Summary
The earliest step in Escherichia coli cell division consists of the assembly of FtsZ protein into a proto-ring structure, tethered to the cytoplasmic membrane by FtsA and ZipA. The proto-ring then recruits additional cell division proteins to form the divisome. Previously we described an ftsZ allele, ftsZL169R, which maps to the side of the FtsZ subunit and confers resistance to FtsZ assembly inhibitory factors including Kil of bacteriophage λ. Here we further characterize this allele and its mechanism of resistance. We found that FtsZL169R permits the bypass of the normally essential ZipA, a property previously observed for FtsA gain-of-function mutants such as FtsA* or increased levels of the FtsA-interacting protein FtsN. Similar to FtsA*, FtsZL169R also can partially suppress thermosensitive mutants of ftsQ or ftsK, which encode additional divisome proteins, and confers strong resistance to excess levels of FtsA, which normally inhibit FtsZ ring function. Additional genetic and biochemical assays provide further evidence that FtsZL169R enhances FtsZ protofilament bundling, thereby conferring resistance to assembly inhibitors and bypassing the normal requirement for ZipA. This work highlights the importance of FtsZ protofilament bundling during cell division and its likely role in regulating additional divisome activities.
Introduction
The earliest known event in Escherichia coli cell division involves assembly of the highly conserved prokaryotic tubulin-homolog FtsZ into a ring structure at midcell (Bi and Lutkenhaus, 1991; Ma et al., 1996). This formation of a proto-ring is followed by the recruitment of essential and non-essential proteins to the FtsZ scaffold in a partially step-wise fashion to form a mature divisome (Adams and Errington, 2009; Lutkenhaus et al., 2012; Rico et al., 2013). The divisome contains all the components necessary to divide the cell through a combination of membrane constriction, a switch from lateral cell wall growth to septum (cross-wall) formation, and cell separation.
Each FtsZ subunit binds GTP and assembles in a head-to-tail fashion into single stranded protofilaments (Erickson and Stoffler, 1996; Oliva et al., 2004; Mingorance et al., 2005). These protofilaments can associate with each other via the sides of the subunits to form sheets or bundles, depending on buffer conditions or molecular agents (Erickson et al., 1996; Yu and Margolin, 1997; Hale et al., 2000; Lan et al., 2008). By conventional fluorescence microscopy, FtsZ appears as a continuous, ring-shaped structure around the inner cell circumference when viewed down an E. coli’s long axis or as a linear band across the midcell width when viewed from the side (Ma et al., 1996). Recent super-resolution fluorescence microscopy studies of labeled FtsZ in multiple model organisms suggest that the FtsZ ring is not a continuous structure, but instead an irregular clustering of FtsZ filaments and bundles along a narrow circular region around the periphery of the cytoplasmic membrane (Biteen et al., 2010; Fu et al., 2010; Strauss et al., 2012; Rowlett and Margolin, 2014). Results using fluorescence polarization are consistent with this model (Si et al., 2013). Together, these loose assemblages of clustered FtsZ filaments compose the ring-like structure that establishes the site of cytokinesis. However, this view has been challenged by a recent cryo-electron tomography study of dividing cells, which shows long continuous filaments of what is probably FtsZ (Szwedziak et al., 2014).
Within an E. coli population growing in rich media, over 90% of cells have a midcell band of FtsZ localization visible by fluorescence microscopy (Addinall and Lutkenhaus, 1996a). Although an ~100 nm wide FtsZ ring is visible for the majority of the cell cycle under these growth conditions (Fu et al., 2010; Piro et al., 2013), its initial formation is preceded by formation of a broader spiral-shaped FtsZ localization that rapidly coalesces along the long cell axis into a tightly coiled band of clustered filaments (Thanedar and Margolin, 2004; Fischer-Friedrich et al., 2012). These FtsZ spiral structures can be artificially stabilized or prolonged by increasing cellular ftsZ expression, by expressing particular ftsZ mutant alleles, or by perturbing other cellular organizing proteins (Addinall and Lutkenhaus, 1996b; Sun et al., 1998; Stricker and Erickson, 2003; Michie et al., 2006). FtsZ spirals are also observed quite easily during the natural cell differentiation process of Bacillus subtilis sporulation, when FtsZ assembly transitions from a midcell to polar localization (Ben-Yehuda and Losick, 2002).
In E. coli, FtsZ assembly is stabilized and linked to the periphery of the cell at the membrane through interactions with two essential proteins: membrane-anchored ZipA and membrane-associated FtsA (Hale and de Boer, 1997; Pichoff and Lutkenhaus, 2002; Pichoff and Lutkenhaus, 2005). Based on its bundling activity on purified FtsZ in vitro (Raychaudhuri, 1999; Hale et al., 2000; Loose and Mitchison, 2014), ZipA is thought to likewise aid FtsZ filament clustering in vivo, in addition to its role as membrane anchor. This proposed in vivo bundling activity for ZipA is a function shared with three of the nonessential Zap proteins (ZapA, ZapC, and ZapD) found in E. coli, based on their similar direct effects on FtsZ assembly (Buss et al., 2013; Durand-Heredia et al., 2011; Gueiros-Filho and Losick, 2002; Hale et al., 2011; Mohammadi et al., 2009; Monahan et al., 2009; Small et al., 2007). However, the differing phenotypes associated with loss of ZipA or any of the Zap proteins singly or in combination suggest that they have overlapping, but mechanistically distinct ways of clustering FtsZ filaments in vivo (Buss et al., 2013; Dajkovic et al., 2010).
Although ZipA is normally essential for E. coli division, numerous gain-of-function mutants of FtsA permit cell survival despite the complete loss of zipA. We previously isolated the first, and most potent, of these zipA-bypass ftsA mutant alleles (ftsAR286W or ftsA*), and work from the Lutkenhaus lab has subsequently identified numerous others (Geissler et al., 2003; Pichoff et al., 2012). While we originally proposed that FtsA* had increased self-interaction (Shiomi and Margolin, 2007a), Pichoff et al. used alternative assays independent of the FtsZ ring to suggest that all FtsA mutants able to bypass ZipA, including FtsA*, actually display decreased intrinsic self-interaction (Pichoff et al., 2012). These findings, along with an atomic structure of the FtsA oligomer (Szwedziak et al., 2012), led to a new model in which ZipA normally functions to antagonize FtsA self-interaction, thereby enhancing FtsA’s recruitment of downstream divisome proteins such as FtsN. According to this model, FtsA mutants deficient in self-interaction would thus no longer require ZipA activity. It was recently shown that increased levels of FtsN can also bypass ZipA, presumably by binding to FtsA directly (Busiek et al., 2012) and mimicking FtsA*-like mutants by forcing FtsA monomerization (Pichoff et al., 2014). However, it was also recently found that a lesion in the periplasmic portion of FtsL, another downstream divisome protein that closely interacts with FtsQ and FtsB, can bypass certain divisome proteins including ZipA and even FtsA itself (Liu et al., 2014; Tsang and Bernhardt, 2014). One interpretation of these findings is that FtsA is not the sole mediator of these effects, and FtsZ assembly itself may be the ultimate target of signaling by the divisome.
Here we describe an ftsZ allele, ftsZL169R, which permits E. coli survival in the absence of zipA. Previously isolated based on its resistance to the Kil peptide of bacteriophage λ and other FtsZ assembly inhibitors, FtsZL169R protein displays aberrant localization as spirals and polar rings that appear to be deficient in disassembly following cytokinesis and/or slower to reorganize into coherent FtsZ rings at midcell. In addition to permitting the bypass of zipA, FtsZL169R compensates for the loss of the non-essential Zap proteins as well as some thermosensitive (ts) essential division components. Purified FtsZL169R shows evidence of strongly enhanced bundling in biochemical assays of FtsZ assembly, suggesting a model in which FtsZL169R stabilizes FtsZ filament clusters in vivo, thereby conferring resistance to FtsZ assembly inhibitors and bypassing the normal requirement for ZipA, even in the presence of an otherwise wild-type (WT) divisome.
Results
FtsZL169R forms aberrant rings and spirals
We previously reported the mechanism by which FtsZ assembly is inhibited by the Kil peptide encoded by the PL operon of bacteriophage λ (Haeusser et al., 2014). In that work we isolated two mutant ftsZ alleles that showed resistance to kil expressed from a modified PL operon of a defective λ prophage. Cells harboring these alleles, ftsZL169R or ftsZV208A, were additionally resistant to overproduction of other FtsZ assembly inhibitors, including SulA or MinC; the cells also displayed abnormal polar FtsZ ring localization, minicell formation, and occasional branching.
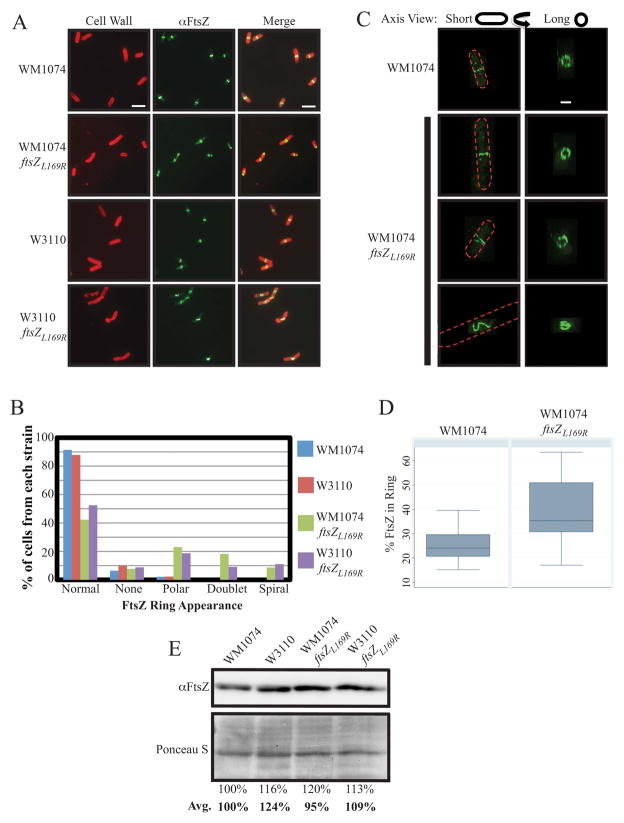
We decided to examine the behavior of these mutant ftsZ alleles more closely, starting with ftsZL169R, the focus of this study. We first transduced the ftsZL169R allele from the defective-prophage harboring strain (AW60) into two of our routinely used WT strains, WM1074 and W3110. Replacement of ftsZWT at its native locus with the ftsZL169R allele did not result in any observable defects in cell growth, but as reported previously for the original isolate, immunofluorescence microscopy (IFM) of FtsZL169R in either WM1074 or W3110 backgrounds revealed aberrant localization compared to the WT parents (Figure 1A). While 91.4 and 87.5% of WM1074 and W3110 cells, respectively, contained clear, solitary midcell bands of FtsZ localization as expected, only 42.3 and 52.5% of their respective ftsZL169R-harboring counterparts displayed this WT FtsZ localization. The percentage of cells without any apparent FtsZ rings did not change, but ftsZL169R cells of either background had an increased frequency of polar FtsZ localization (23.1 and 18.6%) and midcell FtsZ aberrantly formed into apparent doublets (18.3 and 9.1%) or slanted rings/spirals (8.7 and 11.0%) (Figures 1B and S1).
Figure 1.
FtsZL169R localizes aberrantly with a greater concentration in rings compared to FtsZWT, despite equivalent protein levels. (A) Representative IFM images of WT (WM1074 and W3110) and ftsZL169R cells. Cell walls were stained (red) with rhodamine-conjugated wheat-germ agglutinin and FtsZ stained (green) with AlexaFluor 488-conjugated goat α–rabbit recognition of rabbit α-FtsZ. Scale bar = 5 μm. (B) Percentage of mid-log culture cells (n > 100/strain) for indicated strains showing indicated FtsZ localization patterns by IFM as in (A). (C) Representative 3D-SIM images of WT and ftsZL169R cells as imaged from the side (along the short axis) or reconstructed views tilted 90º (along the long axis) with FtsZ signal as in (A), with general cell outlines depicted as red dashes. Scale bar = 1 μm. (D) Estimates of percentage of total FtsZ present in representative 3D-SIM images of wild type or ftsZL169R rings. Boxes show the median with 35th and 75th percentile for each group, and error bars show standard deviation. (E) Representative immunoblot of FtsZ protein levels from mid-exponential cultures of WT or ftsZL169R cells with estimated relative band intensities for the image shown, normalized to a low molecular weight segment of the corresponding SDS-PAGE gel stained with Ponceau S. The average relative band intensities from three separate experiments are also shown in bold.
Despite the aberrant FtsZ assembly, measurements of the average cell length of ftsZL169R cell populations showed no significant deviation from WT cells of either background (Table 1 and data not shown). However, assembled FtsZ occasionally remained visible between two cells attempting to divide, as if persisting at the septum from a decreased ability to disassemble (Figure S1, bottom center panel). Consistent with previous observations, a high proportion (>35%) of ftsZL169R cells generated branches or minicells despite their relatively short length (Table 1).
Table 1.
| Strain | ftsZ | zipA | pDSW210F | IPTG | Cell # | Average length | Median length | Std. dev. | Filaments* | Misshapen** |
|---|---|---|---|---|---|---|---|---|---|---|
| (μm) | (μm) | (μm) | (%) | (%) | ||||||
| DPH861 | WT | + | empty | − | 252 | 3.5 | 3.3 | 1.0 | 0.0 | 0.0 |
| DPH862 | WT | + | ftsA | − | 272 | 3.4 | 3.2 | 1.0 | 0.4 | 1.8 |
| DPH859 | WT | + | ftsAR286W | − | 303 | 2.7 | 2.6 | 0.8 | 0.0 | 1.3 |
| DPH836 | L169R | + | empty | − | 226 | 3.5 | 3.3 | 1.2 | 0.9 | 35.4 |
| DPH837 | L169R | + | ftsA | − | 244 | 3.1 | 2.8 | 1.2 | 0.8 | 36.1 |
| DPH839 | L169R | + | ftsAR286W | − | 167 | 3.5 | 3.4 | 1.3 | 3.0 | 59.3 |
| DPH874 | L169R | − | empty | − | 218 | 9.7 | 6.4 | 8.1 | 45.0 | 2.3 |
| DPH875 | L169R | − | ftsA | − | 264 | 5.4 | 4.5 | 3.9 | 11.7 | 4.5 |
| DPH876 | L169R | − | ftsAR286W | − | 228 | 4.5 | 4.1 | 1.9 | 0.1 | 7.0 |
| DPH861 | WT | + | empty | + | 220 | 3.7 | 3.5 | 1.1 | 2.7 | 0.0 |
| DPH862 | WT | + | ftsA | + | 268 | 9.2 | 7.1 | 5.2 | 51.9 | 0.7 |
| DPH859 | WT | + | ftsAR286W | + | 114 | 2.5 | 2.5 | 0.6 | 0.0 | 1.8 |
| DPH836 | L169R | + | empty | + | 214 | 3.1 | 3.0 | 0.9 | 0.0 | 43.9 |
| DPH837 | L169R | + | ftsA | + | 230 | 3.0 | 3.0 | 0.8 | 0.0 | 17.8 |
| DPH839 | L169R | + | ftsAR286W | + | 172 | 3.8 | 3.7 | 1.3 | 1.2 | 62.2 |
| DPH874 | L169R | − | empty | + | 155 | 8.2 | 6.1 | 7.1 | 38.1 | 3.9 |
| DPH875 | L169R | − | ftsA | + | 207 | 5.3 | 4.8 | 2.9 | 11.6 | 2.9 |
| DPH876 | L169R | − | ftsAR286W | + | 111 | 4.9 | 4.2 | 2.2 | 13.5 | 31.5 |
Filaments defined as cells greater than 7.00 μm
Misshapen defined as cells with minicells, bulges, branching, etc.
We next employed Three-Dimensional Structured Illumination Microscopy (3D-SIM) to visualize aberrant FtsZL169R localization with greater resolution and clarify if the FtsZL169R doublets observed by conventional IFM were each actually separate assemblies or a single compact spiral. Similar to images by conventional IFM, WT WM1074 cells imaged by 3D-SIM (n = 23) predominantly (91.3%) showed a straight band of FtsZ localization when viewed from the side along the cell’s short axis. Rotation of a given image by 90° to reconstruct a view down the length of the cell’s axis revealed the annular structure of FtsZ foci and variable small gaps as previously reported for WT E. coli (Rowlett and Margolin, 2014) (Figure 1C, top row).
Similar to results obtained by conventional IFM (Figure 1A & B), isogenic ftsZL169R cells contained a variety of FtsZ structures when visualized by 3D-SIM (Figure 1C, bottom three rows). Among the WM1074 ftsZL169R cells visualized (n=55), only a minority (38.2%) still contained FtsZ ring structures comparable to WT cells (Figure 1C, second row). The remaining percentage of aberrant FtsZ structures consisted of spirals with varying pitch and length. These ranged from short, kinked V-shape structures (Figure 1C, third row) to elongated helices (Figure 1C, fourth row) when viewed down the cell’s short axis, and an irregular tangle when viewed down the cell’s long axis.
FtsZL169R staining at division sites seemed consistently more intense than for FtsZWT in both standard IFM and 3D-SIM images with comparable exposure times. We therefore used 3D-SIM images to quantify FtsZ ring signal intensity in both WT (n=22) and ftsZL169R-harboring (n=19) cells to estimate the percent of total cellular FtsZ assembled into rings present in each background. Normal FtsZWT rings contained an estimated average 25.5 ± 6.4% total cellular FtsZ, a figure comparable to previous estimates (Stricker et al., 2002). In contrast, FtsZL169R rings showed greater signal intensity, with an estimated average of 39.2 ±12.7% of total cellular FtsZ (Figure 1D). Importantly, this increased intensity did not result from significant changes in total FtsZ levels between the WT and mutant cell populations (Figure 1E).
To maintain consistent sampling areas of ring signal versus background, only relatively normal or close-to-normal shaped FtsZL169R rings were measured for signal intensity. Abnormal, elongated spiral assemblies that would be difficult to accurately measure because of their shape were excluded from analysis and the calculated values for FtsZL169R may therefore be an underestimate. Nonetheless, the measured increased amount of FtsZL169R present in division rings compared to FtsZWT is significant by Student’s t-test (p = 0.0001) and by Wilcoxon rank-sum analysis.
ftsZL169R permits the loss of normally essential zipA
The broad resistance of FtsZL169R to assembly inhibitors, its abnormal formation of spirals that do not ‘collapse’ into narrow rings, and its apparent persistence at cell poles following division led us to the hypothesis that FtsZL169R assembly is somehow more stabilized relative to FtsZWT. We therefore asked whether the presence of FtsZL169R could replace the need for FtsZ assembly stabilization factors, such as the normally essential ZipA.
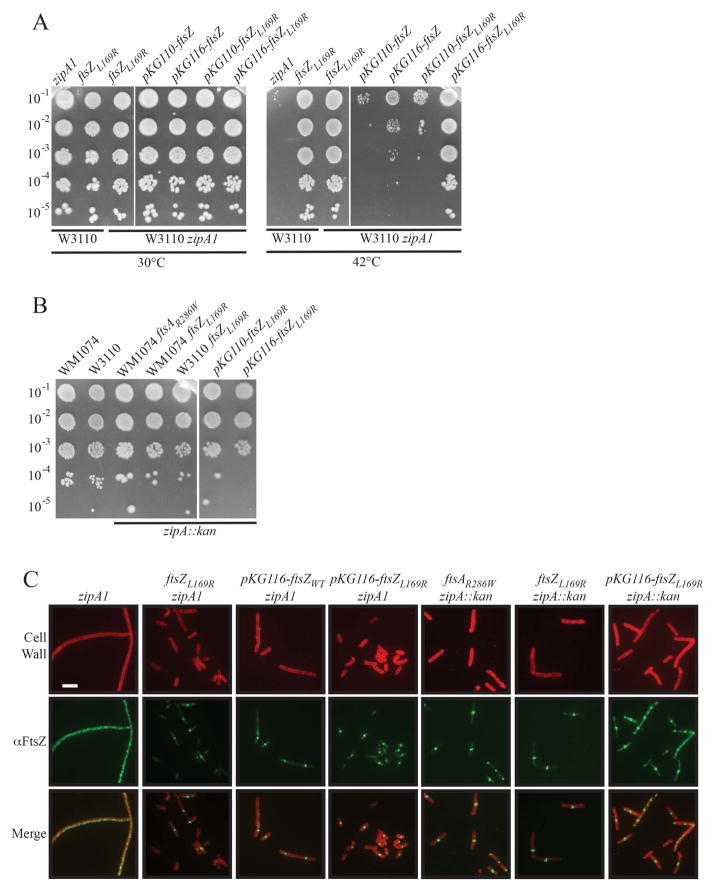
We first tested the ability of ftsZL169R to suppress the ts zipA1 allele. W3110 zipA1 cells divide normally at 30ºC, but at 42ºC many FtsZ rings fail to assemble properly or recruit downstream proteins, leading to cell filamentation and decreased viability (Pichoff and Lutkenhaus, 2002). Spot dilutions of exponentially growing cells showed that while W3110 zipA1 cells failed to survive at 42ºC, an isogenic strain carrying ftsZL169R at the native locus grew as well as cells with wild-type zipA, even at elevated temperature (Figure 2A, left panels, 30ºC and 42ºC). IFM of these strains following a shift of exponentially growing cells to 42ºC verified that zipA1 cells were unable to form coherent FtsZ rings at that temperature, leading to cell filamentation after several mass doublings (Fig. 2C, column 1). In contrast, ftsZL169R suppressed these phenotypes, restoring FtsZ ring assembly and normal cell lengths (Figure 2C, column 2).
Figure 2.
FtsZL169R cells survive without the normally essential ZipA. (A) Spot dilutions of indicated strains in WT (W3110) or zipA1 ts backgrounds at 30º or 42ºC. Mid- exponential phase cultures of strains without plasmids were plated on plain LB plates and those with plasmids were plated on LB plates with chloramphenicol and 0.1 μM sodium salicylate to induce expression of ftsZ derivatives. Note that pKG110 includes an unorthodox ribosome-binding site, leading to relatively modest overexpression, while pKG116 has a strong ribosome binding site, leading to high overexpression. (B) Spot dilutions of indicated strains in WT backgrounds or successfully transduced with zipA::kan. Plasmids induced with sodium salicylate as in (A) (C) Representative IFM images of indicated strains. Cells in the zipA1(ts) background were imaged during mid-exponential growth, ~60 minutes following shift to the nonpermissive temperature of 42ºC. Cells permitting bypass of zipA (zipA::kan) were grown at 37ºC and sampled as normal during mid-exponential growth. Signals and scale as in Figure 1A; plasmids induced with sodium salicylate as in (A).
To ensure that the suppression of zipA1 was caused by ftsZL169R and not by another mutation, we expressed it at relatively low levels from a plasmid in the presence of chromosomal wild-type ftsZ. Low induction of ftsZL169R from pKG116 with 0.1 μM sodium salicylate completely suppressed zipA1 thermosensitivity. Induction from the related pKG110 plasmid, which has a weaker ribosome-binding site, displayed partial suppression (Figure 2A, right panels, ftsZL169R, 30ºC and 42ºC).
As controls, we also included isogenic strains that carried ftsZWT on pKG110 or pKG116. Interestingly, we found that slight overexpression of ftsZ can partially suppress zipA1 viability at 42°C (Figure 2A, right panel), an observation previously not reported. This suggests that the ts ZipA1 has residual activity at 42ºC. Nonetheless, with both weaker pKG110 and stronger pKG116 expression, ftsZL169R suppressed zipA1 thermosensitivity more efficiently than ftsZWT in spot dilutions (Figure 2A). Furthermore, in the zipA1 strain at 42ºC, ftsZL169R expressed from pKG116 reduced the average cell length and restored FtsZL169R ring assembly (Figure 2C, columns 3 & 4, and data not shown).
To further confirm that FtsZL169R confers cell survival in the absence of functional ZipA, we attempted to transduce a zipA::kan deletion into E. coli cells harboring chromosomal ftsZL169R. Normally zipA::kan will only transduce into cells carrying another copy of zipA on the chromosome or on a covering plasmid, or when ZipA bypass mutations are present (Geissler et al., 2003; Pichoff et al., 2012). Whereas no zipA::kan transductants were obtained with ftsZWT unless a ZipA-bypass mutant (ftsAR286W, a positive control) was present, ftsZL169R alone allowed introduction of zipA::kan by transduction and resulted in normal viability of either W3110 or WM1074 strain backgrounds (Figure 2B). Notably, levels of overexpression of ftsZWT from a plasmid that could partially suppress zipA1 thermosensitivity (Figure 2A) did not permit zipA::kan transduction. In contrast, expression of ftsZL169R from plasmids pKG110 or pKG116 into an otherwise WT strain did permit zipA::kan transduction at 30°C (Figure 2B). This also provided further evidence that ftsZL169R is a dominant allele.
IFM of zipA::kan transductants showed that both chromosomal ftsZL169R or its expression from pKG116 in cells with native ftsZWT permitted FtsZ ring formation in the absence of ZipA. However, although these cells could form FtsZ rings and survive, cell division was not completely normal, leading to increased cell lengths and filamentation in the population (Figure 2C, columns 6 & 7). Thus, although the presence of FtsZL169R permits ZipA bypass, it is less effective than the originally described ZipA-bypass mutant FtsAR286W (FtsA*) (Figure 2C, column 5). We also tested the Kil-resistant allele FtsZV208A, which unlike FtsZL169R does not map to the lateral interface, for its ability to bypass ZipA. Notably, FtsZV208A did not bypass ZipA, so we did not study it further here.
ftsZL169R suppresses subtle division defects of zap mutants and ts divisome components
The preceding data, including the bypass of normally essential ZipA by FtsZL169R, argues that FtsZL169R could generally compensate for divisome destabilization. To further test this hypothesis, we explored the ability of cells harboring ftsZL169R to suppress defects associated with the loss of other putative FtsZ stabilization factors (ZapA and ZapC) or the thermosensitivity of various divisome alleles.
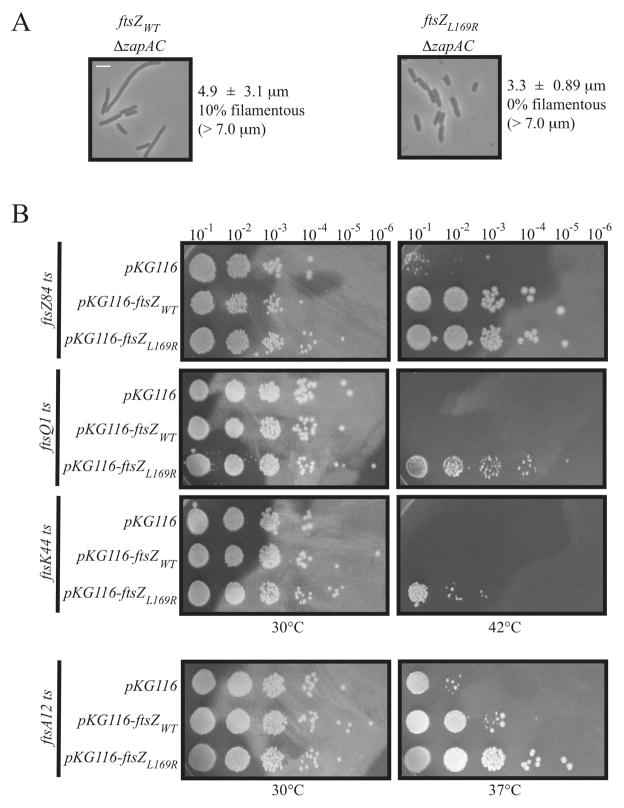
The Zap family of proteins potentially stabilize the FtsZ ring via FtsZ protofilament bundling activity (Huang et al., 2013). A ΔzapA ΔzapC double deletion strain (ΔzapAC) displays a subtle phenotype in which a significant number of cells grow as filaments, particularly during early exponential phase, due to delayed division (Gueiros-Filho and Losick, 2002; Small et al., 2007; Durand-Heredia et al., 2012). In our hands ~10% of ΔzapAC ftsZWT cells were filamentous (> 7.0 μm) with a population average cell length of 4.9 ± 3.1 μm. Replacement of the native ftsZWT allele with ftsZL169R completely suppressed the ΔzapAC phenotype, eliminating cell filamentation and reducing population average cell length to 3.3 ± 0.89 μm. This demonstrates that FtsZL169R is able to compensate for the loss of multiple factors proposed to stabilize FtsZ through bundling, including ZipA and ZapC.
If FtsZL169R has greater assembly stabilization compared to FtsZWT, we hypothesized that the mutant allele might also be able to suppress ts defects arising from mutations in various essential divisome components. As expected from its ability to replace ftsZWT in the chromosome (Figure 1), expression of ftsZL169R from a plasmid is able to complement the ts ftsZ84 allele at 42ºC as effectively as ftsZWT (Figure 3B, top row).
Figure 3.
(A) ftsZL169R suppresses the cell division defects of a strain lacking ZapA and ZapC. Representative phase contrast micrographs of a ΔzapAC strain in an ftsZWT or ftsZL169R background. Scale as in Figure 1A. Average cell length ± standard deviation are indicated for both strains. (B) ftsZL169R suppresses defects in divisome components. Spot dilutions of indicated ts strains with empty pKG116 or with plasmid expressing (0.1 μM sodium salicylate) ftsZWT or ftsZL169R at permissive (30ºC) or restrictive (37º or 42ºC) temperatures.
More interestingly, and in contrast to ftsZWT, plasmid-based expression of ftsZL169R suppressed the thermosensitivity of the ftsQ1 allele quite well, and could very weakly suppress ftsK44 thermosensitivity (********Figure 3B, middle rows). While unable to suppress ftsA12 thermosensitivity at 42ºC (data not shown), ftsZL169R fully suppressed ftsA12 defects at an intermediate temperature of 37ºC. By comparison, ftsZWT under the same conditions could only weakly suppress ftsA12 at 37°C (Figure 3B, bottom row). Together these data support the idea that FtsZ rings formed by FtsZL169R are more stabilized compared to FtsZWT, helping to counterbalance instability elsewhere in the divisome.
Cells are more sensitive to FtsZL169R levels compared with FtsZWT
E. coli cell division is particularly sensitive to abnormally low or high levels of divisome proteins such as FtsA and FtsZ. Overproduction of FtsZ beyond a certain threshold leads to aberrant FtsZ assembly, including visible spiral structures, and reduces sensitivity to assembly inhibitors (Dai et al., 1994; Ma et al., 1996), similar to what is seen with FtsZL169R at baseline expression levels. At high enough levels, FtsZ ends up interfering with cell division, causing cell filamentation (Ward and Lutkenhaus, 1985; Dai and Lutkenhaus, 1992), possibly because of titration of other essential division proteins at limiting concentration, stabilization of filaments from high protein concentration, or both.
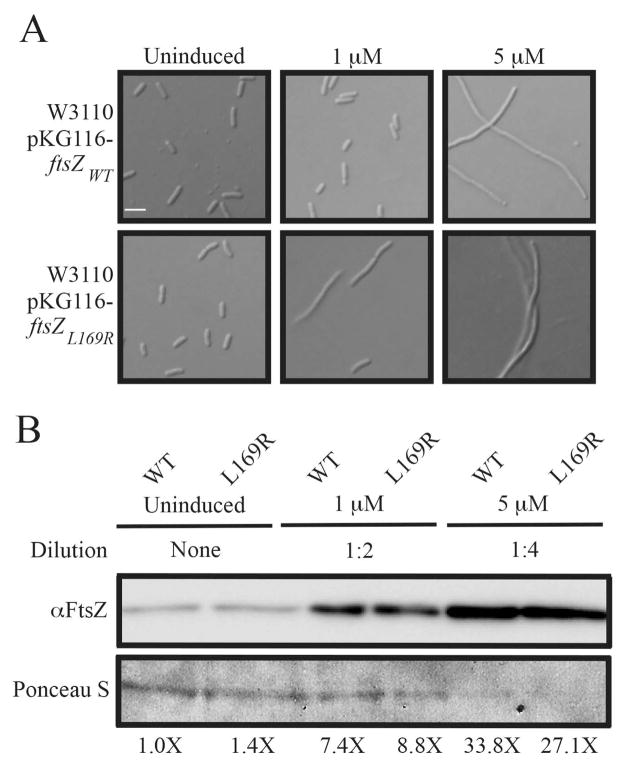
Given its localization and evidence of stabilized assembly, we therefore reasoned that cells would be even more sensitive to FtsZL169R levels than they are to FtsZWT. Overexpression of ftsZWT from pKG116 in a WT strain background caused no discernible defects in division at moderate induction conditions (1 μM sodium salicylate), but caused cell filamentation at higher induction (5 μM). In contrast, similar overexpression of ftsZL169R in a strain also containing ftsZL169R at the native locus caused cell filamentation even at moderate induction levels (1 μM) (Figure 4A). Immunoblots verified that FtsZWT and FtsZL169R levels in the cell population were comparable at each sodium salicylate concentration and were proportionately overproduced as expected (Figure 4B).
Figure 4.
Cells are more sensitive to FtsZL169R levels compared to FtsZWT. (A) Representative DIC micrographs of indicated strains grown to mid-exponential phase in the presence of indicated sodium salicylate concentrations to overexpress the given ftsZ allele. Scale as in Figure 1A. (B) Immunoblot of FtsZ protein levels from mid-exponential cultures of WT or ftsZL169R cells in the presence of the indicated sodium salicylate concentrations, with estimated relative band intensities. Uninduced and 1 μM induced samples were normalized to Ponceau-S-stained loading controls as in Figure 1E. Note that induced samples required 2 and 4-fold dilution to maintain the FtsZ immunostaining in the linear range; because of this, the intensity of Ponceau staining of protein in the 5 μM samples is low.
Excess WT FtsA is not toxic in cells with FtsZL169R and suppresses division defects
The FtsAR286W (FtsA*) allele was the prototype for what seems now to be a number of different pathways to bypass the requirement for ZipA, and remains one of the strongest ZipA bypass alleles (Geissler et al., 2003; Pichoff et al., 2012). Like FtsZL169R, FtsAR286W suppresses the thermosensitivity of the ftsQ1(ts) and ftsK44(ts) alleles (Geissler and Margolin, 2005). Because of their similar suppression abilities, we chose to investigate the effects of these two similar gain-of-function mutations in combination.
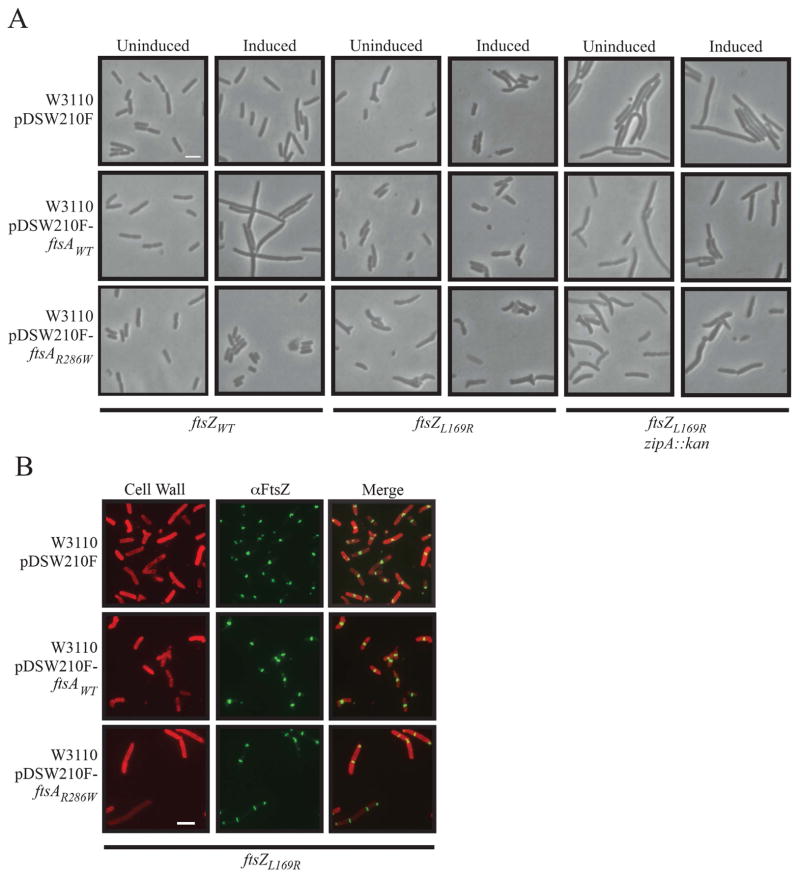
In WT E. coli (ftsZWT), overexpression of ftsA has the same phenotype as ftsZ overexpression, a cell division block leading to filamentation and death (Figure 5A, left pair of columns; Table 1). This is because the proper ratio of WT FtsZ to WT FtsA in the cell is important for divisome function (Dai and Lutkenhaus, 1992; Dewar et al., 1992). In contrast, ftsAR286W overexpression was nontoxic, with normal cell division, as expected (Figure 5A, left pair of columns; Table 1) (Shiomi and Margolin, 2007a). In striking contrast, in the presence of chromosomal ftsZL169R, cells became resistant to overexpression of ftsA and sensitive to ftsAR286W (Figure 5A, middle pair of columns; Table 1). This suggests that FtsZL169R stabilizes FtsZ rings and protects them from the deleterious effects of ftsA overexpression.
Figure 5.
Effects of FtsZ or FtsZL169R on different expression levels of ftsA or ftsAR286W in the presence or absence of zipA. Scale bars as in Figure 1A. (A) Representative phase contrast micrographs of WT (W3110), ftsZL169R, or ftsZL169R zipA::kan backgrounds with empty pDSW210F or with plasmid expressing ftsAWT or ftsAR286W under uninduced (no IPTG) or induced (0.5 mM IPTG) conditions. See Table 1 for quantification of these images. (B) Representative IFM images of W3110 ftsZL169R cells with empty pDSW210F or with plasmid expressing ftsAWT or ftsAR286W (0.5 mM IPTG). Staining and scale are the same as in Figure 1A.
Intriguingly, the delayed cell division/filamentation phenotype observed following zipA::kan transduction of an ftsZL169R background was also partially suppressed by overexpression of ftsAWT, but not ftsAR286W (Figure 5A, right pair of columns; Table 1). This indicates that not only is FtsZL169R stabilized against ftsA overexpression effects, but also the additional FtsA actually improves the partly deficient cell division of ftsZL169R zipA::kan cells. Higher levels of WT FtsA were also able to suppress the cell shape abnormalities of ftsZL169R cells that contained zipA, whereas higher levels of FtsAR286W significantly worsened those abnormalities, even in the absence of zipA (Table 1).
To determine if the effects of excess FtsA or FtsAR286W on cell length and shape correlated with effects on FtsZL169R ring morphology we repeated IFM on ftsZL169R cells with overexpressed ftsAWT or ftsAR286W. Whereas overexpression of ftsAR286W caused increased cell length with the abnormal FtsZL169R localization (Figure 5B, bottom) previously observed for this FtsZ mutant (Figure 1, Figure 5, top), overexpression of ftsAWT suppressed a significant amount of abnormal FtsZL169R localization (Figure 5B, middle). For example, only 53.4% of ftsZL169R cells with empty plasmid contained normal FtsZ rings, whereas 76.2% of ftsZL169R cells with overexpressed ftsAWT displayed normal FtsZ ring localization and shape.
Evidence that self-interaction of FtsA is not significantly disrupted when bound to FtsZL169R
Thus far, we have assumed that the observed phenotypes for the ftsZL169R allele are a result of altered behavior of the FtsZ mutant protein. It remains formally possible, however, that FtsZL169R interacts differently with FtsA, causing FtsA to behave more like the FtsAR286W allele. This would be consistent with the ZipA bypass and suppression of ts mutants. Such a situation could conceivably occur if the FtsZL169R mutant interacted particularly strongly with FtsA monomers, upsetting any balance with FtsA polymers and favoring a monomeric cellular FtsA status as FtsAR286W is believed to manifest (Pichoff et al., 2012).
To test whether FtsZL169R disrupted normal FtsA polymer balance in vivo, we used a mutant of ftsA, ftsAW408E, that has a defective C-terminal amphipathic helix (Pichoff and Lutkenhaus, 2005). FtsAW408E retains its ability to localize at the FtsZ ring, but has a reduced capacity to bind to the cytoplasmic membrane and fails to function in cell division (Shiomi and Margolin, 2008). When overproduced, FtsAW408E and other FtsA proteins with defective C-terminal amphipathic helix domains form large cytoplasmic bundled polymers inside cells, visible as axial rods or bars (Pichoff and Lutkenhaus, 2005; Herricks et al., 2014). In contrast, FtsAWT or FtsA derivatives that are defective in both membrane targeting and in self-interaction no longer form visible bars. Therefore, FtsA bar formation has served as a useful in vivo assay for FtsA-FtsA interactions (Pichoff et al., 2012).
In this case, if FtsZL169R induced FtsA to become more monomeric (like FtsAR286W), then the idea is that overproduced FtsAW408E should not form bars, or form them to a lesser extent, in ftsZL169R cells. Using IFM with α-FtsA, we found that ftsAW408E overexpression led quickly to FtsAW408E self-assembly into bars and eventually to cell curling as previously seen in the presence of WT FtsZ (Rico et al., 2004) (Figure S2A, top two rows). Notably, this behavior was indistinguishable in FtsZL169R-harboring cells (Figure S2A, bottom two rows), suggesting that FtsA polymer balance was not significantly affected by the presence of FtsZL169R. As expected, control cells without IPTG induction of ftsAW408E showed normal FtsA bands at midcell (Fig. S2A), and no bars or cell curling were detected upon overexpression of a W408E derivative of ftsAR286W (data not shown).
Though not previously reported, this aberrant FtsA self-assembly forces both FtsZWT and FtsZL169R (which are at native levels) into similar bar shapes (Figure S2B) that were easier to see than the FtsA bars and clearly distinct from the FtsZ rings in uninduced cells. The recruitment of FtsZWT or FtsZL169R to the FtsAW408E bars was strong enough to disrupt normal FtsZ rings, which were rarely observed once the bars were present. This indicates that these FtsA bars, while defective for membrane binding, are still able to interact efficiently with FtsZ. Although this assay does not directly measure FtsA-FtsA interactions at the FtsZ ring, it does argue against the possibility that FtsA-FtsA interactions are affected significantly by contact with FtsZL169R.
Purified FtsZL169R demonstrates evidence of enhanced bundling activity
Taken together, our in vivo data are consistent with the hypothesis that FtsZL169R stabilizes FtsZ assembly, which protects it from cellular disassembly factors and allows cells to survive or function properly in the absence of other divisome stabilizing factors. As mentioned above, these stabilizing factors include ZipA and the Zap proteins, all of which are proposed to contribute to the stabilization of FtsZ filaments by increasing protofilament bundling.
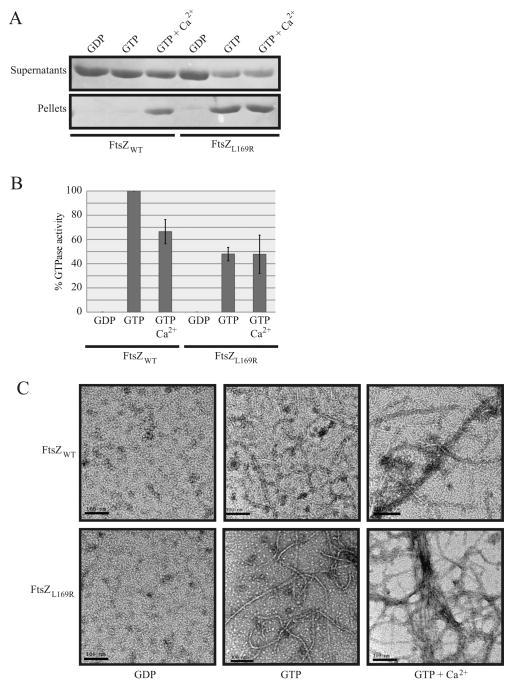
To test this idea directly, we expressed and purified native FtsZL169R to assay its in vitro assembly for evidence of enhanced bundling compared to purified FtsZWT. As expected, in our buffer conditions, significant sedimentation of bundled FtsZWT polymers only occurred in the presence of GTP to induce FtsZ polymerization plus millimolar calcium as a bundling agent (Figure 6A, left). Strikingly, under the same conditions, FtsZL169R polymer bundles formed and sedimented with the addition of GTP alone (Figure 6A, right), and addition of calcium led to no discernable increase in sedimentation.
Figure 6.
FtsZL169R displays evidence of enhanced bundling in vitro compared to FtsZWT. (A) Coomassie-stained gel of supernatant or pellet fractions from sedimentation reactions at 30ºC containing 5 μM purified FtsZWT or FtsZL169R assembled with added components as indicated. (B) Relative rate (%) of GTP hydrolysis activity for purified FtsZWT or FtsZL169R assembled with given components at 30ºC. The FtsZWT rate in GTP is normalized to 100%. Error bars indicate standard deviation between three replicate experiments. (C) Representative electron micrographs of purified, negatively-stained FtsZWT or FtsZL169R in the presence of 1mM GDP, GTP, or GTP plus 10 mM CaCl2. Scale bars = 100 nm.
It is thought that packing of individual FtsZ protofilaments into a bundle lowers FtsZ’s GTP hydrolysis by reducing subunit turnover into new protofilaments (Yu and Margolin, 1997; Mukherjee and Lutkenhaus, 1998; Mukherjee and Lutkenhaus, 1999). If FtsZL169R has increased bundling capability, then it should have a lower GTP hydrolysis rate compared to the WT protein. Measurements of GTP hydrolysis by purified FtsZ variants using a regeneration assay system support this idea (Buske and Levin, 2012). As expected, addition of millimolar calcium to FtsZWT in the presence of GTP led to a lower baseline hydrolysis rate (Figure 6B). However, the GTP hydrolysis rate for FtsZL169R was about half of the FtsZWT rate, and as seen with sedimentation reactions, addition of calcium to induce bundling did not further reduce FtsZL169R GTPase activity (Figure 6B). This supports the idea that FtsZL169R protofilaments become highly bundled in the presence of GTP alone.
Finally, the bundling state of FtsZL169R compared to FtsZWT was addressed directly by observing purified proteins assembled on grids by transmission electron microscopy. As expected, FtsZWT did not form detectable negatively stained filaments in GDP, but assembled into mostly single protofilaments in GTP and visible bundles of protofilaments in the presence of millimolar calcium (Figure 6C). Like FtsZWT, FtsZL169R did not form detectable filaments with GDP. With GTP, however, FtsZL169R mostly formed filaments that were double (~10 nm) the width of the ~5 nm wide single protofilaments formed by FtsZWT (Figure 6C). As with FtsZWT, calcium induced significant bundling of FtsZL169R filaments. Taken together with the sedimentation data, these results strongly support the idea that FtsZL169R promotes lateral interactions between FtsZ protofilaments.
Discussion
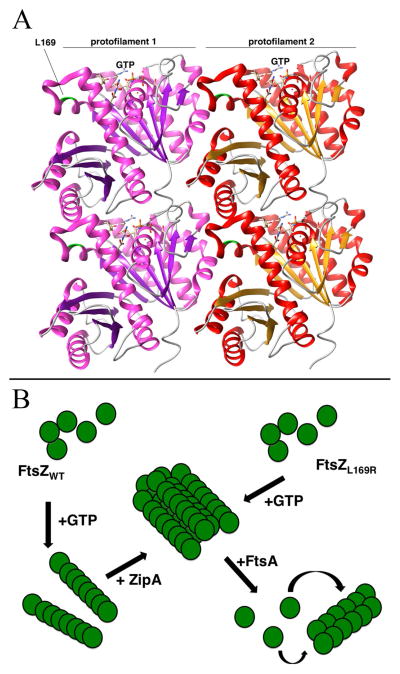
The ability of FtsZL169R to bypass ZipA, suppress several divisome deficiencies, and resist the effects of FtsZ inhibitors such as Kil and MinCD suggests that this allele stabilizes FtsZ assembly. The increased sedimentation and decreased GTP hydrolysis of purified FtsZL169R, along with its strong tendency to assemble into double filaments and polymer bundles as visualized by electron microscopy, suggest that the L169R lesion stabilizes FtsZ assembly by promoting lateral interactions between FtsZ subunits (Figure 7B). The location of L169 at the side of an FtsZ subunit, away from the GTP binding site and protofilament interface (Haeusser et al., 2014), is consistent with this idea (Figure 7A). We propose that the change of L169 to arginine may form a new electrostatic bridge with an acidic residue on an FtsZ subunit within an adjacent protofilament. The trans-dominant phenotype of FtsZL169R supports this model, as a relatively small fraction of FtsZ subunits capable of increased lateral interaction would be predicted to increase bundling when incorporated into FtsZWT protofilaments. Increased FtsZ bundling in vivo is also supported by the higher levels of FtsZL169R observed in the FtsZ ring compared with the cytoplasm, relative to FtsZWT.
Figure 7.
Models for the effects of the L169R lesion on FtsZ protofilament interactions. (A) Potential structure of an FtsZ double protofilament, highlighting the position of L169 near the lateral interaction surface between two protofilaments and distal from the GTP binding site that is near the longitudinal interaction surface. The crystal structures from Pseudomonas FtsZ were manipulated with the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera) developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). The protofilament alignment was based on the atomic structures of FtsZ protofilaments (Li et al., 2013). (B) Scheme to compare assembly of FtsZWT or FtsZL169R into higher-order structures at the FtsZ ring. Upon binding to GTP, FtsZWT assembles into single protofilaments that are then bundled by the action of ZipA and Zap proteins (not shown). In contrast, upon binding to GTP, FtsZL169R assembles into protofilament bundles, reducing the need for additional bundling proteins. In both cases, FtsA acts as a counterbalance to FtsZ protofilament bundling, perhaps by destabilizing protofilament bundles, This putative de-bundling activity of FtsA normally inactivates FtsZ rings, but more highly bundled FtsZL169R rings are resistant.
Other FtsZ residues have been implicated in lateral interactions. E83Q and R174D lesions, both also located on the side of the FtsZ subunit, caused decreased FtsZ polymer bundling and poor cell division, suggesting that the respective charged residues are important (Koppelman et al., 2004; Shin et al., 2013). R174, in particular, is very close to L169. Another lesion on the side of FtsZ, E93R, increased lateral subunit interactions in vitro, although it was not characterized in vivo (Jaiswal et al., 2010). To test whether it behaved like L169R, we cloned the mutation encoding E93R into the same plasmids used for our assays and found that it failed to bypass ZipA (Haeusser and Margolin, unpublished data). A chimeric FtsZ, containing all E. coli residues except for the C-terminal four replaced by the C-terminal six residues of B. subtilis FtsZ, is more proficient at protofilament bundling than native E. coli FtsZ (Buske and Levin, 2012). However, production of this chimera in our system did not bypass ZipA (Haeusser and Margolin, unpublished data). Although the sample size is small, this suggests that FtsZ protofilament bundling per se is not sufficient to bypass ZipA, and that the mechanism mediated by the L169R lesion may be specific.
It is notable that overproduction of FtsAWT was not toxic to ftsZL169R cells and improved the ability of FtsZL169R to bypass ZipA. In particular, FtsAWT largely corrects the prevalent abnormal FtsZL169R rings and cell division septa. Although FtsA* and some FtsA*-like mutant proteins can be overproduced with little toxicity (for reasons that are not yet clear), overproduction of FtsAWT normally prevents FtsZ rings from constricting unless FtsZ levels are concomitantly increased (Shiomi and Margolin, 2007a; Pichoff et al., 2012). One possible reason for the resistance of FtsZL169R to excess FtsA might be that FtsA stimulates removal of FtsZ subunits from bundles, which would balance the over-assembly caused by the L169R lesion, particularly when ZipA is also present to bundle FtsZ (Figure 7B). Such an activity for FtsA would be consistent with the ATP-dependent ability of FtsA* to decrease FtsZ polymer mass in vitro (Beuria et al., 2009) and the postulated ATP-dependent stimulation of FtsZ treadmilling by FtsAWT tethered on supported lipid bilayers (Loose and Mitchison, 2014). Nevertheless, it is not clear how increased de-bundling mediated by FtsA could help FtsZL169R ΔzipA cells divide more efficiently, or why FtsA* would not act similarly. Perhaps excess FtsA, which according to recent models oligomerizes more readily than FtsA*, anchors FtsZL169R bundles more efficiently to the membrane via oligomers than the more monomeric FtsA*, leading to more efficient ring constrictions (Szwedziak et al., 2014). As ZipA is normally twice as abundant in cells as FtsA (Rueda et al., 2003) and probably binds FtsZ more strongly than FtsA (Shen and Lutkenhaus, 2009; Rowlett and Margolin, 2014; Herricks et al., 2014), then excess FtsA might mimic the higher ZipA levels and increase membrane-anchoring capacity. Moreover, FtsA* may be deleterious to ftsZL169R cells because it stimulates the FtsZ ring prematurely to activate constriction (Tsang and Bernhardt, 2015), which would be particularly incompatible with a less dynamic FtsZ such as FtsZL169R.
How does FtsZL169R bypass ZipA? In one model, FtsA*-like lesions or overproduction of FtsN are proposed to bypass ZipA by stimulating FtsA monomerization, which allows release of FtsA’s subdomain 1c to recruit downstream divisome proteins and activate septum formation (Pichoff et al., 2014; Pichoff et al., 2012). One possible scenario consistent with this model is that increased lateral interactions between FtsZ protofilaments block FtsA from forming oligomers alongside FtsZ polymers (Szwedziak et al., 2012), which might result in more FtsA monomers able to recruit downstream proteins. However, the model fails to explain why additional FtsA* (but not FtsAWT) exacerbates the FtsZL169R defects and does not further help the ZipA bypass. The lack of any effect of ftsZL169R on the cytoplasmic bar formation by the truncated FtsA derivatives, albeit a negative result, also does not support this model.
In an alternative, but not mutually exclusive “FtsZ-centric” model, FtsZL169R bypasses ZipA simply by increasing protofilament bundling of FtsZ in the ring, which compensates for the bundling normally promoted by ZipA. This model suggests that ZipA’s main unique activity is to bundle FtsZ protofilaments, and that recruitment of downstream divisome proteins is an indirect effect of this bundling (Geissler et al., 2003). This model also suggests that under normal conditions, signaling from an active divisome regulates the level of FtsZ protofilament bundling itself. For example, the behavior of hypermorphic lesions in the FtsQLB complex hints that this complex normally feeds back on FtsZ to promote FtsZ bundling and induce ring constriction (Weiss, 2015; Tsang and Bernhardt, 2015). FtsA*-like lesions and FtsN binding to FtsA would do the same, possibly by changing the nature of FtsA-FtsZ interactions. Although the structure of the FtsZ ring and mechanisms of ring constriction remain controversial, this regulation of protofilament bundling would fit with the model that the ring becomes more condensed as it constricts (Lan et al., 2009). As has been true of FtsA hypermorphic mutants (Geissler et al., 2003; Bernard et al., 2007; Gerding et al., 2009; Dubarry et al., 2010; Potluri et al., 2012; Osawa and Erickson, 2013; Pazos et al., 2013), the FtsZL169R mutant should prove to be a useful tool for dissecting these signaling pathways and for reconstructing the divisome in vitro from minimal components (Martos et al., 2012).
Experimental Procedures
Strains and growth conditions
All E. coli strains used are listed in Table 2. Standard genetic methods including transformation and P1 vir transduction were used for strain construction.
Table 2.
Strains used in this study (grouped by function)
| Strain | Description | Source/reference |
|---|---|---|
| AG1111 | araDl39 Δ(ara-leu)7697 ΔlacX74 galU galK rpsL hsdR F′ proAB+ lacIq lacZM15 Tn10 | A. Grossman |
| WM1074 | TX3772 (MG1655: ilvG rpb-50 rph-1 ΔlacU169) | Lab strain |
| W3110 | rpoSam rph-1 INV(rrnD-rrnE) | Lab strain |
| AW60 | DY330 λ[int::lacZ (sieB-ea10)<>cat cI857 Δ[cro-bio]] [pSIM18] ftsZL169R leuO::Tn10 | (Haeusser et al., 2014) |
| DPH642 | WM1074 ftsZL169R leuO::Tn10 | This study |
| DPH766 | W3110 ftsZL169R leuO::Tn10 | This study |
| PS223 | W3110 zipA1(ts) | (Pichoff and Lutkenhaus, 2002) |
| DPH767 | PS223 ftsZL169R leuO::Tn10 | This study |
| DPH936 | PS223 [pDH156] | This study |
| DPH937 | PS223 [pDH161] | This study |
| DPH764 | PS223 [pDH155] | This study |
| DPH938 | PS223 [pDH159] | This study |
| WM1657 | ftsAR286W zipA::aph (zipA::kan) | (Geissler et al., 2003) |
| DPH752 | DPH642 zipA::kan | This study |
| DPH789 | DPH766 zipA::kan | This study |
| DPH756 | DPH642 [pDH155] | This study |
| DPH762 | DPH756 zipA::kan | This study |
| DPH778 | WM1074 [pDH159] | This study |
| DPH791 | DPH778 zipA::kan | This study |
| JD257 | TB28 zapA::frt | (Durand-Heredia et al., 2012) |
| JD305 | TB28 zapA::frt zapC::aph | (Durand-Heredia et al., 2012) |
| DPH832 | JD257 ftsZL169R leuO::Tn10 | This study |
| DPH851 | JD345 ftsZL169R leuO::Tn10 | This study |
| WM1125 | WM1074 ftsZ84 | (Yu and Margolin, 2000) |
| DPH626 | WM1125 [pKG116] | This study |
| DPH787 | WM1125 [pDH161] | This study |
| DPH779 | WM1125 [pDH159] | This study |
| TOE1 | AB2497 ftsQ1(ts) thyA leu proA his thi argE lacy galK xyl mtl ara tsxr rpsL supE | (Begg et al., 1980) |
| WM4661 | WM1074 ftsQ1(ts) leuO::Tn10 | This study |
| DPH801 | WM4661 [pKG116] | This study |
| DPH802 | WM4661 [pDH161] | This study |
| DPH803 | WM4661 [pDH159] | This study |
| TOE44 | AB2497 ftsK44(ts) thyA leu proA his thi argE lacy galK xyl mtl ara tsxr rpsL supE | (Begg et al., 1995) |
| WM2101 | WM1074 ftsK44(ts) ycaD::Tn10 | This study |
| DPH798 | WM2101 [pKG116] | This study |
| DPH799 | WM2101 [pDH161] | This study |
| DPH800 | WM2101 [pDH159] | This study |
| WM1115 | WM1074 ftsA12(ts) | (Geissler et al., 2003) |
| DPH795 | WM1115 [pKG116] | This study |
| DPH796 | WM1115 [pDH161] | This study |
| DPH797 | WM1115 [pDH159] | This study |
| DPH786 | W3110 [pDH161] | This study |
| DPH783 | W3110 [pDH159] | This study |
| WM3493 | W3110 [pDSW210F] (AKA pWM2784) | This study |
| WM3496 | W3110 [pWM2785] | This study |
| WM3547 | W3110 [pWM2787] | This study |
| DPH836 | DPH766 [pDSW210F] (AKA pWM2784) | This study |
| DPH837 | DPH766 [pWM2785] | This study |
| DPH839 | DPH766 [pWM2787] | This study |
| DPH874 | DPH836 zipA::kan | This study |
| DPH875 | DPH837 zipA::kan | This study |
| DPH876 | DPH839 zipA::kan | This study |
| WM3239 | WM1074 [pWM3239] | This study |
| DPH940 | DPH642 [pWM3239] | This study |
| BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene1 ind1 sam7 nin5]) | Lab strain |
| WM971 | BL21(DE3) [pWM971] | (Corbin et al., 2007) |
| DPH781 | BL21(DE3) [pDH160] | This study |
Cells were grown in Luria-Bertani (LB; 1% tryptone, 0.5% yeast extract, 0.5% NaCl) medium at 30°C for ts strains under permissive conditions and 37°C (ftsA12) or 42°C (all others) under non-permissive conditions. All non-ts strains were grown at 37°C. Antibiotic concentrations were as previously described, and culture growth was monitored by optical density as previously reported (Haeusser et al., 2014) Detailed procedures in preparation for microscopic imaging are provided below.
Expression of cloned genes from vectors derived from pET11a (Novagen – EMD Millipore) was induced with 1 mM isopropyl-β-D-galactopyranoside (IPTG) (Fisher Scientific) and from pDSW-derived vectors with 0.5 mM IPTG. Gene expression from pKG-derived vectors (J.S. Parkinson, University of Utah) was induced with 0.1, 1.0, or 5.0 μM sodium salicylate (Mallinkrodt), as indicated in text and figure legends.
General DNA and protein manipulation and analysis, including spot titers, were performed as previously described (Haeusser et al., 2014).
Plasmid construction
All plasmids are listed in Table 3.
Table 3.
Plasmids used in this study
| Plasmid | Description | Source/reference |
|---|---|---|
| pKG110 | pACYC184 derivative containing the nahG promoter | J. Parkinson |
| pDH156 | pKG110-ftsZWT | This study |
| pDH155 | pKG110-ftsZL169R | This study |
| pKG116 | pKG110 derivative with stronger Shine-Dalgarno sequence | J. Parkinson |
| pDH161 | pKG116-ftsZWT | This study |
| pDH159 | pKG116-ftsZL169R | This study |
| pDSW208 | colE1 plasmid with slightly weakened Ptrc promoter | (Weiss et al., 1999) |
| pDSW210 | colE1 plasmid with significantly weakened Ptrc promoter | (Weiss et al., 1999) |
| pDSW210F | pWM2784 (pDSW210 with N-terminal flag epitope) | (Shiomi and Margolin, 2007b) |
| pWM2785 | pDSW210F-ftsAWT | (Shiomi and Margolin, 2007a) |
| pWM2787 | pDSW210F-ftsAR286W | (Shiomi and Margolin, 2007a) |
| pDSW208F | pDSW208 with N-terminal flag epitope | (Shiomi et al., 2008) |
| pWM3239 | pDSW208F-ftsAW408E | This study |
| pET11a | Expression vector | Novagen |
| pWM971 | pET11a-ftsZ | H. Erickson |
| pDH160 | pET11a-ftsZL169R | This study |
Cloning into plasmids
To clone ftsZWT or ftsZL169R into the salicylate-inducible plasmids pKG110 or pKG116, regions were amplified from chromosomal WM1074 or DPH642 DNA, respectively, using oligonucleotides DPH331 (GAATCATATGTTTGAACCAATGGAACTTACCAATG) and DPH332 (GAATGGATCCGTTCAACTCGTCGATACCGG). The amplified inserts and pKG vectors were digested with NdeI and BamHI (underlined sequence in above oligonucleotides), ligated together, and verified by sequencing. For protein overproduction, the ftsZL169R gene was cloned into pET11a using the same NdeI-BamHI fragment. The ftsAW408E gene was cloned into pDSW208F using the same XbaI-PstI fragments used for cloning other truncated ftsA genes, as described previously (Herricks et al., 2014).
Cell fixation, microscopy, and analysis
Cells were grown as previously reported (Haeusser et al., 2014). Overnight cultures were back diluted and allowed to recover from lag phase into exponential growth before being back diluted a second time to equivalent starting OD600s (~0.05 or 0.1). For experiments that required non-permissive growth or induced expression, these conditions were initiated upon this second back dilution. Following the second back dilution, cells were grown to mid-exponential phase (OD600 0.4–0.6) and then harvested for fixation or immediate live visualization by DIC or phase contrast on an Olympus BX60 microscope with a 100X oil objective.
Cell fixation, preparation for immunofluorescence microscopy, and imaging were done as previously described (Haeusser et al., 2014). Affinity-purified polyclonal rabbit α-FtsA (Herricks et al., 2014) was used as the primary antibody at 1:5000.
Images were processed and analyzed for ring frequency and cell length measurements using the ObjectJ extension (van der Ploeg et al., 2013) of ImageJ (Schneider et al., 2012) as previously described (Haeusser et al., 2014). Minicell lengths were added to the total measurement of corresponding parental cell length when still physically attached, but isolated minicells were not included in measurements.
Cell fixation and preparation for 3D-SIM, imaging, and analysis were done as reported previously (Rowlett and Margolin, 2014).
Immunoblot analysis
Cellular levels of FtsZ protein were measured with affinity-purified rabbit polyclonal α-FtsZ on immunoblots as previously described (Haeusser et al., 2014). Briefly, cell extracts were loaded by normalizing to OD600 at the time of harvest of mid-exponential cultures. Gels were stained with Ponceau S prior to blocking to provide images for loading controls. Immunoblot band intensities were quantified using ImageJ, normalizing loading controls to a low molecular weight segment of the Ponceau-S-stained gel image.
Protein biochemistry and electron microscopy
FtsZWT and FtsZL169R were purified by identical protocols, and sedimentation assays were performed as previously reported (Haeusser et al., 2014). Protein was stored and assays were performed in FtsZ buffer (50 mM MES pH 6.5, 50 mM KCl, 5 mM MgCl2, 1 mM EGTA, 10% sucrose). Relative GTP hydrolysis activities were determined from three replicate experiments on a Synergy Mx Microplate Reader (BioTek) using a continuous and regenerative coupled assay (Ingerman and Nunnari, 2005) as previously described (Small and Addinall, 2003; Buske and Levin, 2012), under the same reaction conditions as used in sedimentation assays.
For electron microscopy, FtsZWT or FtsZL169R (3 μM or 5 μM) were incubated with 1 mM GDP or 1 mM GTP in FtsZ polymerization buffer with 2.5 mM MgCl2 added at 30°C for 10 min. As with the sedimentation assay, 10 mM CaCl2 was added when required. Following incubation, 10 μl of each sample was placed on a glow-discharged formvar carbon coated nickel grid (Electron Microscopy Sciences), and incubated for 1 min. Filter paper was used to wick away excess sample, the grids were washed with a 5 μl drop of 1% uranyl acetate, and stained for 30 seconds with a 5 μl drop of 1% uranyl acetate. Stain was wicked away with filter paper and the grids were allowed to dry. Electron micrographs were captured with a 120-kV JEOL 1400 Transmission Electron Microscope equipped with a Gatan Orius CCD camera. Grids were imaged at 120,000x magnification. For reactions in GTP or GTP + CaCl2, 3 μM or 5 μM protein were used with similar results; corresponding EM images in Figure 6C were from reactions with 3 μM protein. 5 μM protein was used for GDP reactions (also shown in Figure 6C) to increase the chances that any FtsZ assembly could be visualized.
Supplementary Material
Acknowledgments
The authors are grateful to the Department of Microbiology and Molecular Genetics, particularly Peter Christie’s and Kevin Morano’s labs, for shared resources. We wish to thank Donald Court’s lab for permission to continue study of their isolated mutant ftsZ alleles, and Anuradha Janakiraman’s lab for kindly providing the zapAC deletion strain. Danielle Guffey helped in statistical analysis of 3D SIM data, Archna Bhasin and Daisuke Shiomi with strain and plasmid construction, and Rahul Nagvekar with microscopy. This project was funded by grant GM61074 from the National Institutes of Health to W.M. and funds from the Graduate School of Biomedical Sciences to V.W.R.
References
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–53. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996a;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996b;22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Hatfull GF, Donachie WD. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol. 1980;144:435–7. doi: 10.1128/jb.144.1.435-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuria TK, Mullapudi S, Mileykovskaya E, Sadasivam M, Dowhan W, Margolin W. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J Biol Chem. 2009;284:14079–14086. doi: 10.1074/jbc.M808872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Biteen JS, Goley ED, Shapiro L, Moerner WE. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. Chemphyschem. 2010;13:1007–12. doi: 10.1002/cphc.201100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through Its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske PJ, Levin PA. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem. 2012;287:10945–57. doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, Hatem C, Xiao J. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89:1099–120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Geissler B, Sadasivam M, Margolin W. A Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol. 2007;189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;176:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Pichoff S, Lutkenhaus J, Wirtz D. Cross-linking FtsZ polymers into coherent Z rings. Mol Microbiol. 2010;78:651–68. doi: 10.1111/j.1365-2958.2010.07352.x. [DOI] [PubMed] [Google Scholar]
- Dewar SJ, Begg KJ, Donachie WD. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992;174:6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry N, Possoz C, Barre FX. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol Microbiol. 2010;78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol. 2012;194:3189–98. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to α, β, and γ tubulin. J Cell Biol. 1996;135:5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Friedrich E, Friedrich BM, Gov NS. FtsZ rings and helices: physical mechanisms for the dynamic alignment of biopolymers in rod-shaped bacteria. Phys Biol. 2012;9:016009. doi: 10.1088/1478-3975/9/1/016009. [DOI] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Hoashi M, Weaver A, Brown N, Pan J, Sawitzke JA, et al. The Kil peptide of bacteriophage lambda blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 2014;10:e1004217. doi: 10.1371/journal.pgen.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herricks JR, Nguyen D, Margolin W. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol Microbiol. 2014;94:713–727. doi: 10.1111/mmi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KH, Durand-Heredia J, Janakiraman A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol. 2013;195:1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
- Jaiswal R, Patel RY, Asthana J, Jindal B, Balaji PV, Panda D. E93R substitution of Escherichia coli FtsZ induces bundling of protofilaments, reduces GTPase activity, and impairs bacterial cytokinesis. J Biol Chem. 2010;285:31796–805. doi: 10.1074/jbc.M110.138719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppelman CM, Aarsman ME, Postmus J, Pas E, Muijsers AO, Scheffers DJ, et al. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol Microbiol. 2004;51:645–57. doi: 10.1046/j.1365-2958.2003.03876.x. [DOI] [PubMed] [Google Scholar]
- Lan G, Dajkovic A, Wirtz D, Sun SX. Polymerization and bundling kinetics of FtsZ filaments. Biophys J. 2008;95:4045–4056. doi: 10.1529/biophysj.108.132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. Condensation of FtsZ filaments can drive bacterial cell division. Proc Natl Acad Sci U S A. 2009;106:121–126. doi: 10.1073/pnas.0807963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Persons L, Lee L, de Boer P. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol. 2014 doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hsin J, Zhao L, Cheng Y, Shang W, Huang KC, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martos A, Jimenez M, Rivas G, Schwille P. Towards a bottom-up reconstitution of bacterial cell division. Trends Cell Biol. 2012;22:634–43. doi: 10.1016/j.tcb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Monahan LG, Beech PL, Harry EJ. Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J Bacteriol. 2006;188:1680–90. doi: 10.1128/JB.188.5.1680-1690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance J, Tadros M, Vicente M, Gonzalez JM, Rivas G, Velez M. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J Biol Chem. 2005;280:20909–20914. doi: 10.1074/jbc.M503059200. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva MA, Cordell SC, Löwe J. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol. 2004;11:1243–1250. doi: 10.1038/nsmb855. [DOI] [PubMed] [Google Scholar]
- Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci U A. 2013;110:11000–4. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos M, Natale P, Vicente M. A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J Biol Chem. 288:3219–26. doi: 10.1074/jbc.M112.434944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83:151–67. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Du S, Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol Microbiol. 2015;95:971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro O, Carmon G, Feingold M, Fishov I. Three-dimensional structure of the Z-ring as a random network of FtsZ filaments. Env Microbiol. 2013;15:3252–8. doi: 10.1111/1462-2920.12197. [DOI] [PubMed] [Google Scholar]
- van der Ploeg R, Verheul J, Vischer NOE, Alexeeva S, Hoogendoorn E, Postma M, et al. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol Microbiol. 2013;87:1074–1087. doi: 10.1111/mmi.12150. [DOI] [PubMed] [Google Scholar]
- Potluri LP, Kannan S, Young KD. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol. 2012;194:5334–42. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol Microbiol. 2004;53:1359–71. doi: 10.1111/j.1365-2958.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- Rico AI, Krupka M, Vicente M. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J Biol Chem. 2013;288:20830–6. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, Margolin W. Asymmetric constriction of dividing Escherichia coli cells induced by expression of a fusion between two Min proteins. J Bacteriol. 2014a;196:2089–2100. doi: 10.1128/JB.01425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, Margolin W. 3D-SIM super-resolution of FtsZ and Its membrane tethers in Escherichia coli cells. Biophys J. 2014b;107:L17–L20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda S, Vicente M, Mingorance J. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J Bacteriol. 2003;185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol. 2009;72:410–24. doi: 10.1111/j.1365-2958.2009.06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JY, Vollmer W, Lagos R, Monasterio O. Glutamate 83 and arginine 85 of helix H3 bend are key residues for FtsZ polymerization, GTPase activity and cellular viability of Escherichia coli: lateral mutations affect FtsZ polymerization and E. coli viability. BMC Microbiol. 2013;13:26. doi: 10.1186/1471-2180-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol. 2007a;66:1396–1415. doi: 10.1111/j.1365-2958.2007.05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W. The C-terminal domain of MinC inhibits assembly of the Z ring in Escherichia coli. J Bacteriol. 2007b;189:236–43. doi: 10.1128/JB.00666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W. Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol Microbiol. 2008;67:558–569. doi: 10.1111/j.1365-2958.2007.06085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si F, Busiek K, Margolin W, Sun SX. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys J. 2013;105:1976–86. doi: 10.1016/j.bpj.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E, Addinall SG. Dynamic FtsZ polymerization is sensitive to the GTP to GDP ratio and can be maintained at steady state using a GTP-regeneration system. Microbiology. 2003;149:2235–42. doi: 10.1099/mic.0.26126-0. [DOI] [PubMed] [Google Scholar]
- Small E, Marrington R, Rodger A, Scott DJ, Sloan K, Roper D, et al. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J Mol Biol. 2007;369:210–221. doi: 10.1016/j.jmb.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Erickson HP. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J Bacteriol. 2003;185:4796–805. doi: 10.1128/JB.185.16.4796-4805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99:3171–5. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Yu XC, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–504. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanedar S, Margolin W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr Biol. 2004;14:1167–1173. doi: 10.1016/j.cub.2004.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang MJ, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol. 2015;95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang MJ, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, Lutkenhaus J. Overproduction of FtsZ induces minicells in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Weiss DS. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Mol Microbiol. 2015;95:903–909. doi: 10.1111/mmi.12925. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J Bacteriol. 2000;182:6203–6213. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.