Abstract
Protein engineering of microbial rhodopsins has been successful in generating variants with improved properties for applications in optogenetics. Members of this membrane protein family can act as both actuators and sensors of neuronal activity. Chimeragenesis, structure-guided mutagenesis, and directed evolution have proven effective strategies for tuning absorption wavelength, altering ion specificity and increasing fluorescence. These approaches facilitate the development of useful optogenetic tools and, in some cases, have yielded insights into rhodopsin structure-function relationships.
Keywords: proton pumps, synthetic biology, channelrhodopsin, opsins, bioelectricity
Introduction
Optogenetics refers to the ability to control or monitor cellular activities with light (‘opto’) using genetically encoded machinery (‘genetics’). For nearly a decade, a major focus has been neuroscience. Light-activated microbial rhodopsins can be transgenically expressed in neurons to reversibly control and sense neural activity with relevant speed and precision [1]. Coupling targeted perturbations stimulated by light to specific readouts (e.g., behavioral phenotypes or electrical recordings) enables the functional dissection of neural circuits [2-4]. Certain rhodopsins can also function as fluorescent voltage indicators providing optical detection of neuronal activity (and perhaps other electrically active cell types) [5-7]. Unfortunately, rhodopsins have broad activation spectra, making multiplexed control of cells with various light colors challenging, and current fluorescent variants are extremely dim, which limits the scope of a potential “all-optical electrophysiology” [8-11]. Overcoming these challenges by improving rhodopsin-based tools has and will continue to require various elements of protein engineering. In this review, we present examples of how protein engineering has enhanced specific rhodopsin functions for applications in optogenetics. Specifically, we describe how rhodopsin actuators and sensors have been engineered and what limitations remain.
Rhodopsins are a family of light-activated integral membrane proteins that adopt a seven trans-membrane α-helical fold referred to as the G protein-coupled receptor fold. The polyene chromophore retinal is covalently attached to the ε-amino group of a conserved lysine residue on the seventh α-helix through a protonated Schiff base (PSB) linkage [12]. In microbes, rhodopsins can act as receptors that change conformation in response to light to trigger intracellular signaling, as pumps that drive protons or chloride ions across the cell membrane, or as non-specific cation channels [13].
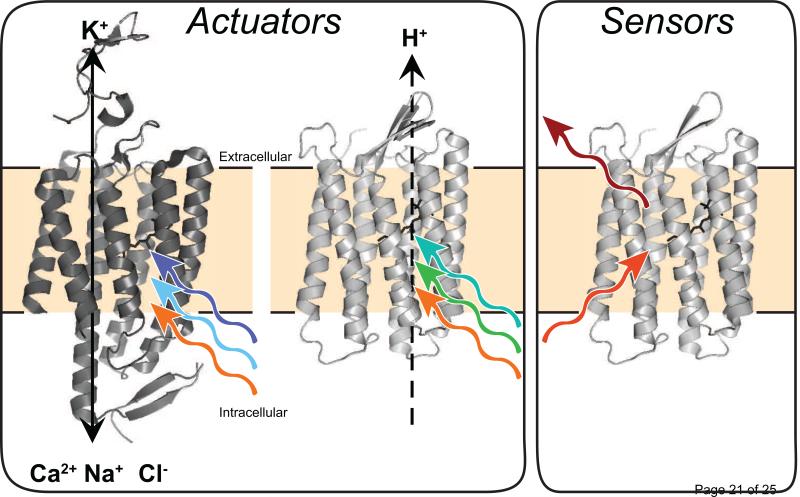
Microbial rhodopsin pumps and channels are widely used for optogenetic applications. Light-triggered isomerization of retinal from all-trans to 13-cis initiates the rhodopsin photocycle and ultimately results in the movement of ions across the membrane [12]. When transgenically expressed in neurons, channelrhodopsins (ChRs) mediate light-dependent transport of cations into the cell, causing depolarization and stimulation of action potentials [1,14-17]. In contrast to the excitatory ChRs, both proton- and chloride-pumping rhodopsins can be used to selectively hyperpolarize the cell and inhibit action potentials through either pumping protons out or pumping chloride into the cell [1,18]. Collectively, these tools facilitate genetically targeted, reversible loss and gain of function experiments in vivo. Since these proteins allow light-dependent ‘actuation’ of neuronal activity, we refer to them as actuators (Figure 1, Table 1).
Figure 1. Rhodopsins can be used as actuators and sensors in optogenetics.
Actuators transport ions across the membrane to activate or repress neuronal activity. ChRs transport positively charged ions into the cell, while proton-pumping rhodopsins (PPRs) move protons out of the cell. In the ideal case, engineered rhodopsin sensors emit light as fluorescence in the farred in a voltage-dependent fashion.
Table 1. Comparison of engineered rhodopsin actuators for a number of relevant characteristics and engineering methods.
Rhodopsin molecules are functionally classified as either ‘excitatory’ or ‘inhibitory’. The rhodopsin actuators are compared for: optimal wavelength for photocurrent excitation (λmax), ion specificity, kinetic off rate (τoff) indicating how quickly the molecule closes once light stimulation is turned off, and reversal potential. The engineering approach is briefly described.
| Rhodopsin | Function | λmax nm | Ion Specificity | Toff ms | Reversal Potential mV | Distinguishing Properties | Engineering Approach | Ref |
|---|---|---|---|---|---|---|---|---|
| ChR2 | Excitatory | 480 | none | ~10 | ~0 | Most commonly used ChR | Native | [17] |
| ReaChR | Excitatory | 589 | none | 137.2±7.1 | 7±4 | Red shifted ChR | Chimera of ChR1, VChR1 and VChR2 with mutation | [27] |
| Chronos | Excitatory | ~500 | none | 3.6±0.3 | NR | Fast kinetics and sensitive to low intensity light | Native: de novo transcriptome sequencing | [45] |
| ChrimsonR | Excitatory | ~600 | none | 15.8±0.4 | NR | Red-shifted ChR | Native: de novo transcriptome sequencing with single amino acid substitution to improve kinetics | [45] |
| CheRiff | Excitatory | ~460 | none | 16±0.8 | NR | Very sensitive to low intensity light | Native: de novo transcriptome sequencing with additional point mutation to improve kinetics and trafficking sequence to improve membrane localization | [9] |
| iC1C2 | Inhibitory | 480 | Cl- | 24 | −64 | ChR with decreased reversal potential and increased Cl- ion specificity | Structure based site-directed mutagenesis of ChR2 | [36] |
| SwiChRCT | Inhibitory | 480 | Cl- | ~7300 | −61 | Slow channel closure version of iC1C2 | iC1C2 with single amino acid substitution | [36] |
| ChloC | Inhibitory | ~480 | Cl- | NR | −61.8±1.0 | ChR with decreased reversal potential and increased Cl- ion specificity | Mutagenesis of C1C2 based off of molecular dynamics simulations based off of structure | [37] |
| Slow ChloC | Inhibitory | ~480 | Cl- | 10500±300 | −68.2±0.8 | Slow channel closure version of ChloC | ChloC with single amino acid substitution | [37] |
| Jaws | Inhibitory | 600 | Cl- | ~7 | NR | Red-shifted inhibitory chloride pump | Native with amino acid substitutions for improved photocurrents and added trafficking sequences for improved localization | [44] |
Over the past few years, several proton-pumping rhodopsins have been identified that exhibit weak fluorescence that is sensitive to changes in the local electronic environment (e.g., changes in pH and trans-membrane voltage) [5-7]. One proton pumping rhodopsin, Archaerhodopsin-3 (Arch) from Halorubrum sodomense, has been extensively characterized in mammalian neurons for both light-activated proton pumping and voltage sensitive fluorescence [5,8,9,19]. Wild-type Arch transports protons in response to light used to excite opsin fluorescence (635 – 655 nm). This activity can be attenuated or eliminated by introducing mutations at residues known to be critical for pumping [5,8,9,19], thereby creating a tool for voltage sensing independent of hyperpolarization. We refer to these rhodopsin variants as sensors (Figure 1, Table 2).
Table 2. Comparison of engineered rhodopsin based fluorescent voltage sensors for a number of relevant characteristics and engineering methods.
Rhodopsin-based voltage sensors are far red-shifted from most fluorescent voltage sensors so we report peak excitation wavelength, λexc, and peak emission wavelength, λem, for each fluorescent voltage sensor. Pumping refers to whether or not the engineered molecule, derived from a functional proton pump, still pumps protons. The on kinetics of the voltage sensors, ton, is the time to the maximum fluorescent level after a step in voltage from −70 to +30 mV. Quantum yield is a metric of the molecule's brightness. The %ΔF/F in response to an action potential (AP) is the maximum change in fluorescence from the baseline observed with a single AP divided by the baseline fluorescence. %ΔF/F is used as a metric for the voltage sensor's sensitivity to voltage changes.
| Rhodopsin | λexc nm | λem nm | Pumping | ton ms | Quantum yield | %ΔF/F in response to AP | Distinguishing Properties | Engineering Approach | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Arch | 556 | 687 | Yes | ~0.5-1 | 1×10−4 | ~2% | First identified rhodopsin with voltage sensitive fluorescence | Native with trafficking sequences to improve membrane localization | [5,49] |
| Archer | 626 | 731 | Minimal | ~1 | 3×10−3 | 25-40% | Minimally active at fluorescence excitation wavelength. Fast fluorescence response to voltage changes, red-shifted excitation and large ΔF/F | Mutagenesis around the retinal binding pocket of Arch with trafficking sequences to improve membrane localization | [8] |
| QuasAr1 | 590 | 715 | None | ~0.1 | 8×10−3 | 21% ± 2% | Strong baseline fluorescence and very fast fluorescence response to voltage changes. No residual pumping activity | Random mutagenesis of Arch with trafficking sequences to improve membrane localization | [9] |
| QuasAr2 | 590 | 715 | None | ~2 | 4×10−3 | 48% ± 3% | Large ΔF/F. No residual pumping activity | Random mutagenesis of Arch with trafficking sequences to improve membrane localization | [9] |
| MacQ-mOrange2 | 530 | 565 | Minimal | ~5 | NR | 7.2±0.4% | Bright baseline fluorescence. Minimal residual pumping activity | Fused Mac to mOrange2 with additional mutations in Mac to eliminate pumping. Improved membrane localization with trafficking signals | [7] |
Spectral Tuning of Microbial Rhodopsins
Microbial rhodospsin actuators from nature are optimally activated by light in the range of 450 – 570 nm. The absorption maximum of rhodopsin is determined by the energy gap between the resting state (S0) and excited state (S1) of the retinal chromophore. Narrowing or increasing the S0-S1 energy gap results in blue or red shifts, respectively. Stabilization of these states is governed by interactions between the protein and retinal, which itself is surrounded by a hydrophobic binding pocket with five conserved aromatic residues in transmembrane helix 3, 5, 6, and 7 [20]. Experimental and theoretical work suggest that the amino acids surrounding retinal affect the S0-S1 energy gap by altering the polarity of the retinal binding cavity [21-23] and the distance between the Schiff base linkage to retinal and its counter-ion [24-26].
For optogenetics, identifying variants with well-separated absorption spectra is of great interest for multiplexed control of excitation and inhibition by different colors of light in a single cell or in a population of cells. Lin et al. reported a variant called ReaChR that is optimally excited by orange-red light with λmax in the range 590 – 630 nm [27]. ReaChR is an engineered chimeric variant of VChR1, a cation-conducting ChR from Volvo carteri, which is maximally excited at 589 nm [27,28]. ReaChR has helix 6 replaced with that of VChR2 (also from Volvo carteri), which improves protein expression, and has the sequence of ChR1 from Chlamydamonas reinhardtii at the N-terminus, which further improves plasma membrane localization. To further improve the chimera's properties a number of single amino acid mutations were tested based on mutations that had previously been shown to alter ChR properties. One such single amino acid mutation (L171I) increased the amplitude of the photoresponse at 610 nm and 630 nm [27]. The L171 position was previously mutated in the ChR chimera ChEF [29] and was targeted because of its position proximal to the retinal binding pocket. ReaChR demonstrates that transferring mutations or even parts of domains between variants can confer desired properties (i.e., improved photostability and membrane localization). More broadly, chimeragenesis has proven to be a good engineering strategy to achieve spectral shifts in ChRs: in an earlier study from Prigge et al., helix swapping between ChR1, ChR2, VChR1, and VChR2 resulted in variants with red- and blue-shifted spectra, though none as red-shifted as ReaChR [30].
Spectral tuning of ChRs using higher throughput approaches has remained a challenge in part due to limited ChR expression in Escherichia coli, a common host for directed evolution [31,32]. The presence of predicted N-glycosylation sites in several rhodopsins suggests that glycosylation, which E. coli does not naturally perform, is required for functional ChR expression [32]. If the lack of glycosylation is limiting expression, then expressing ChRs in E. coli with a re-constituted eukaryotic glycosylation pathway (which was recently reported in [33]) may be possible. ChRs can be expressed in Pichia pastoris [34], suggesting that directed evolution should be possible in this system or in laboratory yeasts such as Saccharomyces cerevisiae.
In contrast to ChRs, proton-pumping rhodopsins (PPRs) can typically be expressed in E. coli. Recently, spectral tuning of a PPR from Gloeobacter violaceus called GR was performed by directed evolution in moderate-throughput (2,000 variants/round of screening) using E. coli to express the variants [23*]. Site-saturation mutagenesis at 19 positions around the retinal chromophore followed by recombination of beneficial mutations and further site-saturation mutagenesis generated large spectral shifts in absorption spectra relative to wildtype GR. Collectively, variants with shifts of +/− 80 nm compared to wildtype GR were achieved. The large shifts, however, came at the cost of proton pumping capacity [23*]. Further characterization of evolved variants revealed that blue-tuning mutations modulate the polarity along the retinal chromophore. Blue-tuning mutations near the PSB generally increased polarity relative to the native residues, while blue-tuning mutations near the beta-ionone ring decreased polarity [23*], consistent with recent theoretical predictions [35]. In contrast, red-tuning mutations occurred near the PSB linkage to retinal and likely disrupted its interaction with the negatively charged counter-ion [23*]. While directed evolution is clearly an effective strategy for spectral tuning, identifying variants with large shifts in absorbance and wildtype activity levels remains a challenge that the screening methods used to date have not been able to address.
Engineering Rhodopsin Ion Selectivity
Currently, inward-pumping chloride-transporting rhodopsins and outward-pumping proton-transporting rhodopsins are widely used for inhibiting neurons [1]. Rhodopsin channels (ChRs) can transport many ions for every photon of absorbed light, while pumps can only move a single ion per photon. Increased efficiency of ion translocation enables targeted perturbations with less light (often advantageous for optogenetics applications) but comes at the cost of transient perturbations of membrane conductance. Engineering potassium- and chloride-selective ChRs would enable selective inhibition in a way that better mimics natural neuronal physiology, with decreased photon flux. While ChRs’ non-specific influx of cations can effectively stimulate neuronal activity, enhanced selectivity for calcium could enable direct control of a number of cellular processes dependent on intracellular calcium ions (e.g., muscle contraction, release of neurotransmitters from nerve terminals, and gene expression). Ion-specific rhodopsins would be invaluable tools for studying downstream physiological responses to specific ions/second messengers both in neuroscience and beyond.
Recently, two groups independently engineered ChR chloride channels that can silence neurons [36**,37] with the aid of the dark state crystal structure of the ChR variant, C1C2 (a chimera of ChR1 and ChR2) [20] (Figure 2). Berndt et al. speculated that since the ion-selectivity pore in C1C2 is less ordered than that in potassium-selective channels [38-40], natural cation-specific activity is driven by the electrostatic potential surrounding the C1C2 pore and vestibule [36**]. By identifying single amino acid mutations in this region that modified the channel reversal potential and combining the single mutations into a variant called inhibitory C1C2 (iC1C2), they created a chloride-specific channel that can silence action potentials in response to light [36]. Wietek et al. took a different approach: using molecular dynamics simulations, they identified 5 residues that form a hydrophobic barrier in darkness to prevent water from entering the protein vestibule [37]. One of these residues, E90, when mutated to lysine or arginine, decreased ChR2's reversal potential and turned ChR2 into a light-activated chloride channel at membrane holding potentials above about −40 mV. Introduction of the T159C mutation improved membrane targeting of the protein in mammalian cells [37]. The resulting variant, ChloC, required two mutations to transform ChR2 into an effective tool for silencing action potentials in neurons in the presence of light [37].
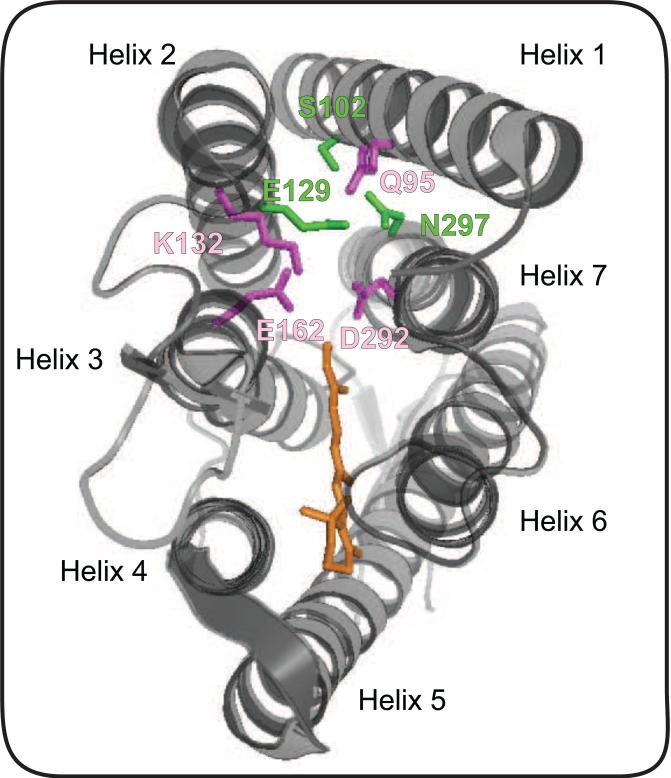
Figure 2. Residues that affect ion selectivity in the channelrhodopsin C1C2.
The illustration shows crystal structure of C1C2, with putative ion gating residues S102, E129 and N297 highlighted in green. Mutation of the gating residue N297 to D results in a significant increase in selectivity for Ca2+, while mutation of E129 to Q or A results in a significant decrease in the channel's Ca2+ selectivity [20]. Mutating the highly conserved gating residue E129 [45] has significant effects on the channel's selectivity for Cl− in both the C1C2 backbone and the ChR2 backbone (position E90 in the ChR2 backbone) [36,37]. Mutation of E90 in ChR2 to R or K increases the reversal potential as a result of increased Cl− selectivity to generate a light activated inhibitory channel [37]. Residues outside of the putative ion gate also influence channel selectivity (residues highlighted in purple). Mutations Q95A, E162 and D292A have all been shown to enhance H+ selectivity. Mutants K132A and Q95A display increased K+ permeability in the C1C2 backbone [20].
Ideally the inhibitory channels would have a decelerated channel closure, which would enable a prolonged ion-conducting state with a brief light stimulation. This has been achieved for the excitatory channel, ChR2, by introduction of a mutation at C128 which significantly decreased the time for channel closure once light is turned off (off kinetics, τoff) of the ChR2 parent [41]. The C128 mutation was introduced into ChR2 by analogy to previous work done with bacteriorhodopsin (bR), a light-driven proton pump, showing that the equivalent position in bR, when mutated, affects kinetics of the photocycle and lifetimes of intermediates [42,43]. The C128 residue is within 4 Å of the 12th carbon of retinal and, based on the C1C2 crystal structure [20], the thiol group is associated with the π-electron system in the retinal molecule [20]. Berndt et al. applied the equivalent mutation in iC1C2, which resulted in an inhibitory channel with slower channel closure that was named SwiChRCT. Wietek et al. engineered a slow-closing version of the inhibitory channel ChloC with mutations at position D156, a residue thought to interact with C128 [37].
Exploring Natural Variants for New Rhodopsin Actuators
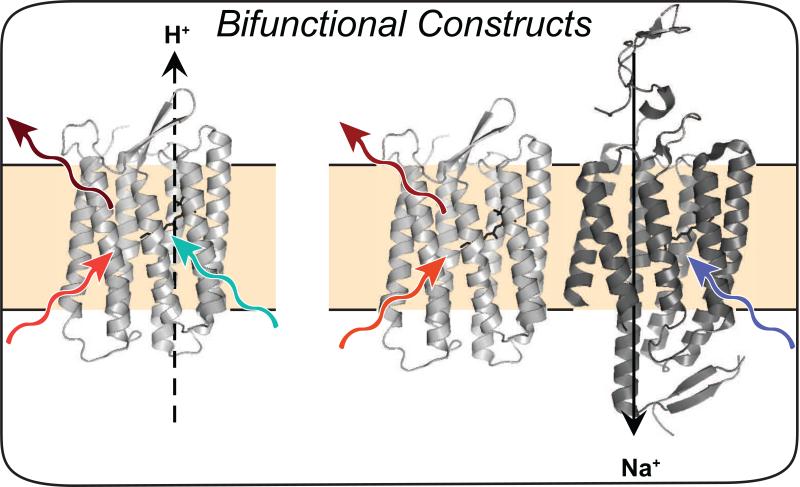
Combining protein engineering with environmental sample mining via de novo transcriptome sequencing has led to the identification of dozens of new rhodopsins [44,45]. Two new valuable ChRs recently identified, Chronos (activated with low intensity blue light) and Chrimson (activated with red light), together enable wavelength specific multiplexed perturbations of neurons [45]. A single mutation, K176R (which was previously shown to enhance photocurrents at the equivalent position in ChR2 [46]), was introduced into Chrimson to improve its slow kinetics to generate ChrimsonR [45]. Screening members of the cruxhalorhodopsin family led to identification of Halo57 from H. salinarum [44]. Introducing two single mutations into Halo57 to boost photocurrents and appending trafficking sequences from [47] resulted in an optimized variant called Jaws, a red-shifted inhibitor of neuronal activity [44]. A major limitation in synchronous sensing and perturbing of neuronal activity for all-optical electrophysiology is that the light used to activate the actuator can perturb the fluorescence readout of the sensor. A highly light-sensitive, blue-shifted channelrhodopsin variant (sdChR, [45**]) identified in a screen of plant genomes was further engineered for faster kinetics and improved membrane localization to produce CheRiff to enable subcellular excitation [9] (Figure 3).
Figure 3. Bifunctional constructs for all-optical electrophysiology.
Archer, an engineered Archaerhodopsin-3 variant, enables optical monitoring of voltage with red light, and perturbation of membrane potential with blue light (left) [8]. Alternatively, one rhodopsin can be used for sensing with red light, while an engineered ChR can be used for perturbing the membrane with blue light (right) [9].
Engineering of Rhodopsin Voltage Indicators
Adam Cohen and colleagues recently discovered that rhodopsins can be used as genetically encoded voltage indicators (GEVIs); however, the natural proteins suffer from extremely low quantum efficiencies (~10−4) [5]. Eliminating pumping activity while retaining fast kinetics also presents an engineering challenge since the relationship between pumping, fluorescence, and kinetics is not completely understood. The photocycle of Arch, a leading candidate for GEVI development, is thought to proceed as follows: absorption of a photon initiates the photocycle (g → M), leading to an equilibrium between the M state (protonated counter-ion) and N state (protonated Schiff base) [48]. Following conversion of N → Q (through absorption of photon at 540 nm) and excitation of the Q-state (absorption of photon at 570 nm), a photon at 710 is emitted as fluorescence as Arch returns to the N intermediate [48]. Retinal thermally isomerizes back to all-trans (N → O) and a proton is released at the extracellular side (O → g). Based on this model, mutants with a longer-lived Q-state should exhibit increased fluorescence.
Directed evolution is an effective strategy for enhancing the brightness of Arch [9,49,50]. For example, introduction of mutations near the lysine that forms the covalent Schiff base linkage to retinal and screening for fluorescence enabled identification of two variants of Arch, one a double mutant, D95E/T99C (Archer) and another containing 5 mutations (referred to as QuasAr1). Both Archer and QuasAr1 show enhanced voltage sensitive fluorescence with emission in the far-red (maximal emission > 680 nm) [8,9,49]. Both of these engineered variants have improved brightness and dynamic range compared to two previously published variants, Arch EEQ and Arch EEN [19].
Directed evolution of Archer revealed two fluorescence enhancing mutations, V59A and I129T [49], that were independently identified at the homologous positions in bR (V49A and I119T) and shown to stabilize the Q-state intermediate [51]. Many mutations at P60 (<5Å from retinal) also increase Arch fluorescence [49]; similarly, many mutations at the homologous bR position (P50) stabilized the Q state [51]. These observations are consistent with the Q state being the fluorescent state in the Arch photocycle [48].
Since their absorbance is sensitive to changes in electric potential [52], rhodopsins can also potentially be used in FRET sensors, assuming the absorbance overlaps with the emission of a bright fluorescent protein. Recently, a FRET-opsin sensor (a fusion between L. maculans [Mac] rhodopsin and mOrange2 [a monomeric orange fluorescent protein [53]]) was developed, achieving a response time of ~5 ms following a step change in membrane voltage and successful detection of sub-threshold events [7]. To eliminate Mac pumping, the PSB counter-ion D139 was replaced with glutamine (Q). While replacing the counter-ion with a neutral residue is a general strategy for eliminating pumping, the D139N variant retained sufficient pumping activity to perturb neural spiking patterns; thus, D139Q or its equivalent is the preferred mutation for Mac-based sensors [7]. However, current Mac-mOrange2 derivatives have a lower dynamic range (defined as voltage-dependent changes with respect to the probe's baseline fluorescence) than recently engineered Arch variants [8,9,49]. Using an expression vector that can drive expression in both prokaryotic and eukaryotic cells, Zou et al. developed a screening strategy in which brighter Arch-mOrange2 variants can be identified in E. coli and subsequently transfected into HEK293 cells to measure their voltage sensitivity [50*]. This engineering strategy accelerates the speed at which brighter, multi-colored, and voltage-sensitive rhodopsins can be identified and has resulted in FRET sensors with rise times in the range 1 – 7 ms [50*].
Engineered rhodopsin-based sensors are still quite dim, with quantum yields of <1%. Alternative voltage sensors have been engineered by fusing the Ciona intestinalis voltage-sensor containing domain (Ci-VSD), a non-rhodopsin protein that undergoes a voltage-dependent conformational change, to a fluorescent protein [54]. The issue of slow kinetics of these non-rhodopsin sensors [55] has been largely overcome [56], but they exhibit non-linear voltage sensitivity, which may limit their capacity for detecting sub-threshold events [56]. Despite being fused to bright fluorescent proteins and increased basal fluorescence over rhodopsins, the spectral overlap between Ci-VSD-based sensors and rhodopsins limits their compatibility for all-optical electrophysiology (Figure 3); furthermore, rhodopsins appear to be less susceptible to photo-bleaching [8].
Conclusion
Rhodopsins are powerful tools for brain research. Identifying actuators with shifted and narrowed spectra would improve the ability to multiplex perturbations with different colors of light, whereas enhancing ion specificity will enable more physiological studies within and beyond neuroscience. Brighter rhodopsin sensors have been engineered, but further improved brightness would facilitate imaging populations of neurons (and perhaps other electrically-active cell-types such as cardiomyocytes) with wide-field microscopy. The development of opsin-FRET sensors could also enable monitoring different cell types with different colors of light [50], a potentially powerful application of all-optical electrophysiology. Future work would greatly benefit from an understanding of how characterized mutations impact the photocycle and the protein structure and thereby contribute to the desirable properties found in engineered rhodopsins. Chimeragenesis, structure-guided mutagenesis, and directed evolution have and will continue to play central roles in the development of improved rhodopsins for optogenetics.
Highlights.
Rhodopsins are useful sensors and actuators of neuronal activity.
Bifunctional rhodopsin constructs facilitate all-optical electrophysiology.
Protein engineering can enhance rhodopsin properties for optogenetics.
Acknowledgements
This work was funded by the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office (to F.H.A.) and 1R21MH103824-01 from the National Institutes of Health (to F.H.A.). R.S.M. acknowledges funding from the Shurl and Kay Curci Foundation and the Life Sciences Research Foundation. C.N.B. acknowledges support from the NIMH of the NIH for the NRSA fellowship under Award Number F31MH102913.
Abbrevations
- Arch
Archaerhodopsin-3
- bR
bacteriorhodopsin
- ChR
Channelrhodopsin
- ChR1
Chlamydomonas reinhardtii channelrhodopsin-1
- ChR2
Chlamydomonas reinhardtii channelrhodopsin-2
- C1C2
chimera of ChR1 and ChR2
- GEVI
genetically encoded voltage indicator
- HEK293 cells
human embryonic kidney 293 cells
- FRET
Förster resonance energy transfer
- GR
Gloeobacter violaceus rhodopsin
- PPR
proton-pumping rhodopsin
- PSB
protonated Schiff base
- sdChR
Scherffelia dubia channelrhodopsin
- VChR1
Volvox carteri channelrhodopsin-1
- VChR2
Volvox carteri channelrhodopsin-2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*of special interest
- 1.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, Magnuson J, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147(7):1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324(5925):354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X. In vivo application of optogenetics for neural circuit analysis. ACS Chemical Neuroscience. 2012;3(8):577–584. doi: 10.1021/cn300065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving caenorhabditis elegans. Nat Methods. 2011;8(2):147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9(1):90–U130. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kralj JM, Hochbaum DR, Douglass AD, Cohen AE. Electrical spiking in escherichia coli probed with a fluorescent voltage-indicating protein. Science. 2011;333(6040):345–348. doi: 10.1126/science.1204763. [DOI] [PubMed] [Google Scholar]
- 7.Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ. Imaging neural spiking in brain tissue using fret-opsin protein voltage sensors. Nature Communications. 2014;5(3674) doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flytzanis NC, Bedbrook CN, Chiu H, Engqvist MKM, Xiao C, Chan KY, Sternberg PW, Arnold FH, Gradinaru V. Archaerhodopsin variants with enhanced voltage sensitive fluorescence in mammalian and C. elegans neurons. Nature Communications. 2014 doi: 10.1038/ncomms5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, Saulnier JL, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014 doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Looger LL, Griesbeck O. Genetically encoded neural activity indicators. Current Opinion in Neurobiology. 2012;22(1):18–23. doi: 10.1016/j.conb.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Looger LL. Running in reverse: Rhodopsins sense voltage. Nat Methods. 2012;9(1):43–44. doi: 10.1038/nmeth.1817. [DOI] [PubMed] [Google Scholar]
- 12.Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chemical Reviews. 2014;114(1):126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: Structures and functions from archaea to humans. Annual Review of Cell and Developmental Biology. 2000;16(365-392) doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 14.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation- selective membrane channel. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: A light-gated proton channel in green algae. Science. 2002;296(5577):2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 16.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neuroscience. 2005;8(9):1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 17.Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O'Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, Yizhar O, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9(2):159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 19.Gong Y, Li JZ, Schnitzer MJ. Enhanced archaerhodopsin fluorescent protein voltage indicators. PloS One. 2013;8(6):e66959. doi: 10.1371/journal.pone.0066959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, Ito J, Aita Y, Tsukazaki T, Hayashi S, Hegemann P, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482(7385):369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houjou H, Inoue Y, Sakurai M. Study of the opsin shift of bacteriorhodopsin: Insight from qm/mm calculations with electronic polarization effects of the protein environment. J Phys Chem B. 2001;105(4):867–879. [Google Scholar]
- 22.Melaccio F, Ferre N, Olivucci M. Quantum chemical modeling of rhodopsin mutants displaying switchable colors. Physical Chemistry Chemical Physics: PCCP. 2012;14(36):12485–12495. doi: 10.1039/c2cp40940b. [DOI] [PubMed] [Google Scholar]
- 23*.Engqvist MK, McIsaac RS, Dollinger P, Flytzanis NC, Abrams M, Schor S, Arnold FH. Directed evolution of gloeobacter violaceus rhodopsin spectral properties. Journal of Molecular Biology. 2015;427(1):205–220. doi: 10.1016/j.jmb.2014.06.015. [A directed evolution strategy enabled shifts of +/− 80 nm in the preferred wavelength of absorbance in a proton-pumping rhodopsin from Gloeobacter violaceus. Though not specifically selected for, a subset of red-shifted variants were found to be highly fluorescent in the near-infrared with a maximal emission of >700 nm.] [DOI] [PubMed] [Google Scholar]
- 24.Wang WJ, Nossoni Z, Berbasova T, Watson CT, Yapici I, Lee KSS, Vasileiou C, Geiger JH, Borhan B. Tuning the electronic absorption of protein- embedded all-trans-retinal. Science. 2012;338(6112):1340–1343. doi: 10.1126/science.1226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butt HJ, Fendler K, Bamberg E, Tittor J, Oesterhelt D. Aspartic acid-96 and aspartic acid-85 play a central role in the function of bacteriorhodopsin as a proton pump. Embo J. 1989;8(6):1657–1663. doi: 10.1002/j.1460-2075.1989.tb03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi S, Tajkhorshid E, Pebay-Peyroula E, Royant A, Landau EM, Navarro J, Schulten K. Structural determinants of spectral tuning in retinal proteins - bacteriorhodopsin vs sensory rhodopsin ii. J Phys Chem B. 2001;105:10124–10131. [Google Scholar]
- 27.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. Reachr: A red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nature Neuroscience. 2013;16(10):1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F, Prigge M, Beyriere F, Tsunoda SP, Mattis J, Yizhar O, Hegemann P, Deisseroth K. Red-shifted optogenetic excitation: A tool for fast neural control derived from volvox carteri. Nature Neuroscience. 2008;11(6):631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophysical journal. 2009;96(5):1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prigge M, Schneider F, Tsunoda SP, Shilyansky C, Wietek J, Deisseroth K, Hegemann P. Color-tuned channelrhodopsins for multiwavelength optogenetics. The Journal of Biological Chemistry. 2012;287(38):31804–31812. doi: 10.1074/jbc.M112.391185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sineshchekov OA, Govorunova EG, Wang J, Li H, Spudich JL. Intramolecular proton transfer in channelrhodopsins. Biophysical journal. 2013;104(4):807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou SY, Govorunova EG, Ntefidou M, Lane CE, Spudich EN, Sineshchekov OA, Spudich JL. Diversity of chlamydomonas channelrhodopsins. Photochemistry and photobiology. 2012;88(1):119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan YY, Reading CA, Chhiba K, Heiss C, Azadi P, Aebi M, DeLisa MP. An engineered eukaryotic protein glycosylation pathway in escherichia coli. Nature chemical biology. 2012;8(5):434–436. doi: 10.1038/nchembio.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. Journal of Molecular Biology. 2008;375(3):686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Sundholm D, Wesolowski TA, Kaila VR. Spectral tuning of rhodopsin and visual cone pigments. Journal of the American Chemical Society. 2014;136(7):2723–2726. doi: 10.1021/ja411864m. [DOI] [PubMed] [Google Scholar]
- 36**.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344(6182):420–424. doi: 10.1126/science.1252367. [The authors demonstrated that a cation-conducting ChR can be converted to a chloride-conducting anion channel by using structure-guided electrostatic modeling and accumulating individual mutations that either modify the reversal potential and/or do not eliminate photocurrents.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wietek J, Wiegert JS, Adeishvili N, Schneider F, Watanabe H, Tsunoda SP, Vogt A, Elstner M, Oertner TG, Hegemann P. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344(6182):409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 38.Yellen G. The voltage-gated potassium channels and their relatives. Nature. 2002;419(6902):35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- 39.Sauer DB, Zeng W, Raghunathan S, Jiang Y. Protein interactions central to stabilizing the k+ channel selectivity filter in a four-sited configuration for selective k+ permeation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(40):16634–16639. doi: 10.1073/pnas.1111688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YS, Sompornpisut P, Perozo E. Structure of the kcsa channel intracellular gate in the open state. Nature Structural Biology. 2001;8(10):883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 41.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nature Neuroscience. 2009;12(2):229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 42.Lanyi JK, Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the k, l, m1, m2, and m2′ intermediates of the photocycle. Journal of Molecular Biology. 2003;328(2):439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
- 43.Peralvarez-Marin A, Marquez M, Bourdelande JL, Querol E, Padros E. Thr-90 plays a vital role in the structure and function of bacteriorhodopsin. The Journal of Biological Chemistry. 2004;279(16):16403–16409. doi: 10.1074/jbc.M313988200. [DOI] [PubMed] [Google Scholar]
- 44.Chuong AS, Miri ML, Busskamp V, Matthews GA, Acker LC, Sorensen AT, Young A, Klapoetke NC, Henninger MA, Kodandaramaiah SB, Ogawa M, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nature Neuroscience. 2014;17(8):1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11(3):338–346. doi: 10.1038/nmeth.2836. [Next-generation sequencing was used to identify and characterize opsins from more than 100 species of algae. Despite the wide activation spectra of all known rhodopsins, two variants were identified that enable independent excitation of neurons with blue and red light.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of caenorhabditis elegans triggers rapid behavioral responses. Current Biology. 2005;15(24):2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 47.Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141(1):154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maclaurin D, Venkatachalam V, Lee H, Cohen AE. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(15):5939–5944. doi: 10.1073/pnas.1215595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McIsaac RS, Engqvist MKM, Wannier T, Rosenthal AZ, Herwig L, Flytzanis NC, Imasheva ES, Lanyi JK, Balashov SP, Gradinaru V, Arnold FH. Directed evolution of a far-red fluorescent rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(36):13034–13039. doi: 10.1073/pnas.1413987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Zou P, Zhao Y, Douglass AD, Hochbaum DR, Brinks D, Werley CA, Harrison DJ, Campbell RE, Cohen AE. Bright and fast voltage reporters across the visible spectrum via electrochromic fret (efret). aRxiV. 2014 doi: 10.1038/ncomms5625. [The authors constructed a panel of Arch-FRET sensors using different color fluorescent proteins as FRET donors. The availability of voltage sensors that span the visible spectrum may enable multiplexed monitoring of voltage in distinct cell types using different colors of light.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner NL, Greco JA, Ranaghan MJ, Birge RR. Directed evolution of bacteriorhodopsin for applications in bioelectronics. Journal of the Royal Society, Interface / the Royal Society. 2013;10(84):20130197. doi: 10.1098/rsif.2013.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolodner P, Lukashev EP, Ching YC, Rousseau DL. Electric-field-induced schiff-base deprotonation in d85n mutant bacteriorhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11618–11621. doi: 10.1073/pnas.93.21.11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5(6):545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mutoh H, Akemann W, Knopfel T. Genetically engineered fluorescent voltage reporters. ACS Chemical Neuroscience. 2012;3(8):585–592. doi: 10.1021/cn300041b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75(5):779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nature Neuroscience. 2014;17(6):884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]