0001. All mice analyzed in these experiments were between 7–16 weeks old. Abbreviations: BM, bone marrow; cKO, conditional knockout; WT, wild type.
Figure 5.
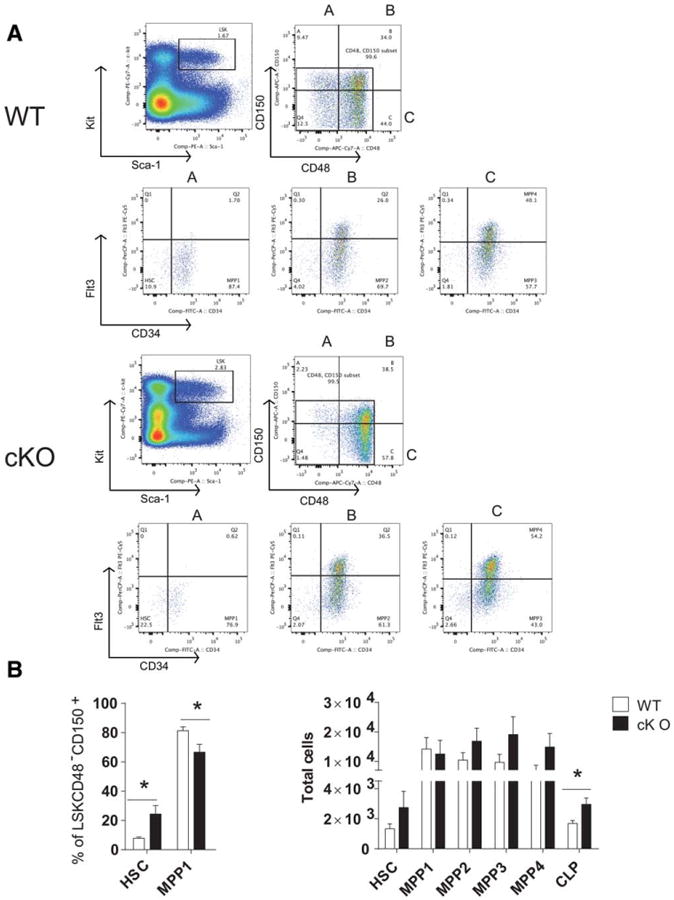
Hematopoietically expressed homeobox (Hhex) cKO mice have skewed HSC and MPP distribution and increased CLPs. (A): Representative flow cytometric analysis of the hematopoietic stem and progenitor populations from the bone marrow of WT and Hhex cKO mice at 7–16 weeks of age. The sizes of gated populations as percentages of the parental populations are shown. (B): Plots show the absolute numbers of each stem and progenitor population in the bone marrow of WT or Hhex cKO mice. Total cells were calculated based on the total number of cells multiplied by the percentage of each population, WT, n = 10; cKO, n = 11. p values were generated by two-tailed Student's t test: *, p ≤ .05. Abbreviations: APC, allophycocyanin; cKO, conditional knockout; CLP, common lymphoid progenitor; FITC, fluorescein isothiocyanate; HSC, hematopoietic stem cell; LK, Lin−Sca-1−Kit+, myeloid progenitors; LSK, Lin−Sca-1+Kit+ HSPCs; MPP, multipotent progenitor; MPP1, LSKCD48−CD150+CD34+Flt3−; MPP2, LSKCD34+CD48+CD150+Flt3−; MPP3, LSKCD34+CD48+ CD150−Flt3−; MPP4, LSKCD34+CD48+CD150−Flt3+; PE, phycoerythrin; WT, wild type.
Hhex cKO BM is Compromised in Competitive Repopulation of Lethally Irradiated Host Mice
To demonstrate a cell-autonomous role for Hhex in B-cell development, we cocultured Lin−Sca-1+Kit+ (LSK) cells from Hhex cKO or WT BMs with OP9 stromal cells. After 1 week of culture, WT LSK cells increased by eightfold and the majority of cells expressed B220 or CD19. In contrast, the Hhex cKO LSK cells increased by twofold, could not be further passaged, and did not express B220 or CD19. We performed RNA-seq on LSK and on passage-1 cells in coculture and discovered that the Hhex cKO LSK did not upregulate canonical B-cell-specific transcripts (Supporting Information Fig. S4).
We next performed competitive repopulation assays where we transplanted lethally irradiated CD45.1 congenic host mice with Hhex cKO or WT B6 bone (donor phenotype, CD45.2+) marrow mixed with B6.CD45.1 congenic BM (i.e., host phenotype). We observed negligible CD45.2+ hematopoiesis in transplanted mice at various input ratios (Supporting Information Table S5). Next, we performed transplants with sixfold excess of Hhex cKO BM compared to CD45.1+ competitor cells (Fig. 3E). We analyzed the peripheral blood of host mice 3–12 weeks post-transplant and found near complete absence of Hhex cKO donor cells (Fig. 3F) whereas WT donor BM contributed to the BM, thymus, and spleen of host mice. Hhex cKO BM cells contributed to the BM of host mice at a markedly reduced ratio than the input of 6:1 and had negligible contribution to the thymi or spleens of host mice 16 weeks post-transplant (Fig. 3G). We determined that Hhex cKO BM cells appropriately homed to recipient host BM (Supporting Information Fig. S9A) and were engrafted 7 days after intravenous injection (Supporting Information Fig. S9B).
Figure 3.
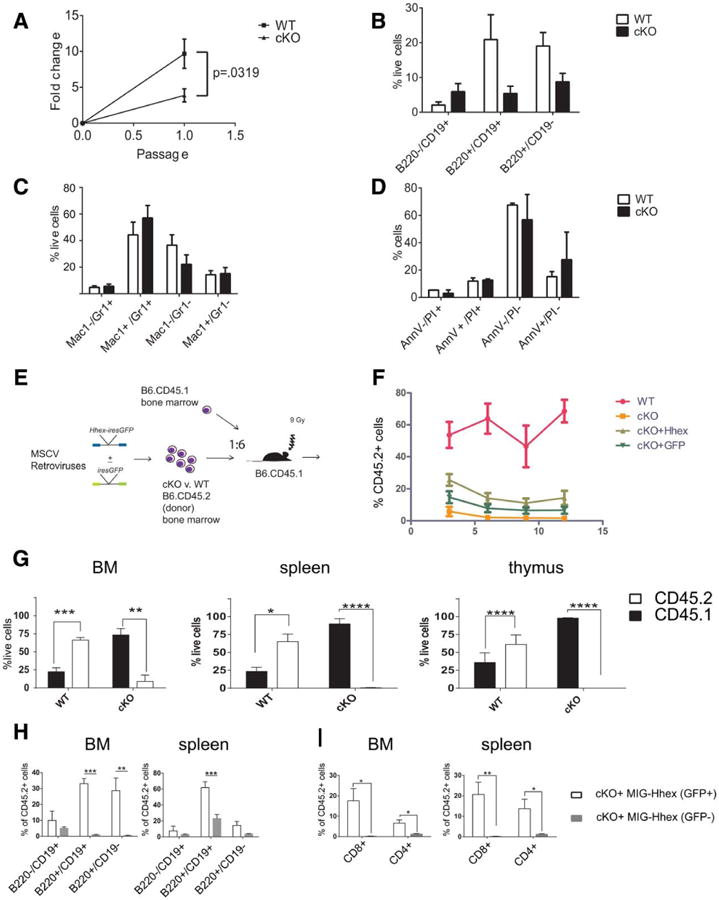
Hhex cKO stem and progenitor cells cannot differentiate into B cells in vitro and are severely compromised in competitive BM transplantation (BMT). (A): Graph shows fold change versus passage number for WT and Hhex cKO Lin−Sca-1+Kit+ cells plated on irradiated OP9 stromal cells. (B): 7 days after the initiation of culture, the remaining cells were stained for B220 and CD19. Bar graphs represent percentage of B-cell populations. Values are means ±SEM. (C): 7 days after the initiation of culture, the remaining cells were stained for CD11b (Mac-1) and Gr-1. Bar graphs represent myeloid populations. Values are means ± SEM. (D): Cell death was measured by flow cytometry 7 days after the initiation of culture, using Annexin V and PI. Bar graphs represent percentage of populations. Values are means ± SEM. The graphs represent a summary of three independent experiments. (E): Schematic of competitive BMT experiments; lethally irradiated recipient mice (CD45.1) were transplanted with whole BM from WT or Hhex cKO mice; sixfold more WT or Hhex cKO (CD45.2) was injected than competitor, CD45.1+. For (D and E), BM was transduced with either MIG-Hhex or MIG-empty retrovirus. Recipient mice were analyzed 16 weeks after injection. (F): Donor chimerism as the percentage of peripheral blood (y-axis) of recipient mice that was CD45.2+ at the time points indicated in weeks on the x-axis. (G): 16 weeks post-transplant, BMs, spleens, and thymi were harvested from BMT mice and analyzed for CD45.1 and CD45.2. (H): Proportion of CD45.2+ B cells in the BM and spleen of host mice 16 weeks after MIG-Hhex transduction and BMT are shown, gating on GFP+ and GFP− cells. (I): Proportion of donor T cells in the bone marrow, spleen, and thymus of host mice 16 weeks after MIG-Hhex transduction and BMT. Values are means ± SEM, n = 7 per group. Two-tailed Student's t tests generated p values are shown: *, p ≤ .05; **, p ≤ .01; ***, p ≤ .001; ****, p ≤ .0001. Abbreviations: BM, bone marrow; cKO, conditional knockout; Hhex, hematopoietically expressed homeobox; WT, wild type.
Next, we repeated the Hhex cKO BM transplantation after transduction with empty retrovirus (MIG) or MIG-Hhex to test whether we could rescue the defect in reconstitution (Fig. 3E). MIG-Hhex-transduced BM showed the highest levels of donor chimerism but did not restore it to the same extent as untransduced WT BM (Fig. 3F). Since both transduced and untransduced BM cells were transplanted into lethally irradiated mice, the MIG-Hhex-transduced GFP+ and untransduced GFP− were compared for their contribution to mature cell lineages. The GFP+ graft (i.e., expressing Hhex mRNA) contributed significantly to the B and T cells of the BM and spleen compared to the GFP− graft (Fig. 3H–3I). Thus, MIG-Hhex was able to partially rescue the T and B cell defects observed after Hhex cKO BM transplantation. To further support the observed T-cell defect, we cocultured double negative (CD4−CD8−) thymocytes derived from Hhex cKO on OP9-DL1 cells. These progenitor cells did not proliferate like WT thymocytes (Supporting Information Fig. S11A) but the proportion of differentiation was comparable to WT (Supporting Information Fig. S11B). The proliferation defect was rescued by MIG-Hhex (Supporting Information Fig. S11A). Interestingly, MIG-Hhex transduced into thymocytes blocked differentiation at DN stages and conferred a growth advantage in WT thymocytes consistent with prior reports (Supporting Information Fig. S11C, S11D) [16]. Although Hhex cKO BM cells competed poorly with CD45.1+ cells, the proportion of mature myeloid progenitor cells, Gr-1+Mac-1+, was the same in GFP+ and GFP− grafts; Mac-1+ cells were increased in cKO BM and spleen and reduced with MIG-Hhex transduction (i.e., GFP+ grafts) (Supporting Information Fig. S8A). Notably, Hhex cKO donor grafts showed similar proportions of stem and progenitor populations (i.e., LK, LSK, LSKFlt3−, and LSKFlt3+ cells) to WT grafts, which were not altered by the transduction of MIG-Hhex (Supporting Information Fig. S8B). Thus, stem and progenitor populations were present post-transplant but were ineffective in reconstituting hematopoiesis.
Sublethal Irradiation Reveals a T-Cell Defect in Hhex cKO Mice
The BM transplantation assay was informative in showing the cell autonomous role of Hhex in hematopoiesis, but the stress hematopoiesis of transplantation revealed a profound myeloid and T-cell defect in Hhex cKO mice that was not apparent at steady state. As an alternative induction of stress hematopoiesis we applied sublethal doses of radiation (650 cGy) to WT and Hhex cKO mice and allowed them to repopulate from endogenous stem and progenitor populations (Fig. 4A). We observed that the BM of Hhex cKO mice repopulated similarly to WT mice at 3, 6, and 9 weeks after radiation (Fig. 4B). In contrast, the spleens and thymi of Hhex cKO showed reduced cellularity as late as 9 weeks after irradiation (Fig. 4B). We analyzed the stem and progenitor cells (Supporting Information Fig. S12) at these same time points and notably, the Hhex cKO mice had significantly increased LSK cells at week 3 compared to WT mice, and no significant difference at 6 or 9 weeks post-radiation (Fig. 4C; Supporting Information Fig. S10). Myeloid progenitor populations, Lin−Kit+ (LK), were significantly increased at 3 weeks and 6 weeks post-radiation (Fig. 4C). Hhex cKO mice showed a decrease in T-cell populations at 3, 6, and 9 weeks post-radiation (Fig. 4D). Hhex cKO thymi had reduced numbers of ETPs at 6 weeks post-radiation (Fig. 4E). The spleens of Hhex cKO mice had reduced numbers of mature T cells when compared to WT mice (Fig. 4F). In summary, Hhex cKO mice have a defect in T-cell differentiation under stress hematopoiesis in addition to the B-cell defect at steady state but no problems with myeloid repopulation.
Figure 4.
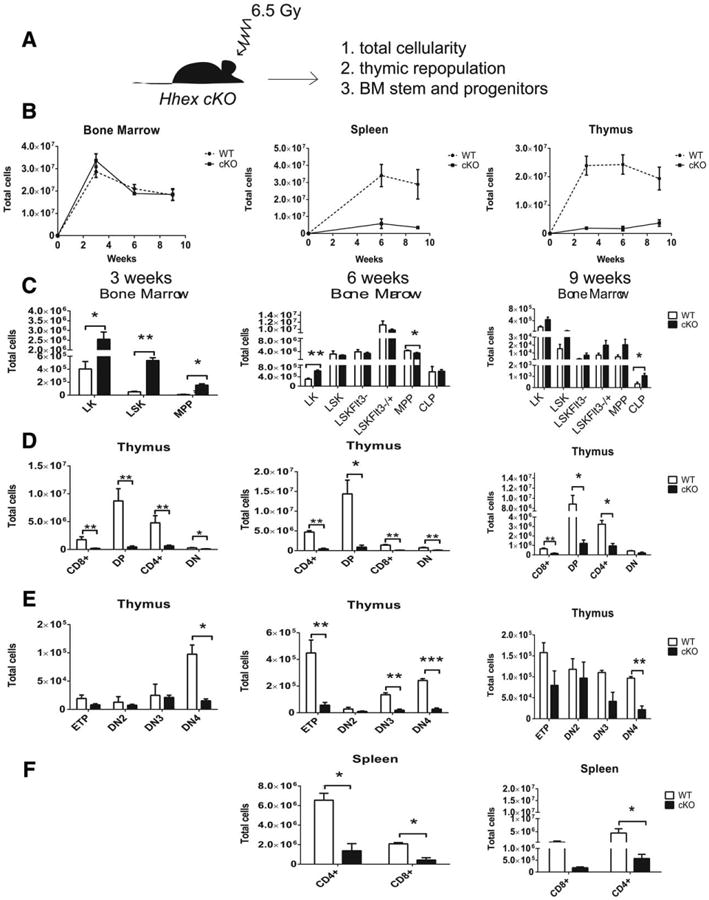
Sublethal irradiation reveals an additional T-cell repopulation defect in Hhex cKO mice. (A): Schematic shows the experiment outline; WT and Hhex cKO mice were irradiated with 6.5 Gy and BMs, spleens, and thymi were analyzed at 3, 6, and 9 weeks. (B): Graph shows total cellularity versus time post-irradiation in weeks for BM, spleen, and thymus of WT and Hhex cKO mice. (C): Bar graphs show the mean numbers of stem and progenitor populations in the BM at 3 weeks (column 1), 6 weeks (column 2), and 9 weeks (column 3). WT in white bars, Hhex cKO in black bars. Total cells were calculated based on the total number of cells multiplied by the percentage of each population. (D, E): Bar graphs represent the sizes of each T cell population in the thymus. (F): Bar graphs show the mean ± SEM of mature T cell populations in the spleen; WT; n = 3, cKO; n = 5. Two-tailed Student's t test was done to generate p values: *, p ≤ .05; **, p ≤ .01; ***, p ≤ .001; ****, p ≤ .0001. Abbreviations: BM, bone marrow; cKO, conditional knockout; CLP, common lymphoid progenitor; ETP, early T-cell precursor; LK, Lin−Kit+; LSK, Lin−Sca-1+Kit+; MPP, multipotent progenitor; WT, wild type.
Hhex cKO Mice Have Reduced Number of Hematopoietic Stem Cells
Next, we used established markers to quantify by FACS the stem and progenitor populations in Hhex cKO and WT mice that may account for the results shown above. The competitive repopulation assay suggested a defect in Hhex cKO hematopoietic stem cells (HSCs) whereas the post-radiation recovery experiments suggested a defect in lymphoid development or multipotent progenitors (MPP). The stem and progenitor populations are within the LSK compartment but the absolute numbers of these were no different between Hhex cKO and WT mice. Similarly, the Lin−Sca-1−c-Kit+ (LK) or myeloid progenitor cells were not significantly different between Hhex cKO and WT BMs (Supporting Information Fig. S12B). The HSC has been defined by LSKCD48−CD150+CD34−Flt3−. A more mature MPP that can rescue lethally irradiated mice without self-renewal potential is defined as LSKCD48− CD150+CD34+Flt3− (MPP1), whereas more mature MPP are defined by MPP2: LSKCD34+CD48+CD150+Flt3−; MPP3: LSKCD34+CD48+ CD150−Flt3−; MPP4: LSKCD34+CD48+CD1502 Flt3+ (Supporting Information Table S2) [36]. Direct comparisons of BMs with these markers showed a skewed distribution in Hhex cKO mice. Hhex cKO BM had increased proportions of HSCs, reduced proportions of MPP1, and proportions of MPP2– 4 that were similar to WT (Fig. 5B). The absolute number of HSC and MPP1–4 were not significantly different between WT and Hhex cKO; however, the absolute number of common lymphoid progenitor (CLP) cells was increased in Hhex cKO versus WT (Fig. 5B). Thus, proportions of HSC and MPP cells were skewed in Hhex cKO but the CLP was the only significantly increased progenitor subset, suggesting a differentiation arrest at this stage in Hhex cKO mice.
Transcriptional Profiling Reveals a Defect in Lymphoid Priming in Hhex cKO MPPs
Next, we considered whether Hhex's role in regulating lymphoid development may be reflected in gene expression. Thus, we sorted HSCs and MPPs based on LSKFlt3+/− and sequenced their cDNA libraries (RNA-seq) [37]. LSKFlt3− cells are immature HSC/MPP populations whereas LSKFlt3+ cells are MPP with commitment toward lymphoid lineages and limited self-renewal [38, 39]. We analyzed the expression of genes that varied by at least twofold in pairwise comparisons. LSKFlt3+ are biased toward lymphoid lineages and do not have megakaryocyte and erythrocyte progenitor (MEP) potential [37, 39]. Consistent with this finding, LSKFlt3+ sorted from WT and Hhex cKO mice downregulated mRNAs associated with MEP lineages such as Gata1, Epor, Gypa, and Klf1 (Fig. 6B) compared to LSKFlt3− cells. LSKFlt3+ MPPs upregulate a lymphoid transcriptional program prior to lineage commitment, a phenomena known as lymphoid priming [40]. Genes within this transcriptional program were markedly upregulated in the differentiation of WT LSKFlt3− to LSKFlt3+ cells such as Blk, Blnk, Rag1, Rag2, Cd79a, Dntt, Vpreb2, and Il7r among others (Fig. 6C). The upregulation of these mRNAs was markedly attenuated in Hhex cKO LSKFlt3− to LSKFlt3+ differentiation. We analyzed the mRNAs encoding Ebf1, Pax5, Ikzf1, and Tcf3, transcription factors required for B-cell commitment and B-cell progenitor cell differentiation. The loss of function of any one of these transcription factors results in differentiation arrests and loss of mature B cells [41, 42]. Hhex cKO MPPs had detectable transcripts for all four of these factors although Ebf1 mRNA was not upregulated in Hhex cKO LSKFlt3+ differentiation to the same extent as WT (Fig. 6C). Transcriptional regulators concordantly expressed with Hhex in hematopoiesis (Fig. 1A) were not differentially expressed in the HSCs but in Hhex cKO LSKFlt3+, Lyl1 expression was increased (cKO vs. WT, log2Δ= 1.38) and Mef2c expression was decreased (log2Δ = −1.16) compared to WT.
Figure 6.
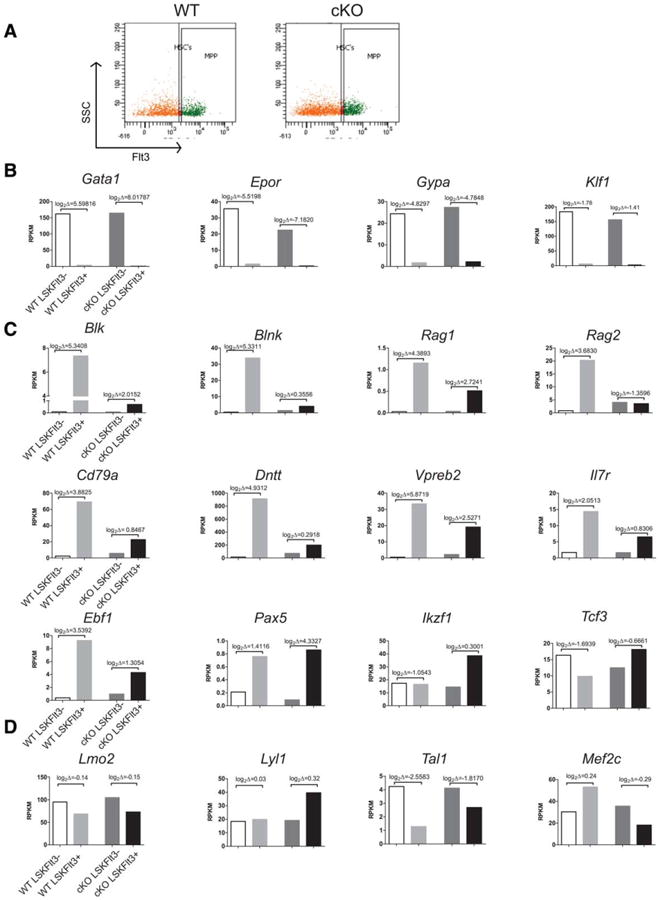
Hematopoietically expressed homeobox (Hhex) is required for lymphoid priming in MPP cells. (A): Representative flow cytometric analysis of the HSCs and MPPs sorted based on Flt3 expression from the bone marrow of WT and Hhex cKO mice. (B): RNA-seq analysis of sorted LSKFlt3− and LSKFlt3+ cells from WT and Hhex cKO bone marrows. Genes in (B) are specific to the megakaryocytic-erythroid lineage; genes in (C) are lymphoid-specific transcripts that prime MPPs for lymphoid differentiation; and, genes in (D) have a normal expression pattern that is closely correlated with Hhex (Fig. 1A). y-axes values are in RPKM. Brackets show log2 changes between LSKFlt3− and LSKFlt3+ differentiation. Abbreviations: cKO, conditional knockout; HSC, hemtopoietic stem cell; LSK, Lin−Sca-1+Kit+; Lmo2, LIM domain only-2; MPP, multipotent progenitor; RPKM, reads per kilobase of gene per million reads total; SSC, side-scattered light; WT, wild type.
Hhex cKO Stem and Progenitor Cells Have Enhanced Proliferative Activity Compared to WT Cells
To analyze cell proliferation, we analyzed in vivo BrdU labeling of the stem and progenitor cell populations 2 hours after intraperitoneal injection [28, 29]. Hhex cKO LSK cells showed higher BrdU labeling than their WT counterparts (Fig. 7A). The WT LSKFlt3−, LSKFlt3−SLAM, LSKFlt3int, and LSKFlt3+ cells showed a mean of 10% BrdU labeling which increased to 15% in CLP cells (Supporting Information Fig. S12A for gating). The same populations in Hhex cKO had double the level of BrdU labeling. Hhex cKO and WT CLP showed comparable levels of BrdU labeling (Fig. 7B). We analyzed the RNA-seq data to see if there were differences in the mRNA abundance of cell cycle regulators between Hhex cKO and WT. A prior report on HHEX in U937 AML cells showed that HHEX regulated the EIF4E-directed transport and stability of CCND1 mRNA such that loss of HHEX is predicted to increase CCND1 translation [23]. Ccnd1 mRNA was rare (compared to Ccnd2/3) and decreased in Hhex cKO HSC and MPP compared to WT mice. A decrease in Ccnd1 mRNA was also observed in Hhex−/− ES cell-derived definitive hematopoietic colonies [43]. Other transcripts for cell cycle regulators implicated in stem and progenitor cell proliferation and quiescence were comparably expressed between WT and Hhex cKO mice except for Cdkn1a and Rb1, which were reduced in Hhex cKO HSCs and MPPs (Fig. 7C) [44].
Figure 7.
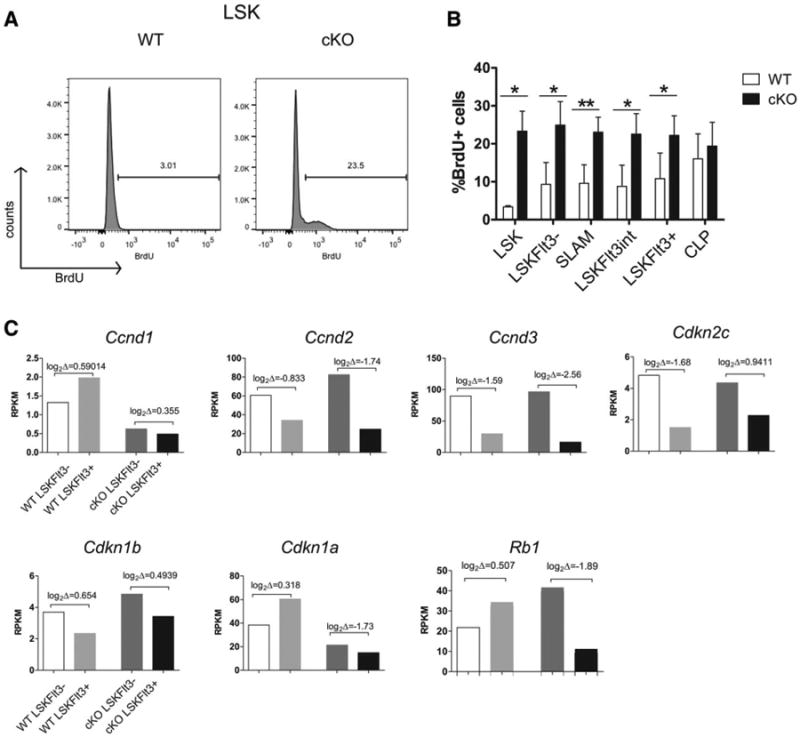
Hhex cKO stem and progenitor populations have increased proliferative activity than WT cells. (A): Representative flow cytometric analysis of BrdU labeling of LSK cells in the bone marrow of WT and Hhex cKO mice. (B): Bar graphs representing the proportion of BrdU in each stem and progenitor populations: values are means ± SEM for WT, n = 3; cKO, n = 5. Two-tailed Student's t test was done to generate p values: *, p ≤ .05; **, p ≤ .01; ***, p ≤ .001; ****, p ≤ .0001. (C): Bar graphs show RPKM values for RNA-seq of hematopoietic stem cell (HSC) and multipotent progenitor (MPP) cells sorted from WT or Hhex cKO mice for genes encoding cell cycle regulators. Brackets show log2 fold changes with HSC to MPP differentiation. Abbreviations: BrdU, bromodeoxyuridine; cKO, conditional knockout; CLP, LSK, Lin−Sca-1+Kit+; RPKM, reads per kilobase of gene per million reads total; WT, wild type.
Based on ingenuity pathway analysis on the RNA-seq of WT and Hhex cKO LSKFlt3− and LSKFlt3+, the top upstream pathway implicated in Hhex cKO in both LSKFlt3− and LSKFlt3+ was Tgfb1; however, 23/56 target genes were regulated in a manner suggesting Tgfb1 pathway activation and 27/56 suggested inhibition (Supporting Information Table S3). In contrast, four upstream regulators were significantly enriched in Hhex cKO LSKFlt3− cells and their downstream targets consistently regulated. As shown in Supporting Information Table S4, Hoxa7 (p = 2.04 × 10−8), Hoxa9 (p = 3.08 × 10−6), E. coli LPS (p = 5.35 × 10−8), Runx1 (p = 1.01 × 10−6), and Ifnb1 (p = 4.56 × 10−6) had a total of 56 target genes, which were regulated in a pattern consistent with activation of these transcriptional pathways. Hoxa7 (cKO vs. WT, log2Δ = 0.958) and Hoxa9 (cKO vs. WT, log2Δ = 0.808) mRNAs were increased in abundance in LSKFlt3− compared to WT.
Discussion
In this article, we present studies characterizing adult hematopoiesis in Hhex cKO mice induced by the vav-Cre transgene. Our mice were clinically normal at birth in SPF conditions but exhibited three remarkable phenotypes. At steady state, the mice had reduced cellularity of the BM and spleen due to a severe defect in B-cell development, specifically in the formation of pro-B cells. BM transplantation revealed a more severe phenotype, Hhex cKO BM competed poorly with normal BM in reconstituting hematopoiesis in lethally irradiated host mice. In the setting of sublethal irradiation, Hhex cKO showed normal myeloid repopulation of BM from endogenous stem and progenitor cells but severely compromised T-cell reconstitution. The HSC and MPP1 populations were skewed in their distribution but other progenitor cell types were similar to WT levels; and, stem and progenitor populations had increased proliferative activity in Hhex cKO compared to WT mice.
The profound loss of B-cell progenitors and mature B cells at steady state implicates Hhex in the development of this lineage at the point of early commitment from MPP (or CLP) to pro-B cells. However, the BM transplantation and sublethal irradiation experiments show that Hhex has a role in T-cell differentiation as well. Transcriptionally, Hhex cKO LSKFlt3+ or lymphoid-primed MPP appropriately downregulated megakaryocytic and erythroid transcripts but failed to upregulate lymphoid transcripts compared to WT, underscoring a problem with lymphoid specification [37]. Hhex cKO LSKFlt3+ showed high Cd2 (cKO vs. WT: log2Δ = 7.7) gene expression suggesting that there may be some progenitor cells that are T-cell biased compared to WT, which may account for the thymic progenitors being intact at steady state in Hhex cKO mice compared to B-cell progenitors. Most interestingly, mRNAs for known transcription factors (e.g., Sfpi1, Lyl1, Mef2c, Tcf3, Ikzf1) required in lymphoid specification were not differentially expressed to an extent suggestive of complete loss of function [45–48]. Thus, Hhex's role in lymphoid development does not appear related to the mRNA abundance of known regulators. Still, these quantitative changes in mRNA abundance may be sufficient to induce the phenotypes observed. It is an open question whether Hhex transcriptionally regulates key factors or interacts by protein binding or redistributing cofactors. Currently, commercial antibodies against Hhex are inadequate for chromatin or coimmunoprecipitations but work addressing these questions is ongoing. Hhex cKO LSKFlt3− cells showed activation of transcriptional targets Hoxa7, Hoxa9, and Runx1, and upregulation of Cebpa (cKO vs. WT: log2Δ = 2.2), a transcriptional pattern consistent with myeloid lineage bias [49–51], however numbers of mature and immature myeloid cells were the same in Hhex cKO and WT mice at steady state (and with ageing) and, Hhex cKO LSK cells did not show increased myeloid cells at the expense of B-cell lineage commitment in OP9 cocultures (Fig. 3C). The data do suggest a more subtle myeloid bias. For example, the sublethally irradiated Hhex cKO BM overcompensated with increased numbers of myeloid progenitors at 3–6 weeks post-radiation (Fig. 4C). Our findings support an MPP population that is heterogeneous that requires Hhex for proper lineage allocation especially in states of hematopoietic stress.
The increased proliferation of Hhex cKO stem and progenitor populations was impressive and could explain some of the observed phenotypes. Enhanced cycling or loss of quiescence can exhaust LT-HSC numbers, as seen in Cdkn1a KO mice, and stem and progenitor cells in G1 phase of the cell cycle are unable to engraft compared to those in G0 (i.e., quiescent) [52, 53]. Our transcriptional analysis showed that Cdkn1a (or Rb1) mRNA abundance was decreased in Hhex cKO LSKFlt3− cells, which could be a direct or indirect effect of Hhex loss. There is precedent for homeodomain proteins directly regulating cell cycle genes and at least in one case (Hoxa10) activating Cdkn1a expression [28, 54, 55]. Despite the diminished function of HSCs, we did not observe overt BM failure in aged Hhex cKO mice; however, aged Hhex cKO BM did not show a decrement in cellularity seen in WT BM, which may be due to enhanced progenitor cell activity compensating for HSC loss. Also, despite the marked increase in cycling of Hhex cKO stem and progenitor populations, the absolute numbers were equivalent except for the CLP population that was increased perhaps signifying the point of lymphoid differentiation block. Interestingly, a recent study using in situ lineage tracing based on transposon tags suggests that steady state hematopoiesis is maintained by multipotent progenitors rather than HSCs [56]. Thus, the increased cycling of progenitor cells may explain why Hhex cKO have essentially normal T-cell and myeloid compartments at steady state. In contrast, Sun et al. report that hematopoiesis does emanate from HSCs after transplantation. Therefore, the assays we used in this study were probably assessing two different stem and progenitor cell populations. Importantly, Hhex's effects on proliferation are highly dependent upon the cell context [8, 23]. The loss of Hhex caused increased cycling in stem and progenitor populations but T- and B-cell progenitors from Hhex cKO had reduced proliferation in in vitro assays (Fig. 3A; Supporting Information Fig. S11). Enforced Hhex expression conferred proliferative advantage in T-cell progenitor cells consistent with its role as an oncogene in this lineage immediately downstream of Lmo2 [16, 20–22].
In the course of revising this manuscript, a study appeared in press describing hematopoiesis in an independently engineered Hhex cKO mouse [57]. Jackson et al. used Mx1-Cre to induce an acute deletion of Hhex by poly-dI-dC. Their analyses after deletion corroborate most of our findings that Hhex is required for B lymphopoiesis whereas most other compartments can exist in its absence. Mature B-cell populations, such as marginal zone, follicular, and plasma cells, were unaffected or subtly perturbed after poly-dI-dC induced deletion compared to the severe reduction in numbers observed in our vav-Cre model (Fig. 2E). Similarly, sorted LSK cells from poly-dI-dC treated floxed Hhex mice reconstituted myeloid compartments but not T or B cells. In contrast, vav-Cre-deleted Hhex contributed poorly to the BM and not at all to T- and B-lymphopoiesis in our competitive repopulation assays (Fig. 3G). These results imply functional impairment in HSC or MPP cells due to Hhex loss but with differing levels of severity in the two mouse models. These dissimilar results may be due to different Cre drivers, incomplete Cre-mediated deletion in the Mx1-Cre model, or, the presence of a residual spliced mRNA in Jackson et al.'s floxed allele that may confer some residual function. Vav-Cre deletes in utero at E12.5 and steady state hematopoiesis in Hhex cKO may be supported by other factors and may be uniquely susceptible to the effects of transplantation stress [26]. In fact, network analysis showed upregulation of the popolysaccharide (LPS) and the interferon β pathways in Hhex cKO LT-HSCs, which could be drivers of proliferation and differentiation [58, 59]. The pathway analysis suggests constitutive stress within Hhex cKO stem and progenitor cells making them intolerant of additional stress signals (i.e., transplant or irradiation). Finally, there are clinical scenarios that are modeled by Hhex cKO mice such as the inadequate recovery of T- and B-cell immunity after transplantation or cytotoxic chemotherapy or antiretroviral therapy in AIDS patients. It will be interesting to see if variants within the human HHEX gene or its expression play a role in T-cell and B-cell repopulation in these clinical settings or in explaining other immunodeficiency syndromes.
Conclusion
In this study, we analyzed a conditional knockout of Hhex induced by vav-Cre. At steady state, we observed a profound defect in B-cell development. Hematopoietic stress revealed a requirement for Hhex in early T-cell development and in the maintenance of normal stem and progenitor cell populations.
Supplementary Material
Acknowledgments
We thank Drs. Stephen Brandt, Scott Hiebert, Sandy Zinkel, Neal Copeland, Nancy Jenkins, and Mark Koury for helpful discussions; Dr. Travis Clark for technical assistance with RNA-seq. This work was supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development (BX001799-01A1), the American Society of Hematology, and the Vanderbilt Ingram Cancer Center (P30 CA68485) (U.P.D.). C.G. was supported by the Initiative for Maximizing Student Development (R25GM062459), the Microenvironmental Influences in Cancer training grant (T32CA009592), and an F31 award (F31HL117624). Flow Cytometry experiments were performed in the VMC Flow Cytometry Shared Resource. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Van-derbilt Digestive Disease Research Center (DK058404). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Author contributions: C.G.: conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing; E.S., E.M., N.E., S.C., R.T., J.L., X.C., Y.G., Y.S., and R.H.: collection and/or assembly of data; Y.D.: provision of study material and data interpretation; U.P.D.: conception and design, financial support, data analyses and interpretation, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflict of Interest: The authors indicate no potential conflicts of interest to disclose.
See www.StemCells.com for supporting information available online.
References
- 1.Keng VW, Yagi H, Ikawa M, et al. Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem Biophys Res Commun. 2000;276:1155–1161. doi: 10.1006/bbrc.2000.3548. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y, Chan R, Ramsey H, et al. The homeoprotein Hex is required for hemangioblast differentiation. Blood. 2003;102:2428–2435. doi: 10.1182/blood-2003-02-0634. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Barbera JP, Rodriguez TA, Beddington RS. The homeobox gene Hesx1 is required in the anterior neural ectoderm for normal forebrain formation. Dev Biol. 2000;223:422–430. doi: 10.1006/dbio.2000.9757. [DOI] [PubMed] [Google Scholar]
- 4.Hallaq H, Pinter E, Enciso J, et al. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development. 2004;131:5197–5209. doi: 10.1242/dev.01393. [DOI] [PubMed] [Google Scholar]
- 5.Brickman JM, Jones CM, Clements M, et al. Hex is a transcriptional repressor that contributes to anterior identity and suppresses Spemann organiser function. Development. 2000;127:2303–2315. doi: 10.1242/dev.127.11.2303. [DOI] [PubMed] [Google Scholar]
- 6.Bedford FK, Ashworth A, Enver T, et al. HEX: A novel homeobox gene expressed during haematopoiesis and conserved between mouse and human. Nucleic Acids Res. 1993;21:1245–1249. doi: 10.1093/nar/21.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manfioletti G, Gattei V, Buratti E, et al. Differential expression of a novel proline-rich homeobox gene (Prh) in human hematolymphopoietic cells. Blood. 1995;85:1237–1245. [PubMed] [Google Scholar]
- 8.Kubo A, Chen V, Kennedy M, et al. The homeobox gene HEX regulates proliferation and differentiation of hemangioblasts and endothelial cells during ES cell differentiation. Blood. 2005;105:4590–4597. doi: 10.1182/blood-2004-10-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soufi A, Jayaraman PS. PRH/Hex: An oligomeric transcription factor and multifunctional regulator of cell fate. Biochem J. 2008;412:399–413. doi: 10.1042/BJ20080035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swingler TE, Bess KL, Yao J, et al. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J Biol Chem. 2004;279:34938–34947. doi: 10.1074/jbc.M404488200. [DOI] [PubMed] [Google Scholar]
- 11.Denson LA, Karpen SJ, Bogue CW, et al. Divergent homeobox gene hex regulates promoter of the Na(+)-dependent bile acid cotransporter. Am J Physiol Gastrointestinal Liver Physiol. 2000;279:G347–G355. doi: 10.1152/ajpgi.2000.279.2.G347. [DOI] [PubMed] [Google Scholar]
- 12.Kasamatsu S, Sato A, Yamamoto T, et al. Identification of the transactivating region of the homeodomain protein, hex. J Biochem. 2004;135:217–223. doi: 10.1093/jb/mvh025. [DOI] [PubMed] [Google Scholar]
- 13.Guiral M, Bess K, Goodwin G, et al. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J Biol Chem. 2001;276:2961–2970. doi: 10.1074/jbc.M004948200. [DOI] [PubMed] [Google Scholar]
- 14.Soufi A, Smith C, Clarke AR, et al. Oligomerisation of the developmental regulator proline rich homeodomain (PRH/Hex) is mediated by a novel proline-rich dimerisation domain. J Mol Biol. 2006;358:943–962. doi: 10.1016/j.jmb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka T, Inazu T, Yamada K, et al. cDNA cloning and expression of rat homeobox gene, Hex, and functional characterization of the protein. Biochem J. 1999;339(Pt 1):111–117. [PMC free article] [PubMed] [Google Scholar]
- 16.George A, Morse HC, 3rd, Justice MJ. The homeobox gene Hex induces T-cell-derived lymphomas when overexpressed in hematopoietic precursor cells. Oncogene. 2003;22:6764–6773. doi: 10.1038/sj.onc.1206822. [DOI] [PubMed] [Google Scholar]
- 17.Hansen GM, Justice MJ. Activation of Hex and mEg5 by retroviral insertion may contribute to mouse B-cell leukemia. Oncogene. 1999;18:6531–6539. doi: 10.1038/sj.onc.1203023. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Shen H, Himmel KL, et al. Leukaemia disease genes: Large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 19.Mack DL, Leibowitz DS, Cooper S, et al. Down-regulation of the myeloid homeobox protein Hex is essential for normal T-cell development. Immunology. 2002;107:444–451. doi: 10.1046/j.1365-2567.2002.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith S, Tripathi R, Goodings C, et al. LIM Domain Only-2 (LMO2) induces T-cell leukemia by two distinct pathways. PLoS ONE. 2014;9:e85883. doi: 10.1371/journal.pone.0085883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cleveland SM, Smith S, Tripathi R, et al. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells. 2013;31:882–894. doi: 10.1002/stem.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormack MP, Young LF, Vasudevan S, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- 23.Topisirovic I, Culjkovic B, Cohen N, et al. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 2003;22:689–703. doi: 10.1093/emboj/cdg069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jankovic D, Gorello P, Liu T, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111:5672–5682. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
- 25.Bogue CW, Zhang PX, McGrath J, et al. Impaired B cell development and function in mice with a targeted disruption of the homeobox gene Hex. Proc Natl Acad Sci USA. 2003;100:556–561. doi: 10.1073/pnas.0236979100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Boer J, Williams A, Skavdis G, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 27.Georgiades P, Ogilvy S, Duval Hln, et al. VavCre transgenic mice: A tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- 28.Yan L, Womack B, Wotton D, et al. Tgif1 regulates quiescence and self-renewal of hematopoietic stem cells. Mol Cell Biol. 2013;33:4824–4833. doi: 10.1128/MCB.01076-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer Ma, Moreno-Miralles I, Hunt A, et al. Myeloid translocation gene 16 is required for maintenance of haematopoietic stem cell quiescence. EMBO J. 2012;31:1494–1505. doi: 10.1038/emboj.2011.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 31.Biswas S, Shi Q, Matise L, et al. A role for proapoptotic Bax and Bak in T-cell differentiation and transformation. Blood. 2010;116:5237–5246. doi: 10.1182/blood-2010-04-279687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodings C, Tripathi R, Cleveland SM, et al. Enforced expression of E47 has differential effects on Lmo2-induced T-cell leukemias. Leukemia Res. 2015;39:100–109. doi: 10.1016/j.leukres.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shay T, Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jojic V, Shay T, Sylvia K, et al. Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heng TS, Painter MW, Elpek K, et al. The Immunological Genome Project: Networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 36.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 37.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: A revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, Bryder D, Adolfsson J, et al. Identification of Lin−Sca1+ kit+ CD34+ Flt3– short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 40.Hu M, Krause D, Greaves M, et al. Multi-lineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 41.Lin YC, Jhunjhunwala S, Benner C, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nutt SL, Heavey B, Rolink AG, et al. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 43.Paz H, Lynch MR, Bogue CW, et al. The homeobox gene Hhex regulates the earliest stages of definitive hematopoiesis. Blood. 2010;116:1254–1262. doi: 10.1182/blood-2009-11-254383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi L, Lin Kuanyin K, Boles Nathan C, et al. Less is more: Unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stehling-Sun S, Dade J, Nutt S, et al. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat Immunol. 2009;10:289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- 46.Zohren F, Souroullas GP, Luo M, et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–769. doi: 10.1038/ni.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 48.Rothenberg EV. Transcriptional control of early T and B cell developmental choices. Ann Rev Immunol. 2014;32:283–321. doi: 10.1146/annurev-immunol-032712-100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alharbi RA, Pettengell R, Pandha HS, et al. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2012;27:1000–1008. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 50.Hsu CL, King-Fleischman AG, Lai AY, et al. Antagonistic effect of CCAAT enhancer-binding protein-alpha and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc Natl Acad Sci USA. 2006;103:672–677. doi: 10.1073/pnas.0510304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 52.Passegue E, Wagers AJ, Giuriato S, et al. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 54.Bromleigh VC, Freedman LP. p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 2000;14:2581–2586. doi: 10.1101/gad.817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins C, Wang J, Miao H, et al. C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc Natl Acad Sci USA. 2014;111:9899–9904. doi: 10.1073/pnas.1402238111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson JT, Nasa C, Shi W, et al. A crucial role for the homeodomain transcription factor Hhex in lymphopoiesis. Blood. 2015;125:803–814. doi: 10.1182/blood-2014-06-579813. [DOI] [PubMed] [Google Scholar]
- 58.King KY, Goodell MA. Inflammatory modulation of HSCs: Viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11:685–692. doi: 10.1038/nri3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takizawa H, Regoes RR, Boddupalli CS, et al. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J Exp Med. 2011;208:273–284. doi: 10.1084/jem.20101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


