Abstract
Over 50 million hogs are raised annually in the United States for consumption, mostly on industrial hog operations (IHOs). Workers at IHOs are exposed to airborne particulates, zoonotic pathogens, and other workplace hazards, but lack of access to IHOs can hinder exposure assessment in epidemiologic studies. Here, we demonstrate the utility of pig-specific Bacteroidales (Pig-2-Bac) as a biomarker of exposure to pigs and pig waste and to help identify sources of Staphylococcus aureus carriage among IHO workers.
Keywords: Pig-specific fecal Bacteroidales, Quantitative polymerase chain reaction, Livestock-associated Staphylococcus aureus, Occupational exposure, Industrial hog operation
1. Introduction
Over 50 million hogs are raised annually in the United States for consumption, mostly on industrial hog operations (IHOs) (USDA, 2015). Workers at IHOs may be exposed to a variety of hazards in the workplace, including airborne particulates and zoonotic pathogens such as certain strains of antibiotic-resistant and livestock-associated Staphylococcus aureus (Nadimpalli et al., 2015b; Rinsky et al., 2013) and swine-origin influenza viruses (Gray et al., 2007). In Europe and other regions of the world, members of the public health research community have been able to sample livestock production workplace environments, animal herds and workers on-the-job to investigate conditions that can expose workers to zoonotic pathogens. However, limited access to IHOs in the US impedes direct exposure assessment and epidemiologic investigations of the sources of zoonotic pathogen exposure among US IHO workers. Instead, IHO workers’ self-reports of exposure are often used as indicators in epidemiological studies, however, workers’ perception of the frequency, intensity, and duration of microbial exposure may vary between workers and within workers over time, leading to exposure misclassification. An alternative method to identify occupational microbial exposure burdens is to develop exposure biomarkers using existing microbial source tracking methods. Microbial source tracking has been used to identify the sources of human and animal fecal pollution in the environment, primarily in water (Boehm et al., 2013). One biomarker that has been used to track swine fecal microbial contamination in surface water is Pig-2-Bac, a pig-specific Bacteroidales marker (Heaney et al., 2015; Mieszkin et al., 2009). Such biomarkers could provide a more objective estimate of microbial exposure than IHO workers’ self-reports and therefore could improve exposure assessment even in settings with complete access to the workplace.
We estimated associations between Pig-2-Bac isolated from nasal swabs collected among IHO workers and time since last work shift to determine whether Pig-2-Bac could be a useful biomarker of occupational exposure to pigs and pig waste. We then assessed associations between Pig-2-Bac and workers’ nasal carriage of a S. aureus gene (femA), a methicillin resistance gene (mecA), and culture-based livestock-associated vs. human-associated S. aureus nasal carriage status (persistent, intermittent, non-carrier).
2. Methods
Twenty-two IHO workers were enrolled between June–August, 2012 in a 14-day repeated measures study described previously (Nadimpalli et al., 2015b). Workers self-collected a dual-tipped nasal swab (CultureSwab™ Liquid Stuart, BD Diagnostics) on the evening of day 1, and each morning and evening on days 2–7 and 14. Up to 15 swabs were collected per IHO worker. Each time participants collected a nasal swab, they recorded collection time (morning or evening) and whether they worked that day or not. One swab of each pair was tested for culturable S. aureus using previously described methods (Nadimpalli et al., 2015b) and S. aureus isolates from nasal swabs were assessed for antibiotic susceptibility, spa type and absence of the scn gene to assess livestock vs. human association. Livestock association was defined by absence of scn (Price et al., 2012; Stegger et al., 2013).
2.1. Molecular analysis
DNA was extracted from nasal swabs using a Qiagen Viral RNA Mini kit (Valencia, CA), which co-purifies RNA and DNA. The spin protocol was used according to the manufacturer’s instruction with the exception that clipped nasal swabs were immersed in the recommended volume of lysis buffer during the lysis step (Nadimpalli et al., 2015a). Nucleic acid extracts were assessed by real-time qPCR for the S. aureus specific gene femA, the methicillin resistance gene mecA (Francois et al., 2003), and Pig-2-Bac (Mieszkin et al., 2009). Two μL of each nucleic acid extract were used as template in a 20 μL qPCR reaction containing 10 μL 2X reaction mix (TaqMan® Fast Advanced Master Mix, Applied Bio-systems, Foster City, CA), 500 nM forward and reverse primers and 250 nM dual-labeled probe. All samples were analyzed on a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA) using 40 amplification cycles. Each cycle consisted of a denaturation step (20 s at 95 °C), an annealing step (20 s at 56 °C for mecA and 60 °C for femA and Pig-2-Bac) and an amplification step (20 s at 60 °C). A quantitative standard curve using femA or mecA containing plasmid standards with known gene copy numbers was included in each femA and mecA qPCR. In each Pig-2-Bac qPCR we included Bacteroidales DNA isolated from a NC pig lagoon sample in duplicate as a positive control (CT = 31.0 ± 0.1). Additionally, non-template negative controls (nuclease-free water) were included in each qPCR. femA and mecA were quantified using the StepOne™ software version 2.3. For Pig-2-Bac, relative qPCR estimates were calculated based on the observed CT (cycle threshold) using the formula in Eq. (1).
| (1) |
2.2. Statistical analysis
Samples below the limit of detection for femA and mecA were assigned a value of ½ the lowest detectable qPCR estimate and for Pig-2-Bac were assigned a value of ΔCT = 1 (i.e. CT = 40). All qPCR estimates were log10 transformed. Unity-based normalization of the log10 transformed qPCR estimates was used to scale each participant’s qPCR estimates to a range between 0 and 1 using the formula in Eq. (2) (Aksoy and Haralick, 2001).
| (2) |
This normalization reduces the potential influences introduced by workers at the extremes of the qPCR target distribution – e.g., persistent S. aureus carriers (with high femA qPCR estimates) compared to non-carriers (with low femA qPCR estimates). We graphed associations between time since last IHO shift and Pig-2-Bac qPCR estimates by plotting the mean ± standard error (SE) of the log10 and unity-normalized qPCR estimates by days since last IHO shift. We also graphed associations of Pig-2-Bac with femA and mecA qPCR estimates by plotting the log10 mean ± SE and unity-normalized mean ± SE femA and mecA qPCR estimates by tertile of the Pig-2-Bac qPCR estimates. Graphs were produced with Matlab R2015a (The MathWorks, Inc., Natick, MA).
We used fixed effect linear regression models, which account for non-independence of repeated measurements within person (Allison, 2005), to estimate beta coefficients and SEs of associations between time since last IHO shift and Pig-2-Bac qPCR estimates and between Pig-2-Bac and femA and mecA qPCR estimates. Finally, we used multinomial logistic regression models to estimate odds ratios (OR) and 95% confidence intervals (CI) of the associations of (1) mean Pig-2-Bac qPCR estimates and (2) mean time since last work shift with livestock-vs. human-associated S. aureus carriage status (persistent and intermittent compared to non-carriage of culture-based S. aureus). For each type of S. aureus (i.e., livestock-associated S. aureus; human-associated S. aureus), respectively, IHO workers who carried at each or each minus one sampling point were classified as persistent carriers, workers who never carried were classified as non-carriers, and remaining workers were classified as intermittent carriers. The non-carrier category was used as the referent group in multinomial logistic regression analyses. Regression analyses were completed using SAS version 9.3 (SAS Institute, Cary, NC).
3. Results and discussion
Twenty-two North Carolina IHO workers provided 327 nasal swabs; 316 were dual swabs with one swab available for qPCR analysis. Swabs were collected each morning and evening, regardless of whether the participant worked that day. Most samples (76%) were collected <1 day after an IHO work shift (n= 221). Of those, 55% were collected in the evening after an IHO work shift and 45% were collected the following morning. Thirteen percent of samples (n= 39) were collected ≥1 and <2 days after an IHO work shift (after one day off), 9% (n= 26) were collected ≥2 and <3 days after an IHO shift (after two days off), and 2% of samples (n= 6) were collected ≥3 days after an IHO shift (after 3 or more days off).
3.1. Association of Pig-2-Bac with time since last work shift
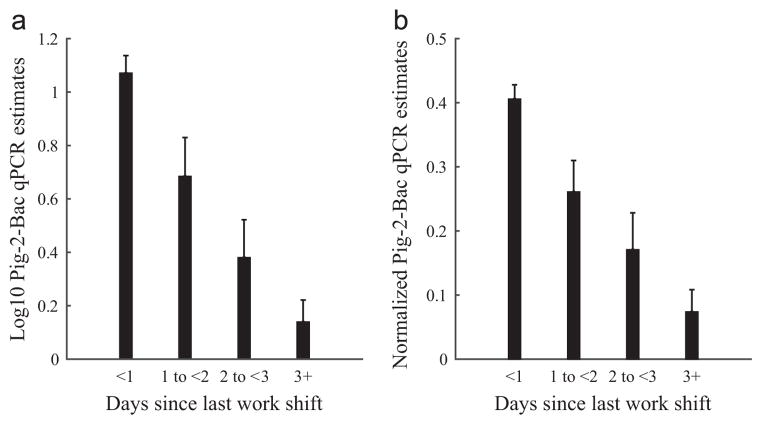
Pig-2-Bac was detected in 61% of nasal swab samples (mean ± SE Pig-2-Bac log10 ΔCT/swab = 1.5 ± 0.8). Pig-2-Bac mean log10 and normalized qPCR estimates declined with increasing time since IHO shift (Fig. 1). Fixed-effects linear regression modeling also showed that log10 Pig-2-Bac decreased 0.35 (SE: 0.07) ΔCT/swab per day since last work shift (Table 1a). This negative association suggests that Pig-2-Bac is a useful biomarker for human exposure to pigs or pig waste during IHO work.
Fig. 1.
Relation of time since last work shift with (a) log10 transformed and (b) normalized by participant qPCR estimates of Pig-2-Bac DNA ΔCT per nasal swab collected from 22 industrial hog operation workers in North Carolina, 2012. Note. qPCR= quantitative polymerase chain reaction. Error bars represent the standard error of the mean of the log10 transformed or normalized data, respectively.
Table 1.
Relation of days since last work shift with Pig-2-Bac qPCR estimates and of Pig-2-Bac with femA and mecA qPCR estimates among 22 industrial hog operation workers in NC, 2012.
| Na | β (SE) | t | p | |
|---|---|---|---|---|
| (a) Pig-2-Bac DNA ΔCT per nasal swab | ||||
| Days since last work shift | 289 | −0.35 (0.07)b | −5.14 | <0.01 |
| (b) femA DNA copy # per nasal swab | ||||
| Pig-2-Bac DNA ΔCT per nasal swab | 313 | 0.14 (0.06)c | 2.23 | 0.03 |
| (c) mecA DNA copy # per nasal swab | ||||
| Pig-2-Bac DNA ΔCT per nasal swab | 313 | 0.09 (0.04)c | 1.97 | 0.06 |
Beta coefficients are derived from fixed effects linear regression models. SE = standard error. CT = cycle threshold. ΔCT = 2(40 – swab CT). qPCR = quantitative polymerase chain reaction.
Ns represent the total number of observations included in the analysis for the days since last work shift (N missing = 24) and qPCR (N missing = 3) variables.
The beta coefficient is the one log10 unit change in the Pig-2-Bac ΔCT per nasal swab qPCR estimate for every one unit change in the days since last work shift variable (0–3 days).
The beta coefficient is the one log10 unit change in the femA or mecA qPCR estimate for every one log10 unit change in the Pig-2-Bac ΔCT per nasal swab qPCR estimate.
3.2. Association of Pig-2-Bac with S. aureus and methicillin resistance
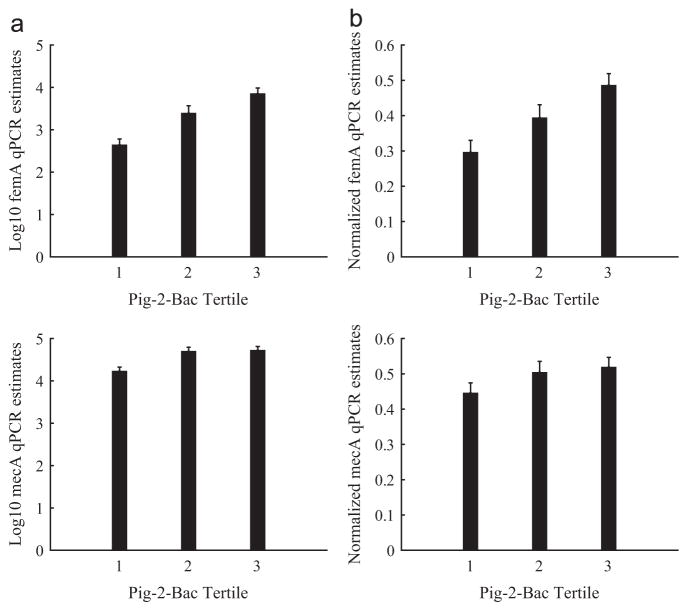
We next estimated associations of Pig-2-Bac with femA and mecA qPCR estimates. The femA gene was present in 67% of nasal swab samples (mean ± SE log10 copy number/swab = 4.2 ± 1.3) and mecA was present in 93% of nasal swab samples (mean ± SE log10 copy number/swab= 4.7 ± 0.9). Both mean log10 and unity-normalized femA and mecA qPCR estimates increased by Pig-2-Bac tertile, although mecA qPCR differences were less pronounced between the 2nd and 3rd tertile than femA differences (Fig. 2). Fixed-effects linear regression modeling also showed that femA and mecA increased by 0.14 (SE:0.06) and 0.09 (SE:0.04), respectively, for every one-unit change in Pig-2-Bac (Table 1b–c). These associations suggest that intensive contact with pigs or pig waste (reflected by high Pig-2-Bac estimates in the nares) may be the source of IHO worker exposure to S. aureus (femA) and methicillin resistance (mecA is carried by diverse bacteria).
Fig. 2.
Relation of Pig-2-Bac qPCR estimates with log10 transformed (a) and normalized (b) qPCR estimates of femA and mecA gene copy number per nasal swab collected from 22 industrial hog operation workers in North Carolina, 2012. Note. qPCR= quantitative polymerase chain reaction. Error bars represent the standard error of the mean of the log10 transformed or normalized data, respectively.
3.3. Association of Pig-2-Bac with livestock-versus human-associated S. aureus nasal carriage status
Finally, we examined whether mean Pig-2-Bac qPCR estimates (as a proxy for intensity of contact with pigs or pig waste during IHO work) and mean time since last work shift (as a proxy for time off from work) was associated with nasal carriage status of livestock- or human-associated S. aureus. In this study, 10 workers carried livestock-associated S. aureus strains persistently, and two carried human-associated S. aureus strains persistently. Analyses of associations of mean Pig-2-Bac qPCR estimates with livestock-associated S. aureus nasal carriage status (persistent, intermittent, non-carrier) showed higher odds ratios for intermittent (OR: 1.8; 95% CI: 0.2, 19.8) and persistent (OR: 12.8; 95% CI: 1.0, 160.6) carriage compared to non-carriage (Table 2a). We did not observe associations between mean Pig-2-Bac and human-associated S. aureus carriage status (Table 2a). We also did not see associations between mean time since last work shift and either livestock- or human-associated S. aureus carriage status (Table 2b). These findings indicate that nasal carriage of livestock-associated S. aureus may be related to intensity of contact with pigs or pig waste during IHO work (as reflected by associations in Table 2a) and that not all IHO work may contribute equally as a source of exposure to livestock-associated S. aureus (as reflected by no association with increasing mean time since last work shift; Table 2b). Although the ORs presented in Table 2 are based on 22 people and therefore are imprecise, the directions of the associations found are informative.
Table 2.
Relation of mean Pig-2-Bac qPCR estimates and mean time since last work shift with S. aureus nasal carriage status among 22 industrial hog operation workers in North Carolina, 2012.
| a) Pig-2-Bac
|
b) Days since last IHO shift
|
||||
|---|---|---|---|---|---|
| Nd | OR (95% CI) | p | OR (95% CI) | p | |
| S. aureus nasal carriage statusa | |||||
| Livestock-associatedb | |||||
| Non-carrier | 6 | ref | ref | ||
| Intermittent carrier | 6 | 1.8 (0.2, 19.8) | 0.60 | 1.0 (0.0, 66.6) | 0.99 |
| Persistent carrier | 10 | 12.8 (1.0, 160.6) | <0.05 | 2.0 (0.0, 80.0) | 0.72 |
| Human-associatedc | |||||
| Non-carrier | 16 | ref | ref | ||
| Intermittent carrier | 4 | 0.2 (0.0, 2.1) | 0.18 | 8.1 (0.1, 476.6) | 0.31 |
| Persistent carrier | 2 | 0.7 (0.1, 10.2) | 0.80 | 0.1 (0.0, 79.5) | 0.55 |
Note. CT = cycle threshold. ΔCT = 2(40 – swab CT). qPCR = quantitative polymerase chain reaction. OR = odds ratio. CI = confidence interval. Odds ratios were estimated using multinomial logistic regression models with non-carrier (0) as the referent group.
S. aureus nasal carriage status is the dependent variable, coded as 0=non-carrier, 1=intermittent carrier, 2=persistent carrier.
Defined as non-, intermittent or persistent nasal carriage of scn negative S. aureus strains
Defined as non-, intermittent or persistent nasal carriage of scn positive S. aureus strains
Number of non-, intermittent and persistent livestock- or human-associated S. aureus carriers in each category.
Limitations of this study include the lack of a referent group (non-IHO worker group without exposure to pigs) to evaluate the specificity of Pig-2-Bac as a biomarker of exposure to pigs and pigs waste in human biospecimens. However, the specificity of Pig-2-Bac as a source tracking marker for pig fecal contamination in water has been validated previously and found to be 100% specific (Mieszkin et al., 2009). Other limitations included the lack of the exact nasal swab collection time; exact time the last IHO work shift ended and information about specific IHO work activities performed each day. The time since last work shift variable was therefore binned into 4 categories: <1 day since last work shift, 1–<2 days, 2–<3 days and 3+ days after last work shift. Even though the categories of the time since last shift variable are somewhat broad, Pig-2-Bac estimates declined significantly with days since last work shift.
Although self-reported time since last work shift or job tasks may be useful indicators for recent IHO work exposure, time since last shift alone often may not accurately reflect the intensity of exposure to pigs or to pig waste during specific job duties. Quantitative Pig-2-Bac estimates derived from nasal swab samples provide a more objective measure of exposure to high concentrations of pig microbes on-the-job than self-reported time since last work exposure. Additionally, participants’ perception of the frequency, intensity, and duration of exposure to pigs or pig wastes on a specific day or during a specific job task may vary significantly and may lead to misclassification in epidemiological studies.
In summary, we found that pig-specific fecal Bacteroidales (Pig-2-Bac) is a useful biomarker of recent human occupational exposure to pigs or pig waste during IHO work. We showed that the application of Pig-2-Bac as a biomarker can provide evidence that the source of nasal carriage of S. aureus (femA gene) and methicillin resistance (mecA gene) may be intensive contact with pigs or pig waste at IHOs. We also showed that higher mean Pig-2-Bac qPCR estimates were associated with higher odds of persistent carriage of livestock-associated S. aureus. To our knowledge, this is the first study to apply Pig-2-Bac as a microbial source tracking marker for recent human exposure to pigs.
Although this study was small and the generalizability of its findings is unclear, its major strength is the repeated measures, which facilitate assessment of the temporality of a quantitative measure of a swine-specific microbial source tracking marker with both quantitative molecular and culture-based measures of S. aureus and antimicrobial resistance. The temporal dynamics we observed suggest that intensive contact with pigs can be a source of IHO worker S. aureus exposure relative to other potential S. aureus exposure sources at home or in the community.
Acknowledgments
Funding sources and Irb approval
Funding for this study was provided by the Johns Hopkins National Institute for Occupational Health and Safety (NIOSH) Education and Research Center (T42OH008428), the North Carolina Occupational Safety and Health and Education and Research Center (T42OH00867302), NIOSH Grant 1K01OH010193-01A1, a directed research award from the Johns Hopkins Center for a Livable Future, and NSF Grant 1316318 as part of the joint NSF-NIH-USDA Ecology and Evolution of Infectious Diseases program. NP was supported by the National Institute of Environmental Health Sciences (NIEHS) award no. 5T32ES007141-30. JLR was supported by NIEHS award no. T32ES007018 and the National Institute of Occupational Safety and Health award no. T42OH00867302. MN was supported by a Royster Society fellowship and an EPA Science to Achieve Results fellowship. CDH was supported by NIOSH grant 1K01OH010193-01A1. This study was approved by the UNC Public Health Nursing Institutional Review Board (IRB no. 12–0712).
This study would not have been possible without a strong partnership between researchers and community-based organizations that have the trust of members of communities in areas where the density of industrial hog operations is high. The authors would like to thank the workers who participated in this study. The authors would also like to acknowledge Norma Mejia, Paul Baker, and Sherri Basnight for assistance with data collection.
Footnotes
Competing interests
SW authored a report in a civil rights complaint against the NC Department of Environment and Natural Resources’ 2014 re-permitting of industrial hog operations in NC, and a report on behalf of plaintiffs in a civil case regarding impacts of IHOs on neighbors. He has not received or requested compensation in either matter. All other authors declare no competing interests.
References
- Aksoy S, Haralick RM. Feature normalization and likelihood-based similarity measures for image retrieval. Pattern Recognit Lett. 2001;22:563–582. [Google Scholar]
- Allison PD. Fixed effects regression methods for longitudinal data using SAS®. SAS Institute Inc; Cary, NC: 2005. [Google Scholar]
- Boehm AB, et al. Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study. Water Res. 2013;47:6812–6828. doi: 10.1016/j.watres.2012.12.046. [DOI] [PubMed] [Google Scholar]
- Francois P, et al. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J Clin Microbiol. 2003;41:254–260. doi: 10.1128/JCM.41.1.254-260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GC, et al. Swine workers and swine influenza virus infections. Emerg Infect Dis. 2007;13:1871–1878. doi: 10.3201/eid1312.061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney CD, et al. Source tracking swine fecal waste in surface water proximal to swine concentrated animal feeding operations. Sci Total Environ. 2015;511:676–683. doi: 10.1016/j.scitotenv.2014.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieszkin S, et al. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific bacteroidales 16S rRNA genetic markers. Appl Environ Microbiol. 2009;75:3045–3054. doi: 10.1128/AEM.02343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadimpalli M, et al. Equivalence of influenza a virus RNA recovery from nasal swabs when lysing the swab and storage medium versus storage medium alone. J Virol Methods. 2015a;217:14–17. doi: 10.1016/j.jviromet.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadimpalli M, et al. Persistence of livestock-associated antibiotic-resistant Staphylococcus aureus among industrial hog operation workers in North Carolina over 14 days. Occup Environ Med. 2015b;72:90–99. doi: 10.1136/oemed-2014-102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LB, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;3 doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinsky JL, et al. Livestock-associated methicillin and multidrug resistant Staphylococcus aureus is present among industrial, not antibiotic-free livestock operation workers in North Carolina. PLoS One. 2013;8:e67641. doi: 10.1371/journal.pone.0067641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegger M, et al. Rapid Differentiation between Livestock-Associated and Livestock-Independent Staphylococcus aureus CC398 Clades. PLoS One. 2013;8:e79645. doi: 10.1371/journal.pone.0079645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. Quarterly Hogs and Pigs. Vol. 2015. United States Department of Agriculture; Washington, D.C: 2015. [Google Scholar]