Abstract
Comprehensive genetic testing has the potential to become the standard of care for individuals with hearing loss. In this study, we investigated the genetic etiology of autosomal recessive nonsyndromic hearing loss (ARNSHL) in a Turkish cohort including individuals with cochlear implant, who had a pedigree suggestive of an autosomal recessive inheritance. A workflow including prescreening of GJB2 and a targeted next generation sequencing panel (Illumına TruSightTM Exome) covering 2761 genes that we briefly called as mendelian exome sequencing was used. This panel includes 102 deafness genes and a number of genes causing Mendelian disorders. Using this approach, we identified causative variants in 21 of 29 families. Three different GJB2 variants were present in seven families. Remaining 14 families had 15 different variants in other known NSHL genes (MYO7A, MYO15A, MARVELD2, TMIE, DFNB31, LOXHD1, GPSM2, TMC1, USH1G, CDH23). Of these variants, eight are novel. Mutation detection rate of our workflow is 72.4%, confirming the usefulness of targeted sequencing approach in NSHL.
Introduction
Hearing loss (HL) is the most frequent sensory deficit with an incidence of 0.1–0.2% within the newborn population. In developed countries, genetic causes are the most important etiological factors leading to HL. Hereditary hearing loss (HHL) can be syndromic (SHL) (25%) or non-syndromic (NSHL)(75%). While SHL is accompanied by other systemic manifestations, in the non-syndromic form (NSHL), there are no additional findings [1, 2]. Depending on the age of onset and severity of HL, HHL is classified as congenital, prelingual, and postlingual, and mild to profound, respectively.
The genetic transmission of NSHL is autosomal recessive in 75–85% of all cases (ARNSHL), and autosomal dominant in 15–25% of cases (ADNSHL). Small proportion of cases show X-linked or mitochondrial inheritance (1–2%)[3].
Considering the complexity of hearing, many genes play roles in hearing function. It is estimated that, about 1% of human genes (200 to 250 genes) are involved in HHL [4, 5]. To date, more than 80 genes, with more than 1000 mutations, and 140 loci have been identified to be associated with NSHL (http://hereditaryhearingloss.org/). With the exception of few mutations detected recurrently in several genes, such as GJB2 (MIM 121011) and SLC26A4 (MIM 605646), most deafness mutations are extremely rare and only seen in either a single or a very few families [6]. Additionally, more than 400 syndromes have been described in the OMIM (Online Mendelian Inheritance in Man®) as relating to HL. In some cases, differential diagnosis of SHL and NSHL is very difficult, or even not possible. Furthermore, it has been shown that, both SHL and NSHL can be caused by the mutations in the same gene. This overwhelming genetic and clinical heterogeneity makes the identification of genetic etiology challenging, time-consuming and expensive.
Elucidation of the genetic basis of an individual’s deafness might assist in early diagnosis, developing prevention and/or treatment regimens as well as offering improved genetic counseling and future perspective to probands [3]. With the improvement of laboratory approaches in the genomics era, over the last decade, next generation technologies, such as whole genome sequencing (WGS), whole exome sequencing (WES) or targeted next generation sequencing (TNGS), have assumed revolutionary roles in both research or diagnostic areas.
There have been a number of TNGS panels which included known deafness genes. It is not possible to utilize this tool for the discovery of novel NSHL genes or the genes responsible for other rare diseases. Because of this, we used a targeted enrichment approach for genes known to cause a number of Mendelian disorders (Mendelian exome sequencing (MES)). This strategy consisted of 2761 genes including 63 known NSHL and 39 SHL genes.(S1 Table)
The goal of this study was to identify the genetic etiology of deafness in a group of individuals with NSHL using this MES.
Materials and Methods
Patients
Twenty-nine probands who received cochlear implant operation due to congenital or prelingual-onset severe to profound sensorineural HL were included in the study. At least a three-generation pedigree was drawn for each proband. All probands were either born to consanguineous parents or had affected siblings or cousins, suggesting autosomal recessive inheritance. Sensorineural hearing loss was diagnosed with standard audiometric evaluations that included pure tone audiometry, brainstem evoked response, and otoacoustic emissions. No patient exhibited syndromic features during clinical evaluations that included a thorough systemic examination, fundoscopy, EKG, and urinalysis. Written informed consents were provided by all participants. This study was approved by the Ege University Ethics Committee.
Molecular analysis of GJB2
All probands included in the study were screened for mutations in GJB2. Genomic DNA was extracted from peripheral leukocytes according to standard protocols. Sanger sequencing for the GJB2 gene was performed. Each detected variant was then analyzed for cosegregation with deafness in the parents and available family members. A touchdown protocol was used to amplify the targeted DNA region. The amplicons were cleaned up with Sephadex (GE Healthcare). ABI PRISM 3730 DNA analyzer (Applied Biosystems) and Big Dye Terminator Cycle Sequencing V3.1 Ready Reaction Kit (Life Technologies) were used to elucidate the DNA sequence. Variants were named according to NM_004004.5
Targeted Next Generation Sequencing
TruSightTM Exome focuses on a subset of the exome targeting genes with disease causing mutations which have been shown to be important in specific inherited conditions. This commercially available MES was used in the patients with no mutation in GJB2. The coding regions of 2761 genes including total of 63 NSHL genes and 39 SHL genes are covered by the TruSightTM Exome panel (S1 Table). In this gene list, 33 genes have been known to cause purely autosomal recessive nonsyndromic hearing loss (ARNSHL), 19 genes autosomal dominant nonsyndromic hearing loss (ADNSHL), 3 genes X-linked nonsyndromic hearing loss (XNSHL), and 8 genes have been described as being responsible for both ARNSHL and ADNSHL.
Illumina MiSeq platform was used to perform NGS. Sequencing data was analyzed using Illumina VariantStudio variant analysis software and IGV (Integrative Genomics Viewer)[7].
Data analysis
All genes in this panel were annotated. Firstly, 41 ARNSHL causing genes were evaluated. The homozygous or compound heterozygous variants in ARNSHL causing genes with a frequency of less than 0.5% in public databases (e.g. NCBI dbSNP build141 (http://www.ncbi.nlm.nih.gov/SNP/), 1000 Genomes Project (http://www.1000genomes.org/), Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/) and NHLBI Exome Sequencing Project (ESP) Exome Variant Server (http://evs.gs.washington.edu/EVS/)) were selected [8]. The impact of the mutations on the protein structure was identified using several in silico prediction tools such as MutationTaster [9], Polyphen-2 [10], and SIFT [11]. Conservation of residues across species was evaluated by PhyloP algorithm [12] and GERP [13]. Variants identified were categorized according to the ACMG recommendations [14].
Secondly, individuals who had no causative variation in ARNSHL genes were evaluated for known ADNSHL, XNSHL and SHL genes in the MES panel used in this study.
In the third step, patients who had no mutation in the genes related to hearing loss were analysed for all other genes included in the MES panel.
Confirmation and Segregation analysis
The most likely disease-causing variants, identified by data analysis, were confirmed using direct Sanger sequencing on ABI PRISM 3730 DNA analyzer (Applied Biosystems) and Big Dye Terminator Cycle Sequencing V3.1 Ready Reaction Kit (Life Technologies) and segregation analysis was then performed.
Results
GJB2 mutations
With the screening of GJB2, homozygous or compound heterozygous causative mutations were found in 7 probands(24.13%) and all had already been previously reported. Mutation spectrum of GJB2 is shown in Table 1.
Table 1. Causative variants identified in probands.
| Family ID | Consanguinity | Gene | Genotype |
|---|---|---|---|
| Cx1 | No | GJB2 | c.[35delG];[35delG] |
| Cx2 | Yes | GJB2 | c.[299_300delAT];[299_300delAT] |
| Cx3 | Yes | GJB2 | c.[35delG];[35delG] |
| Cx4 | No | GJB2 | c.[35delG];[333_334delAA] |
| Cx5 | No | GJB2 | c.[35delG];[35delG] |
| Cx6 | Yes | GJB2 | c.[35delG;[35delG] |
| Cx7 | Yes | GJB2 | c.[35delG];[35delG] |
| T21 | Yes | MYO7A | c.[487G>A]; [487G>A] |
| T16 | Yes | MYO7A | c.[735G>A];[735G>A] |
| T2 | Yes | MYO7A | c.[6337G>C];[6337G>C] |
| T10 | Yes | MYO7A | c.[6487G>A];[6487G>A] |
| T14 | No | TMC1 | c.[534A>C]; [534A>C] |
| T15 | Yes | TMC1 | c.[2050G>A]; [2050G>A] |
| T3 | No | MYO15A | c.[4642G>A]; [5212-2A>G] |
| T4 | Yes | MARVELD2 | c.[1331+2T>C];[1331+2T>C] |
| T6 | Yes | TMIE | c.[250C>T]; [250C>T] |
| T7 | Yes | DFNB31 | c.[302C>T]; [302C>T] |
| T11 | Yes | LOXHD1 | c.[71delT]; [71delT] |
| T12 | Yes | GPSM2 | c.[832C>T]; [832C>T] |
| T17 | Yes | USH1G | c.[355T>C]; [355T>C] |
| T19 | Yes | CDH23 | c.[5545C>G]; [5545C>G] |
TNGS data
On average, 22 probands had 98.70%, 93.33% and 87.17% of mappable bases represented by a coverage of at least 1X, 10X and 20X reads, respectively. Considering only the 41 ARNSHL genes included in TruSightTM Exome the percentage of mappable bases representing by a coverage of at least 1X, 10X and 20X reads were 99.80%, 99.60% and 98.90%, respectively. An average depth of 140.89 reads was achieved. The average numbers of single nucleotide variations (SNVs) and insertions or deletions (INDELs) were 3497 and 64.54, respectively. 93.99% of SNVs and 60.01% of INDELs were found in dbSNP.
Mutations in known deafness genes
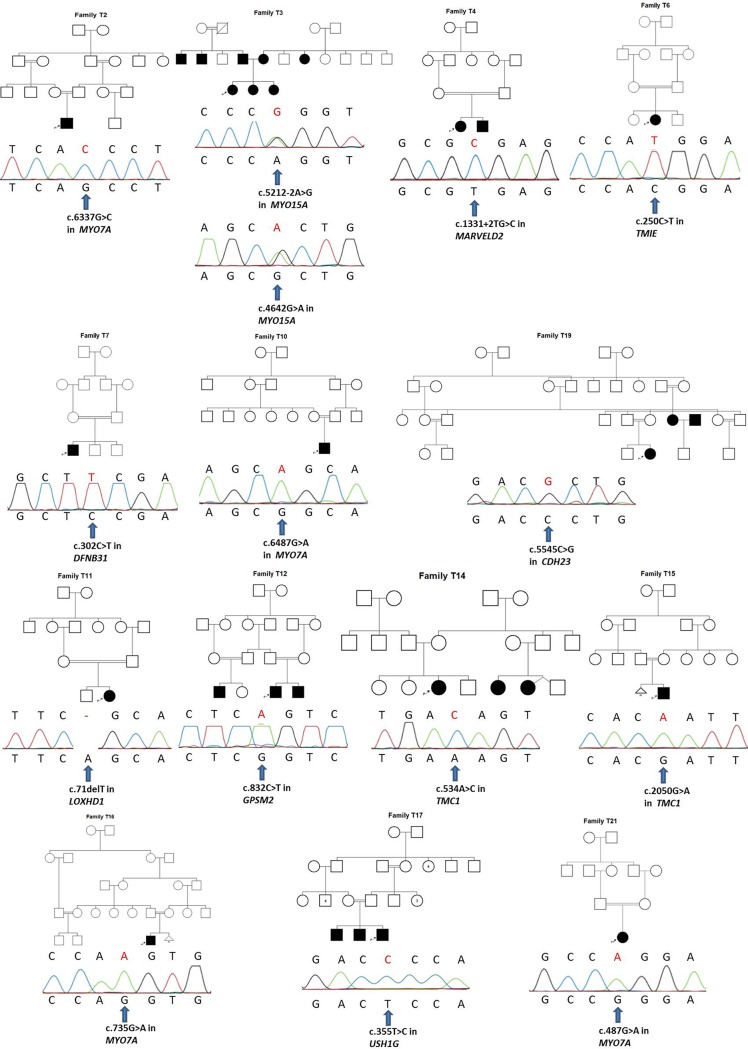
In this study, using TNGS, molecular diagnosis was established in 14 out of 22 probands who had no mutation in their GJB2 gene (63.63%). Tables 1, 2 and Fig 1 shows all mutations found in 10 different known ARNSHL genes and their in silico prediction analysis. Except two cases, all cases were born to consangiouneous parents.
Table 2. Characteristics of the identified variants in the study.
| Gene | Transcript ID | cDNA | Protein | Mutation Type | MT | Polyphen2 score/ranking | SIFT | ExAC* , # (Overall Allele frequency) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| MYO7A | NM_000260 | c.487G>A | p.G163R | M | DC | 1.000/PD | D | - | Known 24 |
| MYO7A | NM_000260 | c.735G>A | - | S | DC | NA | NA | - | Novel |
| MYO7A | NM_000260 | c.6337G>C | p.A2113P | M | DC | 0.980/PD | D | - | Novel |
| MYO7A | NM_000260 | c.6487G>A | p.G2163S | M | DC | 1.000/PD | D | 0.00003312 | Known 23 |
| TMC1 | NM_138691 | c.534A>C | p.E178D | M | DC | 0.992/PD | T | - | Novel |
| TMC1 | NM_138691 | c.2050G>A | p.D684N | M | DC | 0.944/PsD | T | 0.00001647 | rs563322370 |
| MYO15A | NM_016239.3 | c.4642G>A | p.A1548T | M | DC | 1.000/PD | D | 0.00003318 | rs201067821 |
| MYO15A | NM_016239.3 | c.5212-2A>G | - | S | DC | NA | NA | - | rs200760936 |
| MARVELD2 | NM_001038603.2 | c.1331+2T>C | - | S | DC | NA | NA | 0.00004119 | Known 31 |
| TMIE | NM_147196 | c.250C>T | p.R84W | M | DC | 1.000/PD | D | 0.00001658 | Known 34 , rs28942097 |
| DFNB31 | NM_015404.3 | c.302C>T | p.S101F | M | DC | 1.000/PD | D | - | Novel |
| LOXHD1 | NM_144612 | c.71delT | p.L24RfsX74 | F | DC | NA | NA | 0.00005042 | Novel |
| GPSM2 | NM_013296 | c.832C>T | p.R278X | NS | DC | NA | NA | 0.00002473 | Novel |
| USH1G | NM_173477.2 | c.355T>C | p.S119P | M | DC | 0.994/PD | D | - | Novel |
| CDH23 | NM_022124.5 | c.5545C>G | p.P1849A | M | DC | 1.000/PD | D | - | Novel |
| GJB2 | NM_004004.5 | c.35delG | p.G12VfsX2 | F | DC | NA | NA | 0.00604 | rs80338939 |
| GJB2 | NM_004004.5 | c.299_300delAT | p.H100RfsX14 | F | DC | NA | NA | 0.00004124 | rs111033204 |
| GJB2 | NM_004004.5 | c.333_334delAA | p.K112EfsX2 | F | DC | NA | NA | - | Known 16 |
* Exome Aggregation Consortium (http://exac.broadinstitude.org)
# The allele frequency in the ExAC database does not contain representative controls for all ethnic groups.
M: Missense, S: Splice site, F: Frameshift, NS: Nonsense, MT: MutationTaster, DC: Disease causing, PD: Probably Damaging, PsD: Possibly Damaging, D: Damaging, T: Tolerated, NA: Not available
Fig 1. Pedigrees and Sanger sequencing electropherograms demonstrating mutations found in TNGS in the probands.
Discussion
In this study, a TNGS panel including known ARNSHL genes has been used to define the genetic etiology in a group of individuals with autosomal recessive NSHL, all recipients of cochlear implants.
In our study, GJB2 mutations with the direct link to NSHL were found in 7 of 29 probands (24.13%), in line with previous studies, we have confirmed GJB2 mutations as the principal cause of NSHL in our population. Mutations in GJB2 have been shown to cause up to 50% of cases with ARNSHL [15]. Although more than 200 different mutations have been identified in this gene, the c.35delG mutation is the most frequent pathogenic variant, possibly accounting for up to 70% of all GJB2 mutations [15, 16]. In this study, allelic frequency of c.35delG mutation was found to be 78.5% among the patients having GJB2 mutations (Table 1).
In this study, using the TNGS panel which included 63 known NSHL genes, causative mutations were detected in 63.6% of patients with NSHL (14/22)(Tables 1 and 2). When the patients, identified as having mutations in GJB2, were included in the study group, the mutation detection rate increased to 72.4% (21/29). Focusing on familial NSHL cases, Choi et al. (2013) designed a diagnostic pipeline including both prescreening of GJB2, SLC26A4, POU3F4 and mitochondrial DNA, and a targeted next generation sequencing panel covering 80 known NSHL genes. In their study included 32 subjects,the total detection rate of 78.1% (25/32) was established [17]. Shearer et al. (2013) used a panel including 89 known deafness genes in 100 probands with presumed genetic NSHL, 39 of these 100 subjects were classified as having autosomal recessive inheritance and molecular defects have been detected in 22 of them (56%) [18]. Vozzi et al. (2014), using a panel including 96 known NSHL genes, found the causative gene variants in four out of 12 families from Italy and Qatar confirming the usefulness of a targeted sequencing approach. In this study, all probands were completely negative for mutations in GJB2 and GJB6 genes, as well as for the A1555G mitochondrial mutation at the beginning of TNGS. [1]. More recently, Bademci et al. (2015) reported a different approach to detect deafness-causing variants in a multiethnic cohort with ARNSHL. After excluding mutations in GJB2, they performed WES in 160 multiplex families, and sequencing all known NSHL genes, they identified causative mutations in 56% of their cohort [19].
In our study, ExAC database has been used for allelic frequencies of the variants detected. ExAC database may not be ideal for our patients, because it does not contain representative controls from Turkish population. Further studies are needed to exhibit exact allelic frequencies of variants detected in our ethnic group.
MYO7A encodes myosin VIIA and has been reported to be associated with Usher syndrome type 1B (USH1B) (MIM 276900), ARNSHL (DFNB2), and ADNSHL (DFNA11) [20–22]. Among previously reported MYO7A mutations identified in our study, one (c.487G˃A) had been described in two patients having Usher syndrome by Roux et al. (2006)[23, 24].
TMC1 has been identified as the responsible gene for both DFNA36 and DFNB7/B11 deafness [25]. To date, more than 30 TMC1 mutations have been reported to be associated with ARNSHL in families from mostly middle-eastern countries [26]. In two studies conducted in 93 and 86 patients the frequencies of TMC1 mutations were found to be 4.3% [27] and 8.1% [28] respectively. In our study, 6.89% of patients were carried TMC1 mutations.
MYO15A, encoding the 3530-amino acid myosin XV protein, is one of the common causes of ARNSHL. The frequency of MYO15A mutations was found to be 3.3% in a study including 600 consanguineous Pakistani, Indian, and Turkish families [29]. In another study conducted in 140 Iranian deaf families who had no GJB2 mutations, this frequency was found to be 5.71% (8/140) [30]. In our study, the patient carried MYO15A mutations was the only compound heterozygous patient.
Mutations in MARVELD2, which encodes tricellulin, and located at the DFNB49 locus, cause ARNSHL [31]. Seven different pathogenic variants of human MARVELD2 have been identified in patients with moderate to profound hearing loss [31, 32]. Nayak et al. reported that MARVELD2 mutations were responsible for about 1.5% of NSHL in their cohort of 800 Pakistani families[32]. The splice junction mutation (c.1331+2T>C (IVS4ds+2T-C)) detected in our study has been recurrently reported in Pakistani population [31].
At least 9 mutations of TMIE, which encodes 155 amino acid, have been reported to be associated with NSHL [33]. The TMIE (c.250C˃T) mutation found in our study had been previously reported by Naz et. al. [34]. Sirmaci et. al. reported that this TMIE variant is a founder mutation in southeastern Anatolia region of Turkey. In this previous study of 258 subjects, the frequency of c.250C˃T among families with NSHL throughout Turkey was 3.1% (8/258); while its frequency in southeastern Anatolia was higher (12.2% (6/49))[35].
Mutations in LOXHD1, which is responsible for DFNB77, have been extremely rare; there have been only five reports, from six families [36]. In a NSHL patient born to consanguineous parents, we identified a novel homozygous c.71delT (p.Leu24ArgfsTer74) mutation.
One patient was found to carry a novel DFNB31 mutation in our study. Mutations in DFNB31 coding WHRN protein can cause both ARNSHL and Usher syndrome Type 2 (MIM 607928) [37, 38]. Although at least 19 mutations in DFNB31 have been reported in the literature, only 2 were presented as being responsible for ARNSHL [37, 39].
CDH23, encoding a 3354 amino acid protein, is responsible for both Usher syndrome 1D (USH1D) (MIM 605516) and ARNSHL (DFNB12)[40]. Individuals with USH1D usually have truncating mutations affecting the CDH23 protein, whereas those with DFNB12 usually carry missense mutations in any domain [40]. In our study, a novel homozygous missense mutation (c.5545C˃G) in CDH23 has been found in a patient with ARNSHL confirming previous findings.
Mutations in GPSM2 have been previously identified in people with ARNSHL. Although subsequent brain imaging of these neurologically asymptomatic individuals revealed structural brain abnormalities consistent with Chudley-McCullough syndrome (CMS) (MIM 604213), this gene is still accepted as a NSHL gene by http://hereditaryhearingloss.org/ [41, 42]. In our study, a novel homozygous truncating c.832C>T (p.R278X) mutation in this gene was found in a deaf patient. There were no neurological abnormalities other than hearing loss in this patient. Subsequent evaluations have been offered to the family, but they refused any radiological tests.
USH1G encodes the protein SANS and is responsible for Usher syndrome Type 1G (MIM 606943). The first cases of ARNSHL caused by mutations in USH1G have been presented recently by Maria Oonk et al. (2015), but this gene has not currently been accepted as an ARNSHL gene by http://hereditaryhearingloss.org/ [43]. In our study, a novel homozygous c.355T>C mutation in USH1G was found in a NSHL patient born to consanguineous parents. Segregation analysis showed that affected sibling and affected father born to consanguineous parents had this mutation homozygously, while the unaffected mother had the same mutation heterozygously (Family T17 in Fig 1). Neither probands nor other affected family members had abnormality in their ophthalmologic examinations. If we had chosen a custom designed TNGS panel including only known NSHL genes, we wouldn’t have found the USH1G mutation in this family. To the best of our knowledge, this family is the second family with NSHL to have a causative mutation in USH1G.
As a conclusion, by broadening the spectrum of the gene panels, detection rates will continuously improve making the molecular diagnosis of NSHL easier, cheaper and more available. Molecular genetic diagnosis in the patients with NSHL may also help to improve the development and management of individual treatment strategies such as early speech therapy and cochlear implantation.
Supporting Information
(XLSX)
Acknowledgments
This project was supported by Ege University Scientific Research Projects (EGEBAP) with grant number 2014-TIP057 and NIH NIDCD grant R01DC009645 to M.T. T.A. was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) (project no: 1059B191401904).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was supported by Ege University Scientific Research Projects (EGEBAP) with grant number 2014-TIP057 and NIH NIDCD grant R01DC009645 to MT. TA was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) (project no: 1059B191401904).
References
- 1. Vozzi D, Morgan A, Vuckovic D, D'Eustacchio A, Abdulhadi K, Rubinato E, et al. Hereditary hearing loss: a 96 gene targeted sequencing protocol reveals novel alleles in a series of Italian and Qatari patients. Gene. 2014;542(2):209–16. Epub 2014/03/25. 10.1016/j.gene.2014.03.033 . [DOI] [PubMed] [Google Scholar]
- 2. Kemperman MH, Hoefsloot LH, Cremers CW. Hearing loss and connexin 26. Journal of the Royal Society of Medicine. 2002;95(4):171–7. Epub 2002/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atik T, Bademci G, Diaz-Horta O, Blanton SH, Tekin M. Whole-exome sequencing and its impact in hereditary hearing loss. Genetics research. 2015;97:e4 Epub 2015/04/01. 10.1017/S001667231500004X . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59(1):5–15. Epub 2013/11/08. 10.1038/jhg.2013.114 . [DOI] [PubMed] [Google Scholar]
- 5. Finsterer J, Fellinger J. Nuclear and mitochondrial genes mutated in nonsyndromic impaired hearing. International journal of pediatric otorhinolaryngology. 2005;69(5):621–47. Epub 2005/04/27. 10.1016/j.ijporl.2004.12.002 . [DOI] [PubMed] [Google Scholar]
- 6. Diaz-Horta O, Duman D, Foster J 2nd, Sirmaci A, Gonzalez M, Mahdieh N, et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One. 2012;7(11):e50628 Epub 2012/12/12. 10.1371/journal.pone.0050628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology. 2011;29(1):24–6. Epub 2011/01/12. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shearer AE, Eppsteiner RW, Booth KT, Ephraim SS, Gurrola J 2nd, Simpson A, et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet. 2014;95(4):445–53. Epub 2014/09/30. 10.1016/j.ajhg.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7(8):575–6. Epub 2010/08/03. 10.1038/nmeth0810-575 . [DOI] [PubMed] [Google Scholar]
- 10. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–9. Epub 2010/04/01. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4(7):1073–81. Epub 2009/06/30. 10.1038/nprot.2009.86 . [DOI] [PubMed] [Google Scholar]
- 12. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome research. 2010;20(1):110–21. Epub 2009/10/28. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS computational biology. 2010;6(12):e1001025 Epub 2010/12/15. 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. Epub 2015/03/06. 10.1038/gim.2015.30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duman D, Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci (Landmark Ed). 2012;17:2213–36. Epub 2012/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kelley PM, Harris DJ, Comer BC, Askew JW, Fowler T, Smith SD, et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am J Hum Genet. 1998;62(4):792–9. Epub 1998/06/13. 10.1086/301807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi BY, Park G, Gim J, Kim AR, Kim BJ, Kim HS, et al. Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS One. 2013;8(8):e68692 Epub 2013/08/31. 10.1371/journal.pone.0068692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shearer AE, Black-Ziegelbein EA, Hildebrand MS, Eppsteiner RW, Ravi H, Joshi S, et al. Advancing genetic testing for deafness with genomic technology. J Med Genet. 2013;50(9):627–34. Epub 2013/06/28. 10.1136/jmedgenet-2013-101749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bademci G, Foster J 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med. 2015. Epub 2015/08/01. 10.1038/gim.2015.89 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, et al. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet. 1997;16(2):188–90. Epub 1997/06/01. 10.1038/ng0697-188 . [DOI] [PubMed] [Google Scholar]
- 21. Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, et al. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet. 1997;16(2):191–3. Epub 1997/06/01. 10.1038/ng0697-191 . [DOI] [PubMed] [Google Scholar]
- 22. Guilford P, Ayadi H, Blanchard S, Chaib H, Le Paslier D, Weissenbach J, et al. A human gene responsible for neurosensory, non-syndromic recessive deafness is a candidate homologue of the mouse sh-1 gene. Hum Mol Genet. 1994;3(6):989–93. Epub 1994/06/01. . [DOI] [PubMed] [Google Scholar]
- 23. Janecke AR, Meins M, Sadeghi M, Grundmann K, Apfelstedt-Sylla E, Zrenner E, et al. Twelve novel myosin VIIA mutations in 34 patients with Usher syndrome type I: confirmation of genetic heterogeneity. Hum Mutat. 1999;13(2):133–40. Epub 1999/03/27. . [DOI] [PubMed] [Google Scholar]
- 24. Roux AF, Faugere V, Le Guedard S, Pallares-Ruiz N, Vielle A, Chambert S, et al. Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90%. J Med Genet. 2006;43(9):763–8. Epub 2006/05/09. 10.1136/jmg.2006.041954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82(3):300–8. Epub 2003/08/09. . [DOI] [PubMed] [Google Scholar]
- 26. Nakanishi H, Kurima K, Kawashima Y, Griffith AJ. Mutations of TMC1 cause deafness by disrupting mechanoelectrical transduction. Auris, nasus, larynx. 2014;41(5):399–408. Epub 2014/06/17. 10.1016/j.anl.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalay E, Karaguzel A, Caylan R, Heister A, Cremers FP, Cremers CW, et al. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26(6):591 Epub 2005/11/16. 10.1002/humu.9384 . [DOI] [PubMed] [Google Scholar]
- 28. Sirmaci A, Duman D, Ozturkmen-Akay H, Erbek S, Incesulu A, Ozturk-Hismi B, et al. Mutations in TMC1 contribute significantly to nonsyndromic autosomal recessive sensorineural hearing loss: a report of five novel mutations. International journal of pediatric otorhinolaryngology. 2009;73(5):699–705. Epub 2009/02/04. 10.1016/j.ijporl.2009.01.005 . [DOI] [PubMed] [Google Scholar]
- 29. Nal N, Ahmed ZM, Erkal E, Alper OM, Luleci G, Dinc O, et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum Mutat. 2007;28(10):1014–9. Epub 2007/06/05. 10.1002/humu.20556 . [DOI] [PubMed] [Google Scholar]
- 30. Fattahi Z, Shearer AE, Babanejad M, Bazazzadegan N, Almadani SN, Nikzat N, et al. Screening for MYO15A gene mutations in autosomal recessive nonsyndromic, GJB2 negative Iranian deaf population. Am J Med Genet A. 2012;158A(8):1857–64. Epub 2012/06/28. 10.1002/ajmg.a.34411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, Ramzan K, et al. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet. 2006;79(6):1040–51. Epub 2006/12/23. 10.1086/510022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nayak G, Varga L, Trincot C, Shahzad M, Friedman PL, Klimes I, et al. Molecular genetics of MARVELD2 and clinical phenotype in Pakistani and Slovak families segregating DFNB49 hearing loss. Hum Genet. 2015;134(4):423–37. Epub 2015/02/11. 10.1007/s00439-015-1532-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang JJ, Su MC, Chien KH, Hsin CH, Li SY. Identification of novel variants in the TMIE gene of patients with nonsyndromic hearing loss. International journal of pediatric otorhinolaryngology. 2010;74(5):489–93. Epub 2010/03/09. 10.1016/j.ijporl.2010.02.001 . [DOI] [PubMed] [Google Scholar]
- 34. Naz S, Giguere CM, Kohrman DC, Mitchem KL, Riazuddin S, Morell RJ, et al. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet. 2002;71(3):632–6. Epub 2002/07/30. 10.1086/342193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sirmaci A, Ozturkmen-Akay H, Erbek S, Incesulu A, Duman D, Tasir-Yilmaz S, et al. A founder TMIE mutation is a frequent cause of hearing loss in southeastern Anatolia. Clin Genet. 2009;75(6):562–7. Epub 2009/05/15. 10.1111/j.1399-0004.2009.01183.x . [DOI] [PubMed] [Google Scholar]
- 36. Mori K, Moteki H, Kobayashi Y, Azaiez H, Booth KT, Nishio SY, et al. Mutations in LOXHD1 Gene Cause Various Types and Severities of Hearing Loss. The Annals of otology, rhinology, and laryngology. 2015;124 Suppl 1:135S–41S. Epub 2015/03/21. 10.1177/0003489415574067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, Holme RH, et al. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34(4):421–8. Epub 2003/07/02. 10.1038/ng1208 . [DOI] [PubMed] [Google Scholar]
- 38. Ebermann I, Scholl HP, Charbel Issa P, Becirovic E, Lamprecht J, Jurklies B, et al. A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet. 2007;121(2):203–11. Epub 2006/12/16. 10.1007/s00439-006-0304-0 . [DOI] [PubMed] [Google Scholar]
- 39. Tlili A, Charfedine I, Lahmar I, Benzina Z, Mohamed BA, Weil D, et al. Identification of a novel frameshift mutation in the DFNB31/WHRN gene in a Tunisian consanguineous family with hereditary non-syndromic recessive hearing loss. Hum Mutat. 2005;25(5):503 Epub 2005/04/21. 10.1002/humu.9333 . [DOI] [PubMed] [Google Scholar]
- 40. Mizutari K, Mutai H, Namba K, Miyanaga Y, Nakano A, Arimoto Y, et al. High prevalence of CDH23 mutations in patients with congenital high-frequency sporadic or recessively inherited hearing loss. Orphanet J Rare Dis. 2015;10(1):60 Epub 2015/05/13. 10.1186/s13023-015-0276-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doherty D, Chudley AE, Coghlan G, Ishak GE, Innes AM, Lemire EG, et al. GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. Am J Hum Genet. 2012;90(6):1088–93. Epub 2012/05/15. 10.1016/j.ajhg.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diaz-Horta O, Sirmaci A, Doherty D, Nance W, Arnos K, Pandya A, et al. GPSM2 mutations in Chudley-McCullough syndrome. Am J Med Genet A. 2012;158A(11):2972–3. Epub 2012/09/19. 10.1002/ajmg.a.35636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maria Oonk AM, van Huet RA, Leijendeckers JM, Oostrik J, Venselaar H, van Wijk E, et al. Nonsyndromic hearing loss caused by USH1G mutations: widening the USH1G disease spectrum. Ear and hearing. 2015;36(2):205–11. Epub 2014/09/26. 10.1097/AUD.0000000000000095 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.