Abstract
Background
Next generation sequencing (NGS) offers a rapid and comprehensive method of screening for mutations associated with retinitis pigmentosa and related disorders. However, certain sequence alterations such as large insertions or deletions may remain undetected using standard NGS pipelines. One such mutation is a recently-identified Alu insertion into the Male Germ Cell-Associated Kinase (MAK) gene, which is missed by standard NGS-based variant callers. Here, we developed an in silico method of searching NGS raw sequence reads to detect this mutation, without the need to recalculate sequence alignments or to screen every sample by PCR.
Methods
The Linux program grep was used to search for a 23 bp “probe” sequence containing the known junction sequence of the insert. A corresponding search was performed with the wildtype sequence. The matching reads were counted and further compared to the known sequences of the full wildtype and mutant genomic loci. (See https://github.com/MEEIBioinformaticsCenter/grepsearch.)
Results
In a test sample set consisting of eleven previously published homozygous mutants, detection of the MAK-Alu insertion was validated with 100% sensitivity and specificity. As a discovery cohort, raw NGS reads from 1,847 samples (including custom and whole exome selective capture) were searched in ~1 hour on a local computer cluster, yielding an additional five samples with MAK-Alu insertions and solving two previously unsolved pedigrees. Of these, one patient was homozygous for the insertion, one compound heterozygous with a missense change on the other allele (c. 46G>A; p.Gly16Arg), and three were heterozygous carriers.
Conclusions
Using the MAK-Alu grep program proved to be a rapid and effective method of finding a known, disease-causing Alu insertion in a large cohort of patients with NGS data. This simple approach avoids wet-lab assays or computationally expensive algorithms, and could also be used for other known disease-causing insertions and deletions.
Introduction
The genetics of retinitis pigmentosa (RP) is particularly challenging due to the large numbers of genes that can cause similar clinical phenotypes [1–3]. Even though it is usually a monogenic, Mendelian disorder, over 90 genes are associated with RP and related disorders [3]. For this reason, the use of NGS has allowed for more comprehensive analysis of these genes and is becoming more widespread for clinical testing [4,5]. However, recent experience shows that there are some regions of the genome that are difficult to analyze by NGS, due to GC-rich highly repetitive sequence or deep intronic mutations not captured by standard NGS techniques [4,6–8]. In addition, the overall diagnostic success rate for retinitis pigmentosa is about 50% [4,9–11] suggesting unknown disease genes or “missing inheritance” in the known disease-associated genes.
Despite much effort, detection of large deletions and insertions (indels) from next generation sequencing (NGS) data is still a challenging problem. Most methods fail when indels exceed a certain fraction of the read length, and sometimes even miss small indels completely. Some methods rely on whole genome sequencing instead of more efficient targeted sequence capture [12–18]. About 7% of the disease-associated or functional variants in The Human Gene Mutation Database (HGMD Professional 2015.1 release) are gross indels, repeats or complex rearrangements. This is most likely an underestimate, due to difficulties in finding these changes. Nevertheless, it is important to incorporate known indels in genetic diagnostic tests.
One such challenge has been the identification of a 353 bp insertion into the Male Germ Cell-Associated Kinase (MAK) gene (MIM #154235). In 2011, distinct classes of MAK mutations were identified as causative mutations in retinitis pigmentosa by two different groups [19,20]. Ozgül and colleagues identified “traditional” homozygous and compound heterozygous mutations in MAK using whole exome sequencing and bioinformatic variant filtering (aided by gene prioritization from experimental work in mouse retinas) [19]. These variants are expected to be detected in standard NGS analysis pipelines.
However, Tucker et al. reported a fairly unusual class of mutation in which a 353 bp Alu repeat sequence was inserted into exon 9 of MAK, disrupting the gene and resulting in improper splicing and loss of the mature MAK protein [20]. It was only by serendipity that this insertion was discovered using the usual NGS bioinformatics pipelines; physical removal of repeat sequences during library preparation for ABI sequencing, combined with creation of a chimeric read led to the artifactual reporting of a “2 bp” insertion in MAK. After PCR amplification of the “2 bp” insertion, a much larger-than-expected fragment was observed [20]. This fragment, when Sanger sequenced, revealed a 353 bp Alu insertion. The presence of the insertion was missed completely using a GATK-based analysis pipeline on Illumina reads, since the algorithm trimmed the Alu sequence from the ends of the junction fragment reads, creating an artifactually normal MAK sequence [20].
Since this time, efforts have turned toward PCR-based screening of DNA from retinitis pigmentosa patients [21]. Venturini et al. developed a nested PCR strategy using primers to amplify exon 9 followed by an amplification using allele-specific primers, one of which contained an insertion-site junction. Using this assay in a panel of recessive retinitis pigmentosa probands, they identified the MAK-Alu insertion in 5/240 (2%) probands of mixed ancestry and in 9/35 (26%) probands of Jewish ancestry. Haplotype analysis confirmed that this mutation was due to a founder effect [21].
We hypothesized that the computational complexity in detection of this Alu insertion could be simplified by searching the unprocessed sequence reads for the known sequence of the mutant junction. This approach provides an attractive alternative to the complexity and resources required to implement allele-specific, nested PCR testing as part of routine genetic screening for retinitis pigmentosa. Furthermore, this computationally simple approach is appropriate for quickly screening archived NGS reads from past sequencing. These methods are of interest to clinical genetic diagnostics centers using NGS to screen patients with inherited eye diseases. Although the approach presented is very simple from a bioinformatics perspective, it solves a practical problem of missed mutation identification that is clinically relevant in current practice.
Results
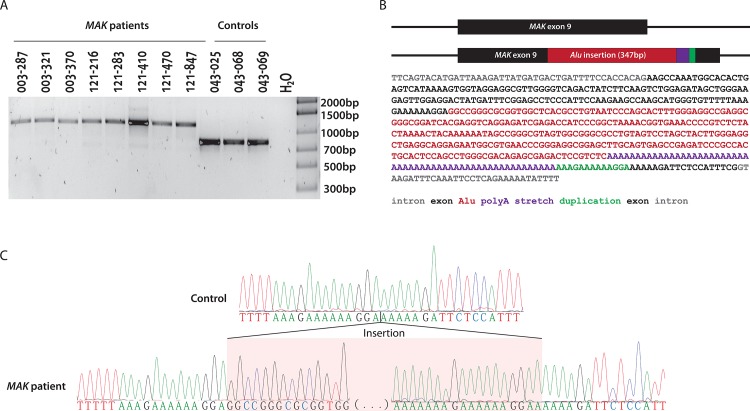
A positive control set of DNAs known to contain the MAK-Alu insertion was validated. This cohort consisted of eleven samples harboring homozygous Alu insertions in MAK exon 9, which were previously reported [21], as well as three negative control samples (Fig 1). PCR amplification and Sanger sequencing of sample OGI412_881 revealed a 280-nucleotide Alu insertion followed by a 58 to 60-nucleotide long polyadenine stretch and a 13 bp target site duplication from MAK exon 9, which is a typical pattern for Alu repeat elements [22,23] (Fig 1). Bioinformatics analysis of the MAK-Alu insertion showed that it belongs to a relatively recently evolved AluYa8 subfamily from the class of the SINE1 non-LTR retrotransposons [24,25]. This sequence was deposited in GenBank (GenBank: KT192064).
Fig 1. Characterization of the test sample set.
A) Samples from previously reported patients with Alu insertion in MAK exon 9 [21] and control samples were PCR amplified to detect homozygous alleles for Alu insertion and WT alleles. B) Sequence of the inserted element (280 bp Alu, 54 bp poly-A and 13 bp duplication of exon 9 sequence). C) Sanger sequence of the exon 9 Alu insertion breakpoints.
In a test sample known to have a MAK-Alu insertion, standard BWA-based alignment of Illumina reads produces a coverage gap in MAK exon 9 but does not clearly identify an insertion (Fig 2). In order to improve the detection of this insertion, the Linux program grep was used to find Alu insertions in the unprocessed NGS reads of the test samples. All of the above test samples were NGS-sequenced using a custom targeted exon capture strategy [4] and three of the samples (003–287, 121–410 and 121–470) were also sequenced using a commercially-available whole exome sequencing protocol. Using a two-stage grep search algorithm (see Methods) on the FASTQ files from the targeted exon capture and the whole exome sequencing, the MAK-Alu insertion was detected in all positive control samples and none of the negative control samples. The reference sequence was detected in all of the negative control samples and none of the positive control samples (Table 1). These results indicate 100% sensitivity and specificity.
Fig 2. Alignment of standard BWA-based Illumina reads of a control (top) and a MAK-Alu homozygous (bottom) sample.
MAK-Alu alignment produces a coverage gap in exon 9 but does not clearly identify an insertion.
Table 1. Specificity and Sensitivity of In Silico Method to Detect the MAK-Alu insertion.
| Capture / run type | Sample genotype | Sample name | Mutant junction | Mutant junction, full match | Reference junction | Reference junction, full match |
|---|---|---|---|---|---|---|
| GEDi | MAK-Alu | BGL003_287 | 21 | 20 | 0 | 0 |
| BGL003_321 | 37 | 30 | 0 | 0 | ||
| BGL003_370 | 27 | 23 | 0 | 0 | ||
| BGL121_216 | 30 | 26 | 0 | 0 | ||
| BGL121_283 | 35 | 34 | 0 | 0 | ||
| BGL121_410 | 43 | 40 | 0 | 0 | ||
| BGL121_470 | 30 | 28 | 0 | 0 | ||
| BGL121_847 | 45 | 39 | 0 | 0 | ||
| Control | BGL043_067 | 0 | 0 | 72 | 58 | |
| BGL043_068 | 0 | 0 | 102 | 73 | ||
| BGL043_069 | 0 | 0 | 91 | 81 | ||
| BGL043_072 | 0 | 0 | 59 | 41 | ||
| WES (Agilent) | MAK-Alu | BGL003_287_WES | 47 | 41 | 0 | 0 |
| BGL121_410_WES | 55 | 50 | 0 | 0 | ||
| BGL121_470_WES | 37 | 35 | 7 | 0 | ||
| Control | BGL038_134 | 0 | 0 | 81 | 75 | |
| BGL038_162 | 0 | 0 | 77 | 73 | ||
| BGL043_002 | 0 | 0 | 85 | 79 | ||
| BGL043_004 | 0 | 0 | 39 | 37 | ||
| BGL043_005 | 0 | 0 | 78 | 70 | ||
| BGL043_007 | 0 | 0 | 74 | 58 | ||
| BGL043_008 | 0 | 0 | 59 | 50 | ||
| BGL043_018 | 0 | 0 | 45 | 36 | ||
| BGL043_059 | 1 | 0 | 62 | 58 | ||
| BGL043_062 | 0 | 0 | 77 | 72 | ||
| BGL121_923 | 0 | 0 | 85 | 75 | ||
| OGI604_001255 | 0 | 0 | 83 | 77 | ||
| OGI604_001264 | 0 | 0 | 82 | 77 |
Testing of in silico method shows 100% sensitivity and specificity using custom selective exon capture data from eight known MAK-Alu insertion samples and four known control samples (top). Testing of in silico method shows 100% sensitivity and specificity using whole exome sequencing (Agilent V5+UTR) from three known MAK-Alu insertion samples and 13 known control samples. A “full match” requires the entire read to match an extended genomic sequence; this step removed the false positive hits in BGL121_470_WES and BGL043_059 seen in the table above (also see Methods).
An expanded version of the grep program was used to investigate a set of 1,847 samples, most of which are from patients with inherited retinal degenerations who were subjected to NGS-based diagnostic testing. The samples contained a mixture of targeted exon sequencing (“GEDi”) [4], whole exon sequencing and whole genome sequencing. In this cohort, five samples (from four families) were found to harbor the MAK-Alu insertion in exon 9 (Table 2). The true population incidence of the insertion cannot be estimated from this study, since the samples tested in this cohort had already been partially depleted of MAK-Alu insertion-containing samples by previous PCR-based screening [21].
Table 2. Identification of Homozygous and Heterozygous MAK-Alu Insertions in a Discovery Sample Set.
| Capture / run type | Sample name | Mutant junction | Mutant junction, full match | Reference junction | Reference junction, full match | Mutant allele frequency, all matches | Mutant allele frequency, full matches | Interpretation |
|---|---|---|---|---|---|---|---|---|
| Gedi | OGI412_881 | 28 | 25 | 0 | 0 | 1 | 1 | homozygous mutant |
| Gedi | D379_148 | 23 | 21 | 59 | 56 | 0.28 | 0.27 | heterozygous mutant |
| WES | C1 (relative of C2) | 13 | 13 | 16 | 16 | 0.45 | 0.45 | heterozygous mutant |
| WES | C2 (relative of C1) | 16 | 16 | 26 | 25 | 0.38 | 0.39 | heterozygous mutant |
| Gedi | D445_255 | 13 | 11 | 15 | 15 | 0.46 | 0.42 | heterozygous mutant |
Analysis of NGS data from 1,847 samples efficiently identifies one homozygous MAK-Alu insertion and four heterozygous insertions. A “full match” requires the entire read to match an extended genomic sequence (see Methods).
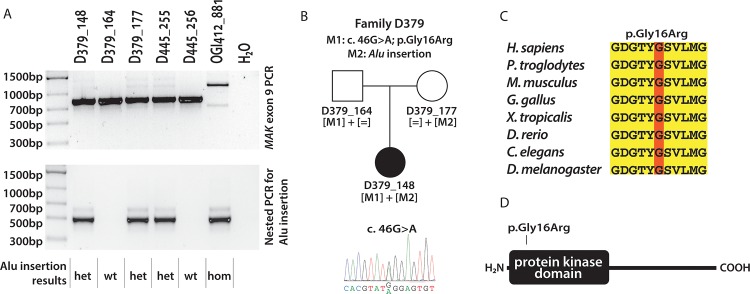
One sample was homozygous and four samples (from three families) were heterozygous for the insertion, which was confirmed in three patients by PCR (Table 2, Fig 3). The patient from family D379 (D379_148) was compound heterozygous for the MAK-Alu insertion and a novel missense change (c. 46G>A; p.Gly16Arg) (Fig 3A and 3B). The missense change affects a highly conserved glycine located in a protein kinase domain (Fig 3C and 3D) and is predicted to be likely pathogenic (using Polyphen-2, SIFT, Provean and MutationTaster [26–29]). The patient OGI412_881 was homozygous for the MAK-Alu insertion (Fig 3A); unfortunately no family members were available for co-segregation analysis. Both patients were of Ashkenazi Jewish descent, which is consistent with the MAK-Alu insertion being a founder mutation in this population [21]. The proband from family D445 (D445_255) carried a heterozygous MAK-Alu insertion (Fig 3A), however no missense changes were found in MAK and this patient was found to be homozygous for the c.144T>G change leading to the p.Asn48Lys substitution in Clarin 1 (CLRN1), which was previously reported as a founder mutation in Usher III in the Ashkenazi Jewish population [30]. Therefore we consider D445_255 to be a heterozygous carrier of the MAK-Alu insertion.
Fig 3. PCR validation of Alu insertion identified by in silico analysis in patients from the discovery cohort.
A) PCR amplification using primers spanning exon 9 (top) and nested PCR using Alu-specific primer (bottom). The 1,194 bp amplicon containing the Alu insertion (arrow) is present strongly in the homozygous sample and weakly in the heterozygous samples (top); the Alu insertion-specific amplification (491 bp, bottom) confirms the presence of the Alu insertion. B) Pedigree of patient D379_148, carrying a missense mutation (p.Gly16Arg) and the Alu insertion mutation. C) Evolutionary conservation of glycine 16, mutated in the patient D379_148. D) Protein domains in MAK and location of the p.Gly16Arg change. The mutation annotations are based on the NM_001242957 transcript, where A from the ATG start codon is designated as a +1 position.
The two newly-identified probands have phenotypes consistent with typical retinitis pigmentosa. The compound heterozygote patient (D379_148) had nyctalopia and constricted visual fields since her teens and was diagnosed with retinitis pigmentosa at age 25 (visual acuity 20/40 OU, visual field to a V4e test stimulus was 20 degrees diameter OU, 30 HZ ERG response of <0.2 microvolts OU). Although at age 57 her visual acuity was slightly worse due to cataracts, progression of her disease was less than expected (potential acuity meter 20/40 OD 20/60 OS, visual fields 20 degrees OU). The patient homozygous for the MAK-Alu insertion (OGI412_881) also had typical retinitis pigmentosa. At age 68, her visual acuity was 20/20- OU. Her visual field to a V4e test light was slightly greater than 20 degrees OU, and her 30 Hz cone ERG was 0.2 microvolts OD and 0.4 microvolts OS.
To extend this technique to other genes, a probe set was developed for the deep intronic mutation (c.2991+1655A>G) in CEP290 [31]. Searching FASTQ file archives for this mutation, which was missed during previous versions of our full analysis pipeline, rapidly identified two samples for further attention.
Discussion
The MAK-Alu grep program is based on knowing the sequence of the junctional insertion site, and therefore is limited to previously detected insertions (or deletions) including founder mutations in the population of interest. As an extension, additional “probe” sets can be validated for other known mutations that are not easily detectable by NGS, such as large insertions, large deletions, or other large rearrangements. Of note, our in-house NGS pipeline has an indel detection limit of approximately 30–50 nucleotides depending on the sequence length and quality.
We have optimized the probe set and reference sequences for the MAK-Alu insertion, which is described in the Methods section and available as part of the downloadable program [32]. Other researchers who find additional non-mapping insertions or deletions of interest are welcome to contact the authors or submit them to the above website.
For mutations that are easily detectable by standard NGS pipelines, this method may occasionally by useful as well. For example, the fact that this method works on.FASTQ or compressed.FASTQ files makes it appropriate for quickly searching archived sequence reads without having to use the computational time and storage to extract archived sequence data, recreate alignments, and recreate variant call files, as shown for the CEP290 deep intronic mutation.
Using this method requires a priori knowledge of the sequence at one of the insertion’s junctions, and that newly formed junction must not already exist in the genome. There has been significant work on more general methods of detection of chromosomal breakpoints and insertions [12]. The MAK-Alu insertion is a particularly difficult subset of “breakpoint” or “chimeric read” to clearly identify, since half of the read is non-mapping due to being a repeat sequence. De novo detection of such sequences is an area for future study. Until those methods are perfected, the simplicity and computational efficiency of searching for the junction sequence is advantageous and effective in practice.
Conclusions
The MAK-Alu insertion was discovered by happenstance [20], as it normally does not show up in typical NGS analysis pipelines, including our own. The need to do a separate PCR to detect this mutation is relatively time-consuming and costly. For this reason it is advantageous to detect the Alu insertion using the MAK-Alu grep program on the NGS data. Using a discovery set of 1,847 samples, the efficient in silico algorithm presented here identified MAK-Alu insertions in five samples and we showed that this technique has high specificity and sensitivity. This approach, while quite simple from a bioinformatics perspective, can be of immediate practical use to clinical diagnostic laboratories that use NGS, until such time as improved NGS processing pipelines no longer miss such clinically-important insertions. The downloadable software is pre-configured to detect the MAK-Alu insertion that is applicable to inherited eye diseases, but is modifiable to detect other known genomic insertions, deletions, and rearrangements from this or other disease areas.
Methods
Patient cohort
The study protocol adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Boards of Massachusetts Eye and Ear Infirmary and Harvard Medical School. The patients harboring the MAK-Alu insertion in the test sample set (Fig 1) were previously reported by Venturini and colleagues [21]. To our knowledge all probands were unrelated. The patients with clinical information included in the study were recruited and clinically examined at the Massachusetts Eye and Ear Infirmary. After patients signed consent forms, blood samples were collected from patients for DNA extraction.
Identification of MAK-Alu insertion in NGS reads
The Linux program grep was used to search FASTQ files for the 5’ junction between the reference sequence of exon 9 and the beginning of the Alu insertion. This is the same sequence used as an allele-specific primer by Venturini et al. [21]. A full software implementation is available online [32]. For the purpose of explanation, at its simplest, the approach can be implemented as follows:
grep–c GAAAAAAGGAGGCCGGGCGCGGT sequence.fastq
This returns the number of reads containing the mutant junction in that sequence file. (An example of the matching raw reads are shown in S1 Fig) Modifications to search compressed files, detect the reverse complement in unoriented reads, and to detect the reference/wildtype sequence are:
zgrep–c ACCGCGCCCGGCCTCCTTTTTTC\|GAAAAAAGGAGGCCGGGCGCGGT sequence.fastq.gz > mutantcount
zgrep–c CGAAATGGAGAATCTTTTTTCCT\|AGGAAAAAAGATTCTCCATTTCG sequence.fastq.gz > wildtypecount
In a MAK-Alu-containing sample, the program returns a positive value depending on the coverage depth in that area (typically 21–55 reads but as low as 13—see Tables 1 and 2). Most files without the insertion return a count of “0” though rarely a false-positive read count of 1 or 2 was detected in a minority of wildtype samples (36/1,847 = 1.9%). In one negative control sample which was run on the same flowcell as a positive sample, seven false positive reads were obtained. A cutoff could be established to exclude such samples with small numbers of hits (e.g. in the grepsearch software, a count of 1 or 2 mutant reads are flagged as probable false positives). Alternatively, the raw read count could be normalized to the total number of reads in the sample; this resulted in a metric that was actually worse at distinguishing true positives from false positives. Instead, a second computational step was implemented to eliminate such false-positive reads automatically. A second level of grep screening was performed where each read matching the probe sequence was further compared to a “reference” sequence spanning the insertion site of ~300 bp of reference genomic DNA. For the mutant “reference” sequence, we included the Alu insertion as described above. The number of reads that match both the probe sequence and the full reference sequence are referred to as a “full match” in the tables. Because using the extended reference sequence eliminated all false positive hits, there is no longer a need to flag/exclude counts of only 1 or 2 mutant reads. The requirement to exactly match the extended reference sequence, as currently implemented, has the disadvantage that, theoretically, a second-site SNP near the junction could prevent matching the full “reference” sequence; this false-negative result was not observed in the current data sets and is probably rare in this haplotype.
Scripts to run these tests on batches of FASTQ files are provided online [32].
PCR validation of MAK-Alu insertion
The validation for MAK-Alu insertion was performed by PCR using the previously reported primers: 5′-TACCGCCCATTTTTGTTCAT-3′ (intron 8, forward) and 5′-ACTGAGAACTGTTACTGTGAG-3′ (intron 9, reverse) [21]. The PCR reaction was performed using a 5x PCR polymerase master mix (5x HOT FIREPol® Blend Master Mix with 7.5 MgCl2, Solis Biodyne, Estonia), using 20ng of genomic DNA and 0.3μM of each primer in a 20μl reaction. The amplification conditions were the following: 95°C for 10 minutes; 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds and 72°C for 1 minute; final extension at 72°C for 5 minutes. Since the PCR reaction preferentially amplifies the shorter WT allele in samples with a heterozygous Alu insertion, a nested PCR was performed using the following primers: 5’-GAAAAAAGGAGGCCGGGCGCGGT-3’(Alu nested [21]) and 5’-CCTGGCCTGTTAAGCAAACT-3’ (reverse nested). The same PCR reaction conditions were used, except for shortening of the extension time to 30 seconds and reducing the cycle number to 25.
Sanger sequencing was performed after PCR cleanup (ExoSap-IT, Affymetrix, Santa Clara, CA, USA) and sequenced (BigDye Terminator v3.1, ABI 3730xl, Life Technologies, Grand Island, NY, USA) using the intron 8 and intron 9 primers described above.
Custom selective exon capture, whole exome sequencing (WES), and next generation sequencing (NGS)
For custom selective exon capture, paired-end/multiplexable SureSelect targeted enrichment capture libraries (Agilent Technologies, Inc.; Santa Clara, CA) were generated on a BRAVO automation workstation (Agilent Technologies, Inc.) according to their standard automation protocol (Pub. No. G7550-90000, Version D.1, April 2012). Targeted enrichment included all currently known monogenic inherited retinal degeneration genes [3,4]. Targeted enrichment sample analysis was performed on a MiSeq NGS platform (Illumina, Inc.; San Diego, CA). A 12-patient sample multiplex was clustered to an average cluster density of ~850 K clusters per mm2 and 2 x 121 bp paired-end sequenced using Illumina’s 300-cycle MiSeq Reagent Kit V2. The average depth-of-coverage (DoC) per-sample was ~100x.
WES targeted enrichment capture libraries were generated on a BRAVO automated workstation using the SureSelect Human V5+UTR All Exon targeted enrichment kit (Agilent Technologies, Inc.) according to their standard automation protocol. NGS analysis was performed using a HiSeq 2500 NGS instrument (Illumina, Inc.) in the High Throughput mode. An 8 picoMolar (pM), 4-sample multiplex sample (i.e. 2 pM per capture library) was clustered in duplicate flow cell lanes at ~700,000 clusters per mm2, followed by 101|7|101 bp paired-end indexed analysis. The average DoC for the sixteen WES samples was 67x; additionally, the average percent on-target coverage for these WES samples at 1x, 10x, and 20x DoC was 99.9%, 92.2% and 80.9%, respectively.
Supporting Information
Using the command “zgrep GAAAAAAGGAGGCCGGGCGCGGT D00379_000148_GCCAAT_L001_R2_001.fastq.gz”, 23 reads were obtained. The reads were aligned manually for display purposes and the sequence matching the probe was underlined. A space was added before the canonical 5’ end of the Alu insertion (GGCCGGG…). The read length of 121 bp was too short to span the entire Alu insertion (even if each read was computationally merged with its mate pair, not shown).
(DOCX)
Acknowledgments
We would like to acknowledge Xiaowu Gai and Eric Pierce for scientific discussions, Elizabeth Engle for contribution of carrier DNA samples, and Aliete Langsdorf for manuscript preparation.
Data Availability
All relevant data are within the paper and its Supporting Information files. The software is freely downloadable at https://github.com/MEEIBioinformaticsCenter/grepsearch. The MAK-Alu sequence was deposited in GenBank (GenBank: KT192064).
Funding Statement
KMB was supported by a fellowship with the Fleming Family Foundation. JC was supported by NEI K12 EY016335 and a Career Development Award from Research to Prevent Blindness. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368: 1795–1809. 10.1016/S0140-6736(06)69740-7 [DOI] [PubMed] [Google Scholar]
- 2. Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. Elsevier Ltd; 2010;29: 335–375. 10.1016/j.preteyeres.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 3.Retinal Information Network (RetNet) [Internet]. Available: https://sph.uth.edu/retnet/home.htm.
- 4. Consugar MB, Navarro-Gomez D, Place EM, Bujakowska KM, Sousa ME, Fonseca-Kelly ZD, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2014;17: 253–261. 10.1038/gim.2014.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bujakowska KM, Consugar MB, Place E, Harper S, Lena J, Taub DG, et al. Targeted exon sequencing in Usher syndrome type I. Invest Ophthalmol Vis Sci. 2014;55: 8488–8496. 10.1167/iovs.14-15169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braun TA, Mullins RF, Wagner AH, Andorf JL, Johnston RM, Bakall BB, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22: 5136–5145. 10.1093/hmg/ddt367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neidhardt J, Glaus ÃE, Barthelmes D, Zeitz C, Fleischhauer J, Berger W. Identification and Characterization of a Novel RPGR Isoform in Human Retina. 2007;28: 797–807. 10.1002/humu [DOI] [PubMed] [Google Scholar]
- 8. Webb TR, Parfitt DA, Gardner JC, Martinez A, Bevilacqua D, Davidson AE, et al. Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23). Hum Mol Genet. 2012;21: 3647–3654. 10.1093/hmg/dds194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Audo I, Bujakowska KM, Léveillard T, Mohand-Saïd S, Lancelot M- E, Germain A, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis. 2012;7: 8 10.1186/1750-1172-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neveling K, Collin RWJ, Gilissen C, van Huet RAC, Visser L, Kwint MP, et al. Next-generation genetic testing for retinitis pigmentosa. Hum Mutat. 2012;33: 963–972. 10.1002/humu.22045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neveling K, Feenstra I, Gilissen C, Hoefsloot LH, Kamsteeg E-J, Mensenkamp AR, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat. 2013;34: 1721–1726. 10.1002/humu.22450 [DOI] [PubMed] [Google Scholar]
- 12. Ratan A, Olson TL, Loughran TP, Miller W. Identification of indels in next-generation sequencing data. BMC Bioinformatics. 2015;16: 1–8. 10.1186/s12859-015-0483-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25: 2865–2871. 10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang Y, Wang Y, Brudno M. PRISM: Pair-read informed split-read mapping for base-pair level detection of insertion, deletion and structural variants. Bioinformatics. 2012;28: 2576–2583. 10.1093/bioinformatics/bts484 [DOI] [PubMed] [Google Scholar]
- 15. Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21: 974–984. 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li S, Li R, Li H, Lu J, Li Y, Bolund L, et al. SOAPindel: Efficient identification of indels from short paired reads. Genome Res. 2013;23: 195–200. 10.1101/gr.132480.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. Nature Publishing Group; 2009;41: 1061–1067. 10.1038/ng.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nord AS, Lee M, King M-C, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. BioMed Central Ltd; 2011;12: 184 10.1186/1471-2164-12-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ozgül RK, Siemiatkowska AM, Yücel D, Myers C a, Collin RWJ, Zonneveld MN, et al. Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet. 2011;89: 253–264. 10.1016/j.ajhg.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tucker B a, Scheetz TE, Mullins RF, DeLuca AP, Hoffmann JM, Johnston RM, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci U S A. 2011;108: E569–E576. 10.1073/pnas.1108918108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Venturini G, Koskiniemi-Kuendig H, Harper S, Berson EL, Rivolta C. Two specific mutations are prevalent causes of recessive retinitis pigmentosa in North American patients of Jewish ancestry. Genet Med. 2014; 1–6. 10.1038/gim.2014.132 [DOI] [PubMed] [Google Scholar]
- 22. Batzer M a, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3: 370–379. 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- 23. Erwin J a, Marchetto MC, a FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. Nature Publishing Group; 2014;15: 497–506. 10.1038/nrn3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price AL, Eskin E, Pevzner P a. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14: 2245–2252. 10.1101/gr.2693004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7: 474 10.1186/1471-2105-7-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7: e46688 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. Nature Publishing Group; 2010;7: 575–576. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 30. Fields RR, Zhou G, Huang D, Davis JR, Möller C, Jacobson SG, et al. Usher syndrome type III: revised genomic structure of the USH3 gene and identification of novel mutations. Am J Hum Genet. 2002;71: 607–617. 10.1086/342098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KEJ, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79: 556–561. 10.1086/507318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.github online software repository [Internet]. 2015. Available: https://github.com/MEEIBioinformaticsCenter/grepsearch. Accessed: 1 Jun 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Using the command “zgrep GAAAAAAGGAGGCCGGGCGCGGT D00379_000148_GCCAAT_L001_R2_001.fastq.gz”, 23 reads were obtained. The reads were aligned manually for display purposes and the sequence matching the probe was underlined. A space was added before the canonical 5’ end of the Alu insertion (GGCCGGG…). The read length of 121 bp was too short to span the entire Alu insertion (even if each read was computationally merged with its mate pair, not shown).
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The software is freely downloadable at https://github.com/MEEIBioinformaticsCenter/grepsearch. The MAK-Alu sequence was deposited in GenBank (GenBank: KT192064).