Summary
Spermatogonial stem cells (SSCs) must balance self-renewal with production of transit-amplifying progenitors that differentiate in response to retinoic acid (RA) before entering meiosis. This self-renewal vs. differentiation spermatogonial fate decision is critical for maintaining tissue homeostasis, as imbalances cause spermatogenesis defects that can lead to human testicular cancer or infertility. A great deal of effort has been exerted to understand how the SSC population is maintained. In contrast, little is known about the essential program of differentiation initiated by retinoic acid (RA) that precedes meiosis, and the pathways and proteins involved are poorly defined. We recently reported a novel role for RA in stimulating the PI3/AKT/mTOR kinase signaling pathway to activate translation of repressed mRNAs such as Kit. Here, we examined the requirement for mTOR complex 1 (mTORC1) in mediating the RA signal to direct spermatogonial differentiation in the neonatal testis. We found that in vivo inhibition of mTORC1 by rapamycin blocked spermatogonial differentiation, which led to an accumulation of undifferentiated spermatogonia. In addition, rapamycin also blocked the RA-induced translational activation of mRNAs encoding KIT, SOHLH1, and SOHLH2 without affecting expression of STRA8. These findings highlight dual roles for RA in germ cell development – transcriptional activation of genes, and kinase signaling to stimulate translation of repressed messages required for spermatogonial differentiation.
Keywords: Translation, spermatogonia, retinoic acid, testis, spermatogenesis, mTOR
Introduction
Retinoic acid (RA) is required for inducing both spermatogonial differentiation and subsequent entry into meiosis [1, 2]. After birth in the mouse, subsets of spermatogonia begin to differentiate in response to RA at ~P3-4, as indicated by their expression of STRA8 and KIT [3-6]. However, germ cells do not enter meiosis in response to RA until ~P10 [7]. This reveals that the processes of differentiation and meiotic entry are temporally separated by ~7 days in the neonate (which lengthens to 8.6 days in the adult). It has been proposed that the reduced time in the neonate may be because some of the steps are skipped or because cell proliferation is accelerated by the increased temperature at which spermatogenesis proceeds in the neonate (37°C in neonates versus 33°C in adults) [8]. During this approximately week-long differentiation period, type A1 spermatogonia successively become type A2, A3, A4, In, and B before entering meiosis as preleptotene spermatocytes [9-11]. Spermatogonia apparently cannot be induced to precociously enter meiosis in response to exogenous RA [3, 4, 12], which implies that these sequential spermatogonial divisions are required. However, little is known about the cellular processes that must occur or the molecular pathways that regulate them in differentiating spermatogonia prior to meiotic initiation. A primary reason for this lack of knowledge is that there are few changes in steady-state mRNA levels during differentiation [13-15]. Without dramatic changes in the transcriptome, scientists have lacked identified targets (pathways, proteins) for focused studies.
A number of recent genome-wide studies have revealed that the transcriptome imperfectly predicts the proteome (estimates range from ~40-80% agreement) [16-18], and there is likely to be considerable variation in this level of disconnect in different cell types and under different conditions. Our previous studies suggest that a majority of gene expression changes required for spermatogonial differentiation occur at the posttranscriptional level [19, 20]. We previously showed that RA treatment leads to increased phosphorylation of the master regulatory kinase mammalian target of rapamycin (mTOR) [19], indicating it is activated in differentiating spermatogonia. This is accompanied by enhanced translation of repressed Kit mRNAs through activation of the PI3K/PDPK1 (also termed PDK1)/AKT signaling network [19, 21]. MTOR exists in functionally distinct protein complexes (mTORC1 and mTORC2), which integrate numerous cues to regulate, in broad terms, cellular growth and differentiation (mTORC1) or the actin cytoskeleton and insulin signaling (mTORC2) (reviewed by [22-26]). A primary role of mTORC1 is to regulate cap-dependent mRNA translation initiation, which is the rate limiting and regulated step of eukaryotic protein synthesis. MTORC1 performs this function in part by phosphorylating downstream targets EIF4EBP1 and RPS6KB1/2 (also termed p70S6K). Enhanced phosphorylation of EIF4EBP1 by activated mTOR releases EIF4E to associate with the 5’-cap of mRNAs, thus allowing recruitment of translationally controlled mRNAs to ribosomes, particularly during germ cell differentiation in lower organisms [27-31]. Activated RPS6KB1/2 phosphorylates the 40S ribosomal subunit RPS6, leading to activation of ribosomes and enhanced mRNA translation. This activity is essential for the translational control of the TOP mRNAs, which have 5’ oligopyrimidine tracts, are activated by changes in cellular metabolism, and often encode components of the translational machinery [32, 33].
The mTOR complexes differ in their sensitivity to rapamycin, a macrolide antifungal compound originally isolated from the soil bacterium Streptomyces hygroscopicus that has an expanding number of clinical uses. Rapamycin acutely inhibits mTORC1 activity, but has also been shown to inhibit mTORC2 in some contexts following prolonged exposure [24, 34, 35]. The effects of rapamycin vary significantly depending on the cell type involved. Some cell types appear to be quite rapamycin-insensitive, while others slow or cease proliferating, fail to differentiate, and/or undergo apoptosis [24, 25, 36]. This has been suggested to be due in part to differential effects on downstream substrates such as EIF4EBP1 and/or RPS6KB1/2 [37]. Rapamycin can be administered in vivo, and actually extends the lifespan of lower organisms and mice ([38, 39], reviewed in [25]). In addition, studies have suggested a link between mTORC1 and spermatogonial cell fate regulation both in vitro and in vivo [19, 20, 40-43].
Studying the effects of mTORC1 inhibition by rapamycin has direct relevance for human male reproductive health. Rapamycin analogs (Sirolimus and Everolimus) are currently used to reduce cellular proliferation as part of immunosuppressive and chemotherapeutic regimens given to organ transplant, cardiology, and cancer patients [24, 25, 44-49]. These drugs can cause reversible human male infertility with unclear etiology [50-54]. Specifically, rapamycin analog treatments caused a block in spermatogonial differentiation in a human patient [55] as well as in a study using rats, although no detailed analyses were performed [56]. Both treated humans and rats exhibit reduced testosterone (T) levels due to inhibition of the hypothalamic-pituitary-gonadal axis. However, this may not cause a defect in spermatogonial differentiation, but rather in progression through meiosis, at least in rodents [57]. Infertility is a significant quality of life concern for reproductive-aged male organ transplant and cancer patients, and to-date no comprehensive studies have explored the mechanism of action of mTOR inhibition during spermatogenesis.
Here, we examine the effects of rapamycin-mediated mTORC1 inactivation on spermatogonial differentiation in vivo in the mouse. Our results reveal that mTORC1 activation is dispensable for the maintenance of undifferentiated spermatogonia, but that it is required for spermatogonial proliferation and differentiation prior to meiotic initiation. In addition, we find that rapamycin inhibition of mTORC1 blocks the RA-induced translation of repressed mRNAs encoding KIT, SOHLH1, and SOHLH2, which are essential regulators of spermatogonial differentiation. However, rapamycin treatment did not block the expression of STRA8, a direct transcriptional target of RA. This reveals that spermatogonia exhibit dual responses to RA in the form of transcriptional activation and kinase signaling-enhanced translation of repressed mRNAs. In addition, these results provide critical insight into the male infertility that can result as an adverse side effect of the clinical use of rapamycin analogs.
Results
Rapamycin treatment reduces testicular size and arrests germ cell development
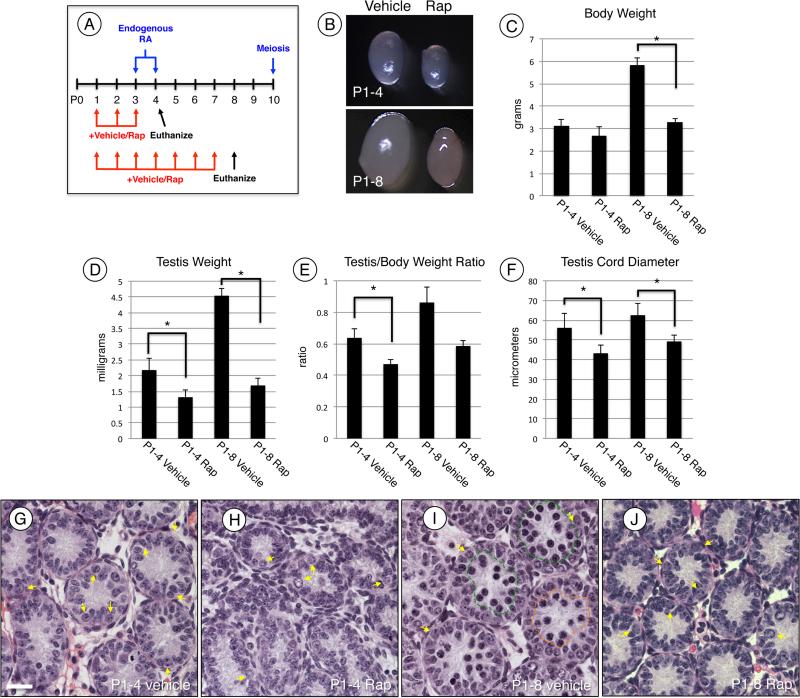
Our previous study revealed that RA activated the PI3K/AKT kinase signaling network in differentiating spermatogonia [19]. This also resulted in phosphorylation (implying activation) of mTOR, which acts in mTORC1 to direct cellular growth, proliferation, and differentiation (reviewed in [24]). Here, we addressed the requirement for mTORC1 in spermatogonial differentiation in vivo by feeding vehicle alone or rapamycin to neonatal mice once daily beginning at P1, which is 2-3 days prior to the onset of normal differentiation. Mice were then euthanized at P4 and P8 (Fig. 1A), and testes were harvested for various analyses. Although animals appeared healthy, there was a significant decrease in body weight and testis size and weight in response to rapamycin treatment at both P4 and at P8 (Fig. 1C-D). Testis weights were normalized to total body weight and there were 26% and 31% decreases at P4 and P8, respectively (Fig. 1E). Since reduced testis weight often results from reduced cellularity, we examined Bouin's-fixed testis sections stained with H&E from vehicle- and rapamycin-treated mice. Germ cell types in the neonatal testis are readily distinguished based on their characteristic size, location, and nuclear morphology [11, 59, 60]. Vehicle-treated control testes from P4 and P8 mice had the germ cells at the expected stages of development (Fig. 1G, I) [9, 59, 61, 62]. At P8, there were abundant spermatogonia and preleptotene spermatocytes and a small number of meiotic leptotene spermatocytes. In contrast, rapamycin-treated P8 testes had significantly reduced testis cord diameters (Fig. 1F) and many fewer germ cells, with the most advanced stage resembling undifferentiated spermatogonia (Fig. 1H and I).
Figure 1. MTORC1 inhibition blocks spermatogonial differentiation.
(A) Experimental design for treating mice with rapamycin in vivo. Mice were treated with vehicle or rapamycin once daily starting at P1 and euthanized 24h after last treatment at P4 or P8. (B) Representative images of testes from vehicle (left) and rapamycin (right) treated mice euthanized at P4 (top) and P8 (bottom). (C-E) Mice treated with vehicle or rapamycin were euthanized at P4 or P8. Prior to euthanasia total body weights were collected (C), following euthanasia total testis weights were collected (D). Testis weights were normalized to body weights and expressed as a ratio (E). (F) Quantitation of testis cord diameter of mice treated with vehicle or rapamycin and euthanized at P4 or P8. (G-J) H&E staining of mice treated with vehicle (G and I) or rapamycin (H and J) and euthanized at P4 (G and H) or P8 (I and J). Yellow arrows indicate spermatogonia, and green and orange lines encircle preleptotene and leptotene spermatocytes, respectively (I). Scale bar = 40 μM. Asterisks indicate statistical significance with P<0.01.
Rapamycin inhibits mTORC1 activity in spermatogonia
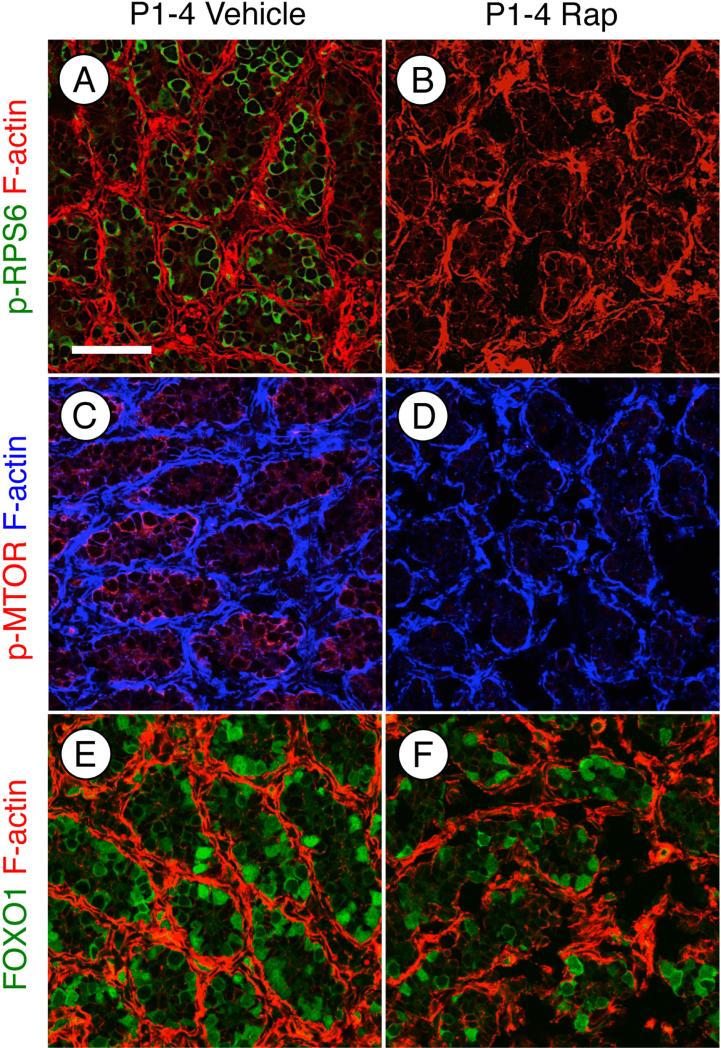
Before carefully examining the phenotype of rapamycin-treated testes, we verified that rapamycin treatment inhibited mTORC1 activity as expected. We examined the phosphorylation of mTOR as well as RPS6, which is an indirect target downstream of activated mTORC1. There was a near-complete loss of p-mTOR and p-RPS6 in response to rapamycin (Fig. 2A-D). We recently reported that RA caused a dramatic nuclear-to-cytoplasmic relocalization of FOXO1 [19], which is indicative of AKT activation [63, 64]. Since this signaling step is generally upstream of mTOR phosphorylation, we hypothesized that rapamycin inhibition would not alter FOXO1 localization. Indeed, there was no appreciable difference in the ratios of cytoplasmic: nuclear FOXO1 in vehicle- or rapamycin-treated germ cells (Fig. 2E-F). Therefore, we conclude that rapamycin treatment inhibited mTORC1 activation in spermatogonia without affecting upstream activation of signaling components such as AKT, which implies that mTORC2 signaling was not perturbed.
Figure 2. Treatment with rapamycin inhibits mTORC1 activity without affecting AKT.
(A-F) Immunostaining of testis sections from mice treated with vehicle (A, C, and E) or rapamycin (B, D, and F) and euthanized at P4. Sections were stained with anti- phosphorylated RPS6 (A and B), anti- phosphorylated EIF4EBP1 (C and D), or total FOXO1 (E and F). F-actin was stained with phalloidin (red) to visualize testis cords. Scale bar = 50 μM.
Undifferentiated spermatogonia accumulate in rapamycin-treated testes
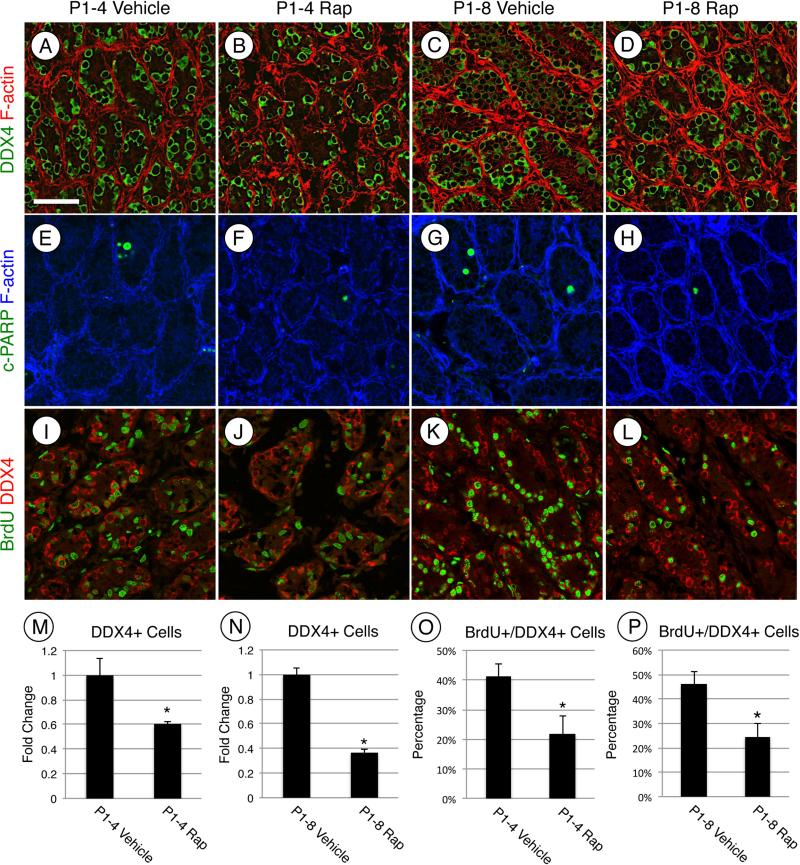
Rapamycin treatment from P1-P4 and P1-P8 resulted in an apparent reduction in germ cells (Fig. 1H, J). We quantified this in vehicle- and rapamycin-treated mice by immunostaining for DDX4, a pan germ cell marker in the neonatal testis. There were reduced numbers of DDX4+ germ cells at P4 (−1.63±0.03-fold) and P8 (−2.74±0.04-fold) (Fig. 3A-D, M and N). The decreased number of germ cells could result from a rapamycin-induced increase in apoptosis, although we did not see evidence for this in H&E-stained sections (Fig. 1G-J, data not shown). We nonetheless assessed this possibility by immunostaining for cleaved PARP1, an accepted marker for apoptotic cells. As expected, there was no significant increase in cleaved-PARP1+ cells in rapamycin-treated testes at P4 or P8 (P1-4 vehicle-treated = 0.17±0.06 cleaved-PARP1+ cells/testis cord, P1-4 rapamycin-treated = 0.13±0.06 cleaved-PARP1+ cells/testis cord, Fig. 3E-H), indicating that apoptosis was not responsible for the reduced germ cell numbers. We next assessed whether there was a reduction in germ cell proliferation in response to rapamycin. We first immunostained using an antibody against MKI67, which marks actively proliferating cells in all stages of the cell cycle except in G0 [65, 66]. There was no difference in MKI67 staining at P4 in response to rapamycin treatment. However, there were significantly fewer MKI67+/DDX4+ cells at P8 in rapamycin treated testes. (−1.85±0.37-fold, Fig. S1A-E). We next injected vehicle- and rapamycin-treated mice with BrdU 10h prior to euthanasia on P4 and P8. There were ≈1.9-fold fewer BrdU+ germ cells in response to rapamycin at P4 and P8 (Fig. 3I-L, O and P). We also assessed effects of rapamycin on Sertoli cell numbers by immunostaining for the Sertoli cell marker GATA4, and found that there was no appreciable change at P4 (P1-4 vehicle = 18.3±0.6 GATA4+ cells/testis cord, P1-4 rap-treated = 19.67±3.3 GATA4+ cells/testis cord) or P8 (P1-8 vehicle = 18.8±4.0 GATA4+ cells/testis cord, P1-8 rap-treated = 17.1±15.5 GATA4+ cells/testis cord, Fig. S2). Therefore, we conclude that rapamycin inhibited germ cell proliferation without increasing apoptosis or changing Sertoli cell numbers.
Figure 3. MTORC1 is required for postnatal expansion of the germ cell population.
(A-L) Immunostaining of testis sections from mice treated with vehicle (A, C, E, G, I, and K) or rapamycin (B, D, F, H, J, and L) and euthanized at P4 (A, B, E, F, I, and J) or P8 (C, D, G, H, K, and L). Sections were stained with anti-DDX4 (A-D), anti-cleaved PARP1 (E-H), or double labeled with anti-BrdU (green, I-L) and anti-DDX4 (red, I-L). F-actin was stained with phalloidin (red, A-D or blue, E-H). Quantitation of the number of DDX4+(M and N) or the number of BrdU+/DDX4+ (O and P) cells from testes treated starting at P1 with vehicle or rapamycin and then euthanized at P4 (M and O) or at P8 (N and P). Asterisks indicate statistical significance with P≤0.01. Scale bar = 50 μM.
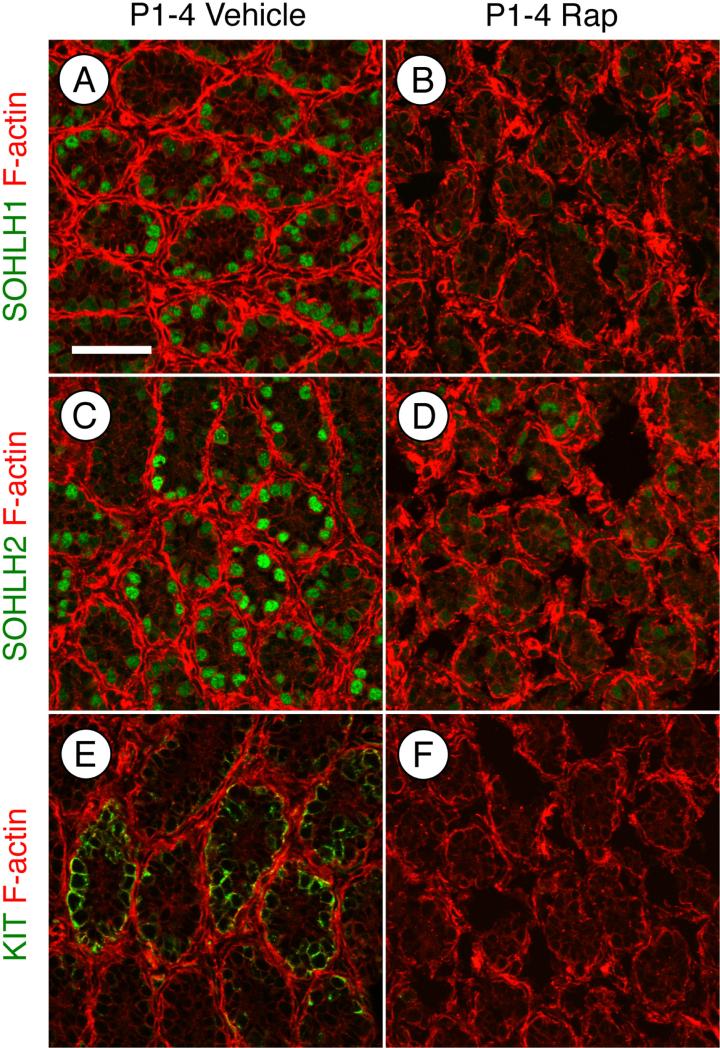
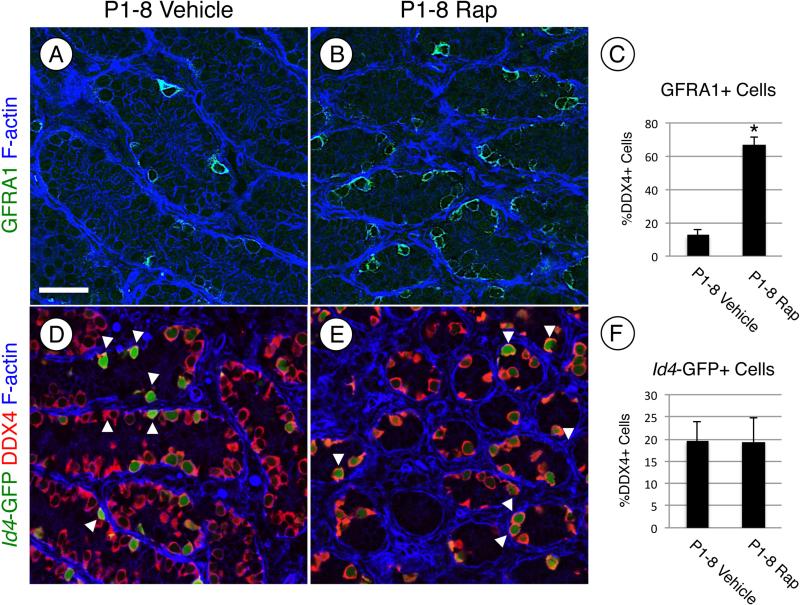
We next tested whether mTORC1 inhibition by rapamycin prevented the normal differentiation of spermatogonia. Based on the apparent accumulation of undifferentiated spermatogonia at P4 and P8 in response to rapamycin (Fig. 1G-J), it appeared that spermatogonial differentiation was blocked by rapamycin treatment. We assessed whether spermatogonia upregulated markers of differentiation (KIT, SOHLH1, and SOHLH2) in the absence of mTORC1 function. Each marker was readily detectable in spermatogonia in vehicle-treated testes at P4 (Fig. 4A, C, and E). In contrast, KIT, SOHLH1, and SOHLH2 were barely detectable in a few spermatogonia in rapamycin-treated testes (Fig. 4B, D, and F). The absence of differentiation markers suggests that the germ cells remained in an undifferentiated state in the absence of mTORC1 activity. We tested this by immunostaining for GFRA1, an established marker of undifferentiated spermatogonia, which together with RET forms the receptor for GDNF [67, 68]. We found that at P8 66% of DDX4+ spermatogonia were GFRA1+ in response to rapamycin, which represented a 5.1-fold increase over vehicle-treated controls (Fig. 5A-C). We further explored this using a recently created transgenic mouse line, in which the spermatogonial stem cell (SSC) population is marked by the expression of eGFP under the control of the Id4 promoter [13]. Recent work has demonstrated that SSC activity resides within the Id4-GFP+ cell population [13]. To test if treatment with rapamycin affects formation or size of the SSC population, we treated Id4-GFP pups with vehicle or rapamycin from P1 through P7 and euthanized them on P8. The results demonstrated that 19% of the total germ cell population was GFP-bright in both the vehicle- and rapamycin-treated testes (Fig. 5D-F), indicating that the size of the SSC pool was not affected by mTORC1 inhibition.
Figure 4. MTOR activation is required for induction of SOHLH1, SOHLH2, and KIT protein.
Immunostaining of mice treated with vehicle (A, C, and E) or rapamycin (B, D, and F) and euthanized at P4. Sections were stained with anti-SOHLH1 (A-B), anti-SOHLH2 (C-D), or anti-KIT (E-F). Phalloidin (red) was added to visualize testis cords. Scale bar = 50 μM.
Figure 5. Inhibiting mTORC1 activation increases the number of undifferentiated spermatogonia.
(A, B, D, and E) Immunostaining was performed on testis sections from mice treated with vehicle (A and D) or rapamycin (B and E) and euthanized at P8. (A-C) Testis sections from CD-1 mice were stained with anti-GFRA1 (green, A and B) and F-actin was stained with phalloidin (blue) to visualize testis cords. The number of GFRA1+ germ cells in vehicle- and rapamycin-treated testes were quantitated and reported as a fold change (C). (D-F) Transgenic Id4-GFP mice were treated with vehicle or rapamycin, and immunostaining was performed on testes. Green represents GFP epifluoresence, and sections were labeled with anti-DDX4 (red). White arrows indicate GFP bright spermatogonia (D and E). The number of GFP bright cells were quantitated and represented as a percentage of the DDX4+ cells (C). Scale bar = 40 μM. Asterisks indicate statistical significance with P<0.01.
Rapamycin blocks RA-enhanced translation of repressed mRNAs
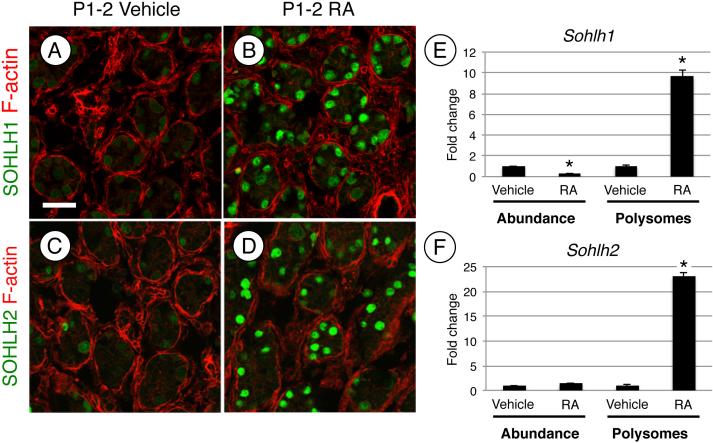
Finally, we investigated how KIT, SOHLH1, and SOHLH2 protein levels were dramatically reduced following inhibition of mTORC1. We previously found that mRNAs for Kit, Sohlh1, and Sohlh2 became associated with heavy polysomes at P4 [20], which coincided with the appearance of detectable protein. In a separate study, we reported that RA activated translation of repressed Kit mRNAs during differentiation by inducing heavy polysome occupancy without a dramatic increase in their abundance [19]. We therefore examined whether Sohlh1 and Sohlh2 mRNAs were regulated similarly in response to RA. In response to RA, both SOHLH1 and SOHLH2 protein levels increased dramatically (Fig. 6A-D), and as previously shown, RA treatment also induced Kit translation (Fig. S3A-B, [3, 19]). This increase in protein was not accompanied by an increase in steady-state mRNA levels (Fig. 6E-F). We then performed polysome gradient analysis to test whether RA induced Sohlh1 and Sohlh2 mRNA heavy polysome occupancy for efficient translation, as we recently showed for Kit [19]. Polysome gradients allow for the fractionation of mRNAs based on their association with ribonucleoprotein particles (RNPs), ribosome subunits, and light and heavy polysomes. The identification of specific mRNAs within sedimenting fractions reflects their translational efficiency, with those in heavy polysomes being most efficiently translated (reviewed in [69]). We pooled heavy polysome fractions in testes from vehicle- and RA-treated mice and discovered that Sohlh1, Sohlh2, and Kit mRNAs became enriched in heavy polysomes in response to RA (Fig. 6E-F and Fig. S3C). We conclude that, like Kit, Sohlh1 and Sohlh2 mRNAs are not efficiently translated in undifferentiated spermatogonia, and become activated at the level of translation in response to RA.
Figure 6. RA induces expression of SOHLH1 and SOHLH2 protein.
(A-D) Immunostaining of testis sections from mice treated at P1 with vehicle (A and C) or RA (B and D) and euthanized 24h later (at P2). Sections were stained with anti-SOHLH1 (A and B) or anti-SOHLH2 (C and D), and F-actin was stained with phalloidin (red) to visualize testis cords. QRT-PCR was performed on RNA isolated from whole testis lysate of mice treated with vehicle or RA, and total Sohlh1 and Sohlh2 mRNA levels were measured (left side, E and F). Messenger RNAs were separated by ribosome occupancy, fractions containing heavy polysomes were pooled, and qRT-PCR was performed to quantify polysome-associated Sohlh1 and Sohlh2 (right side, E and F). Scale bar = 30 μM. Asterisks indicate statistical significance with P<0.01.
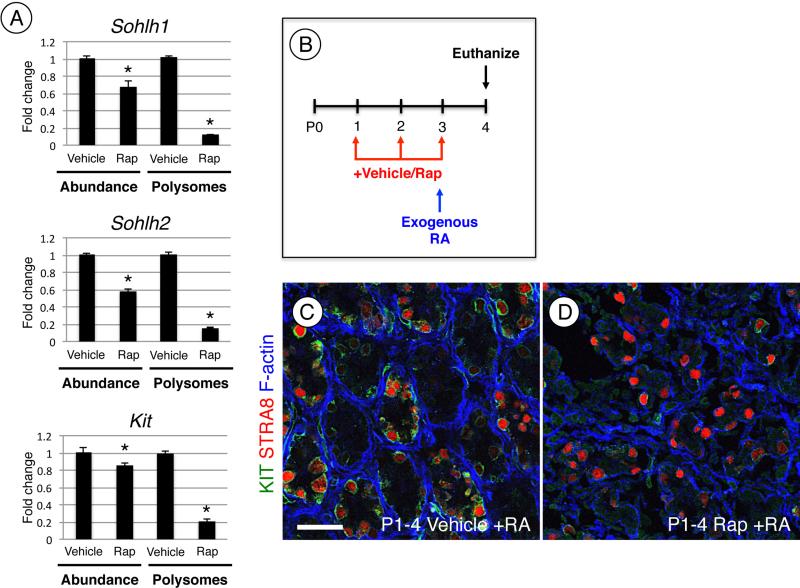
We next examined whether rapamycin treatment would inhibit this RA-induced translational activation. We utilized P4 rapamycin-treated testis, as they contained more similar numbers of germ cells to vehicle-treated controls than at P8 (Fig. 3A-D). In response to rapamycin, there was a small but statistically significant decrease in steady-state mRNA levels for Sohlh1, Sohlh2, and Kit (Fig. 7A), which corresponded closely with the decrease in the germ cell population at P4 (Fig. 3A-B). We found that rapamycin treatment caused a significant decrease in polysome occupancy for each of these mRNAs in comparison with vehicle-treated controls (Fig. 6A).
Figure 7. RA signaling through mTORC1 is required for induction of KIT but not STRA8.
(A) Message levels for Sohlh1, Sohlh2, and Kit were measured by qRT-PCR using whole testis RNA of mice treated with vehicle or rapamycin and then euthanized at P4. Sohlh1, Sohlh2, and Kit mRNAs associated with polysomes were pooled, isolated, and quantitated by qRT-PCR. (B) Mice were treated daily starting at P1 with vehicle or rapamycin. At P3, mice were given a single exogenous injection of RA and euthanized 24h later at P4. (C and D) Immunostaining of testis sections of mice treated with vehicle (C) or rapamycin (D) and RA and then euthanized at P4. Sections were labeled with KIT (green) and STRA8 (red), F-actin was stained with phalloidin to visualize testis cords (in blue). Scale bar = 40 μM. Asterisks indicate statistical significance with P<0.01.
Lastly, because rapamycin treatment could affect other cells in the testis and reduce endogenous RA levels, we tested whether exogenous RA could induce STRA8 a known transcriptional target required for meiotic initiation [3, 6, 70-72], in testes of rapamycin-treated mice. We assessed this using testes from mice euthanized at P4 following treatment with vehicle or rapamycin from P1-4 and injected with RA at P3 (Fig. 7B). We found that RA induced STRA8 and KIT in vehicle-treated control testes, as expected (Fig. 7C). In rapamycin-treated testes, KIT was not induced, as shown above (Fig. 4F and Fig. 7D). However, STRA8 was induced in response to RA. As expected, exogenous RA treatment induced Stra8 mRNA in rapamycin-treated testes (Fig. S3D). This indicates that RA activates expression of STRA8 and KIT in spermatogonia through distinct mechanisms (mTORC1-independent for STRA8, mTORC1-dependent for KIT). Taken together, results from the current study indicate that mTORC1 activation is a critical step downstream of RA in the translational activation of essential regulators of spermatogonial differentiation such as KIT, SOHLH1, and SOHLH2.
Discussion
Summary
Here, we show that inhibition of mTORC1 activity in the testis by rapamycin has profound effects on spermatogonial development. Rapamycin inhibited spermatogonial proliferation and differentiation, which reduced the germ cell population overall, but increased the percentage of undifferentiated spermatogonia (GFRA1+) and did not affect the SSC pool (Id4-GFP bright cells). At the molecular level, we found that rapamycin changed the spermatogonial response to RA. While Stra8 mRNA and protein were still induced in rapamycin-treated testes in response to RA, the enhanced translation of Sohlh1, Sohlh2, and Kit mRNAs was blocked. This supports the concept that RA exerts dual roles in the activation of transcription and translation in spermatogonia, and that blocking mTORC1 activation can functionally decouple these actions. Altogether, our results reveal an essential role for mTORC1 activation in RA induced enhanced translation of genes required for spermatogonia proliferation and differentiation, and provide insight into the male infertility phenotype observed following administration of rapamycin to both rodents and humans.
The diverse effects of rapamycin in various cell types
Rapamycin has wide-ranging cell- and tissue-specific effects. In animal models, rapamycin can exert a variety of positive consequences including lifespan extension, reduction of cancer progression, improved organ transplant retention, neuroprotection from damage caused by diseases such as Alzheimer's, Huntington's, and Parkinson's, and suppression of high fat diet-induced obesity (reviewed in [25]). These diverse outcomes highlight the observations from many laboratories that the molecular signaling through mTORC1 varies in a cell-dependent context. In particular, mTORC1 inhibition can alternatively lead to reduced proliferation, increased apoptosis, and blocked cellular differentiation (reviewed in [24]). In this study, treatment with rapamycin prior to the onset of differentiation (at P3-4) did not result in germ cell loss by apoptosis, but rather a near-complete inhibition of differentiation such that no cells were seen preparing to enter meiosis as preleptotene spermatocytes at P8. Our results also indicate that Sertoli cell numbers were unaffected by the treatment, and there were no significant changes in their position or appearance. The prolonged exposure to rapamycin can also inhibit the function of mTORC2, although this varies widely with cell type [73]. This is not likely occurring in this study; since activated mTORC2 phosphorylates and activates AKT [74], we would expect that its inhibition would lead to an increase in cytoplasmic FOXO1, which we did not observe.
The role of RA in mTORC1 activation and enhanced translation in vivo
During differentiation, spermatogonia respond to RA by proliferating and undergoing largely unknown cellular changes that precede meiosis. A primary reason for this lack of knowledge about spermatogonial differentiation is that there are very few changes in steady-state mRNA levels between undifferentiated and differentiating spermatogonia [13-15]. Without dramatic changes in the transcriptome, scientists have lacked targets (pathways, proteins) for focused studies. The classic mechanism by which RA controls gene expression is through modulating transcription of RA-responsive genes such as Stra8 and Rec8, which are required for entry into and progression through meiosis [70, 72, 75, 76]. Here and in a previous report [19], we identify a novel mechanism by which RA signals through the PI3K/AKT/mTOR signaling pathway to initiate the efficient translation of mRNAs required for spermatogonia differentiation. This reveals that RA can regulate gene expression by multiple mechanisms. In addition to Kit, we report here that the mRNAs for Sohlh1 and Sohlh2, which also encode essential determinants of spermatogonial differentiation, are stimulated by RA to become recruited into polysomes, resulting in a dramatic increase in protein levels without significant increases in steady-state mRNA abundance. This discrepancy between abundant mRNA and undetectable or barely detectable protein was previously alluded to in studies from the Rajkovic laboratory [77, 78]. Additional evidence for the transcription of these genes in undifferentiated spermatogonia comes from whole tubule explant cultures, in which GDNF increased mRNA levels for both Kit and Sohlh1, but did not induce protein expression [79]. Therefore, it is possible that a subset of genes is transcribed in undifferentiated spermatogonia, and that these mRNAs are poorly translated until RA activates the PI3K/AKT/mTOR kinase-signaling pathway to direct their mobilization into heavy polysomes for efficient translation. Our data supports a model whereby mTORC1 activation by RA leads to the translational activation of specific mRNAs required for spermatogonial differentiation (Fig. 8). There is precedent for a similar posttranscriptional regulation downstream of RA in local translation at neuronal dendritic and axonal termini. In that paradigm, RA binds RARA to activate the PI3K/AKT signaling pathway and stimulate translation of repressed mRNAs including Gria1/Glur1 [80-84]. Furthermore, a study found using the F9 cell line that RARG associated with the p-85 regulatory subunit of PI3K, and that PI3K-AKT activation by RA was required for F9 cell differentiation [84, 85].
Figure 8. RA signaling through PI3K/AKT/mTOR is required for spermatogonia differentiation.
Specific mRNAs are inefficiently translated (repressed) in undifferentiated germ cells. RA signaling through a kinase (non-genomic) signaling pathway activates the PI3K/AKT/mTORC1 signaling network to induce efficient translation of genes (e.g. Kit, Sohlh1, and Sohlh2) that are required for differentiation. Rapamycin inhibition of mTORC1 prevents RA induced efficient translation, and blocks spermatogonia differentiation.
It is clear that certain mRNAs are exceedingly sensitive to translational suppression by rapamycin [86]. These disproportionately affected mRNAs may have a stronger reliance on cap-dependent translation. Previous studies have shown that mTORC1-sensitive mRNAs generally contain complex 5’ UTRs that are positively regulated by phosphorylation of EIF4EBP1 [23] or are members of the class of 5’ TOP mRNAs which contain a 5’ terminal oligopyrimidine tract [87]. It is clear that Kit, Sohlh1, and Sohlh2 mRNAs appear to be sensitive to translational repression during spermatogenesis in vivo. This effect is mimicked by mTORC1 inhibition. Future studies will be aimed at identifying features within the UTRs that regulate translational repression and activation during spermatogenesis.
Germ cells are exposed to high levels of RA within discrete segments of the seminiferous cords (in the neonate and juvenile) and tubules (in the adult). In the adult, RA levels are highest along the seminiferous tubules at stages VII-VIII of the epithelial cycle [88]. This provides an explanation for how RA can regulate three distinct events simultaneously in different cell types, as they all occur at these stages: differentiation of type Apr and Aal into A1 spermatogonia, meiotic initiation of preleptotene spermatocytes, and spermiation of condensed spermatids. Indeed, RA has been shown to be required for each of these processes (reviewed in [9, 71, 89, 90]).
One interesting point to consider is that, although RA is required for the initiation of spermatogonial differentiation (to A1 spermatogonia), the subsequent divisions (A2, A3, A4, In, B) occur in levels of low or absent RA [88]. It is possible, then, that an important role of RA in spermatogonia is to stimulate the translation of mRNAs encoding KIT, which binds KITL to signal through the same PI3K/AKT/mTOR signaling pathway in spermatogonia in culture [40, 91]. By doing so, RA may signal through PI3K/AKT initially, and then the newly synthesized KIT receptor will bind KITL and maintain mTORC1 activation during these later differentiation stages when levels of RA are low or absent. This scenario would explain how the mRNAs for Kit, Sohlh1, and Sohlh2 would remain efficiently translated despite low levels of RA.
MTORC1 activity in germ cells
Previous studies have suggested a role for mTORC1 in spermatogonial fate determination. In the first study, rapamycin blocked proliferation (incorporation of BrdU) in cultured spermatogonia and prevented KITL-induced phosphorylation of RPS6KB1, suggesting that PI3K/AKT signaling was required in vivo [40]. In the second study using spermatogonia isolated from juvenile mice, it was concluded that ZBTB16/PLZF repressed mTORC1 activity by maintaining modestly higher steady-state mRNA levels (~3-fold) of an indirect negative regulator, REDD1 [42]. However, it was recently reported that REDD1 KO mice are viable and fertile, with no apparent defects in spermatogenesis [92]. In addition, proliferating/differentiating mTORC1-active neonatal spermatogonia contain abundant ZBTB16 [4], suggesting this model does not fully explain mTORC1 regulation in differentiating spermatogonia in vivo. In a third study, rapamycin was administered to adult testis tubules maintained in hanging drop cultures for 24h [58]. As expected, this reduced levels of p-mTOR, p-RPS6KB1, and p-EIF4EBP1 in spermatogonia and preleptotene spermatocytes. In addition, steady-state levels of PCNA and STRA8 were reduced, although these were assessed by western blot analysis of whole tubule lysates [58]. Since STRA8 levels are highest in preleptotene spermatocytes within stage VIII tubules [6, 12, 88], it is unclear whether the reduction in STRA8 protein levels following short-term rapamycin treatment was from impaired expression in spermatogonia (implying impaired differentiation) or in preleptotene spermatocytes. In a fourth study, Hobbs and colleagues generated germ cell KO mice for Tsc2, which encodes an indirect repressor of mTORC1 activation. Therefore, Tsc2 KO germ cells would be predicted to have elevated mTORC1 activity. When Tsc2 was deleted beginning in fetal prospermatogonia (by Ddx4-Cre), there were fewer undifferentiated spermatogonia (ZBTB16+), and an increased number of atrophic tubules in the adult. However, this incomplete effect suggests that loss of TSC2 either did not increase mTORC1 activation in all undifferentiated spermatogonia, or that a subset of spermatogonia differ in their response to mTORC1 activation. Here, we did not see a change in the Id4-GFP+ SSC-containing population in response to rapamycin treatment, which indicates that mTORC1 activity is low or not required in SSCs. Hobbs et al found that Tsc2 deletion in differentiating spermatogonia (using Stra8-Cre deletion), resulted in no discernable phenotype, presumably because mTORC1 was already activated in these differentiating cells. Taken together, those results complement our findings here that mTORC1 is suppressed in undifferentiated spermatogonia, and that its activation is necessary for differentiation.
In summary, this study provides the first examination of the requirement for mTORC1 activation in spermatogonial differentiation in vivo. Our results indicate that inhibition of mTORC1 blocked the RA-induced translational activation of repressed mRNAs, repressed spermatogonial differentiation, and resulted in an accumulation of undifferentiated progenitor spermatogonia.
Materials and Methods
Animal treatments and tissue collection
All animal procedures were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee of East Carolina University (AUPs #A178a and #A193). Analyses were done using CD-1 mice (Charles River Laboratories) or Id4-GFP mice, which are on a C57Bl/6 background [13]. Rapamycin was dissolved in DMSO and then diluted to a final concentration of 5 μg / μl in a solution of 5% polyethylene glycol 400 (Sigma-Aldrich) and 5% polysorbate 80 (Sigma-Aldrich). Rapamycin was fed daily using a 24-gauge feeding needle, in two regimens: 1) from P1-P3 and then euthanized at P4 or 2) from P1-P7 and then euthanized at P8. Using dosages similar to a previous study [93], we fed CD-1 mice rapamycin at 20 μg/g body weight, and Id4-GFP mice received 10 μg/g. The administration of exogenous RA was done as previously described [94]. Briefly, neonatal mice received one subcutaneous injection of 100 μg all-trans RA (#R2625, Sigma-Aldrich) dissolved in 10 μl dimethyl sulfoxide (DMSO) or DMSO alone at P3, and were euthanized by decapitation 24h later.
Polysome gradient analysis
Polysome gradients were performed as previously described [20]. Briefly, total testis lysates from at least 22 P4 vehicle- or rapamycin-treated mice were loaded onto 15%-45% linear sucrose gradient in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES pH 7.4, 0.5% NP-40, and 100 μg/ml cycloheximide). Gradients were fractionated, and 14 successive fractions were collected. RNA was isolated using TRIzol reagent based on manufacturer's protocol from pooled heavy polysome fractions (9-14).
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) was performed in triplicate on total RNA isolated from pooled polysomal fractions and on RNA isolated from whole testis lysates from at least 3 different mice as before [19, 20]. Briefly, generation of cDNA and amplification were performed in the same reaction tube using One-Step SYBR green and iScript polymerase (Bio-Rad) in an Applied Biosystems ViiA 7 Real-Time PCR System (Life Technologies). Primers were designed to span introns for Kit, Sohlh1, Sohlh2, Stra8, and B2m (Table 1). QRT-PCR was performed to measure the abundance of specific mRNAs within whole testis total RNA. Fold changes were calculated using the delta-delta Ct (ddCt) method using the reference gene B2m. Polysome occupancy of specific mRNAs was determined using qRT-PCR to amplify RNA isolated from pooled polysomal fractions. Relative mRNA levels were assessed using the dCt method from the lowest Ct in the group, and reported as a fold change.
Table 1.
Primer Sequences
| Gene | Upstream primer (5’-3’) | Downstream primer (5’-3’) |
|---|---|---|
| Kit | CATGGCGTTCCTCGCCT | GCCCGAAATCGCAAATCTTT |
| Sohlh1 | GGGCCAATGAGGATTACAGA | AAGTTTGCAGCAGCCACAG |
| Sohlh2 | TCTCAGCCACATCACAGAGG | GGGGACGCGAGTCTTATACA |
| Stra8 | TCACAGCCTCAAAGTGGCAGG | GCAACAGAGTGGAGGAGGAGT |
| B2m | CCGTGATCTTTCTGGTGCTT | CGTAGCAGTTCAGTATGTTCG |
Indirect Immunofluorescence (IIF)
IIF was performed as previously reported. Briefly, testes from at least 3 different mice were fixed in 4% PFA at 4°C. Testes were embedded in O.C.T., frozen, and cut into 5 μm sections. Sections were incubated with primary antibodies (see Table 2) for 1h at room temperature. Following stringency washes, sections were incubated with either Alexa Fluor anti-goat or anti-rabbit secondary antibody (1:2,000, Invitrogen) and phalloidin-635 or −594 (1:1,000, Invitrogen). Coverslips were mounted with Vectastain containing DAPI (Vector Laboratories). Images were captured using a Fluoview FV1000 confocal laser-scanning microscope (Olympus America).
Table 2.
Antibodies
| Protein | Vendor (Catalog Number) | Dilution |
|---|---|---|
| DDX4 | Abcam (ab13480) | 1:250 |
| DDX4 | R&D Systems (AF2030) | 1:800 |
| RET | Cell Signaling Technology (#3223) | 1:200 |
| STRA8 | Abcam (ab49602) | 1:3,000 |
| KIT | Santa Cruz Biotechnology (sc-1494) | 1:1,000 |
| KIT | Cell Signaling Technology (3074) | 1:1,000 |
| SOHLH1 | Alexsandar Rajkovic [95] | 1:200 |
| SOHLH2 | Alexsandar Rajkovic [78] | 1:200 |
| c-PARPl | Cell Signaling Technology (#9544) | 1:100 |
| p-RPS6 | Cell Signaling Technology (#5364) | 1:800 |
| p-MTOR | Cell Signaling Technology (#2880) | 1:100 |
| GFRA1 | R&D Systems (AF560) | 1:800 |
| GATA4 | Santa Cruz Biotechnology (sc-1237) | 1:100 |
Cell Quantitation
Immunostaining was performed on 5 μm frozen testis sections. Quantitation was carried out as previously described [4], and immunostaining was performed for DDX4 to mark all prospermatogonia and spermatogonia in the neonatal testis. Germ cells within 21-30 testis cords were counted from 3 different animals. Cells were identified as positive for a marker if selected by the threshold tool in Image J (U.S. National Institutes of Health) using the default algorithm. Intensity thresholds were as follows: DDX4 = 100-255, KIT = 40-255, SOHLH1 = 90-255, SOHLH2 = 90-255. Testis sections from Id4-Gfp mice were immunostained with anti-DDX4 antibodies. Photomicrographs were captured with an Axio Observer A1 microscope (Carl Zeiss Microscopy, LLC) equipped with an XL16C digital camera and Exponent version 1.3 software (Dage-MTI). Bright Id4-GFP+ cells were selected by Image J software with intensity thresholds set at 0-60. At least 400 DDX4+ cells were selected from 4 testis sections, and the number of GFP-bright cells recorded. Testes were analyzed from at least 3 mice.
Statistics
Statistical analyses of the qRT-PCR results and cell counts were performed using Student's t-test, and the level of significance was set at p≤0.01.
Supplementary Material
Acknowledgements
The authors thank Joani Zary-Oswald for technical assistance, and Brian Hermann (University of Texas at San Antonio) for critically reading the manuscript. Antibodies against SOHLH1 and SOHLH2 were kindly provided by Alexsandar Rajkovic (University of Pittsburgh, Magee-Womens Research Institute), and Id4-GFP transgenic mice were kindly provided by Dr. Jon Oatley (Washington State University). This work was supported by a grant from the NIH/NICHD (HD072552 to C.B.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors do not have any conflicts to disclose.
References
- 1.Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, Hamada H, Koopman P. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 2.Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busada JT, Kaye EP, Renegar RH, Geyer CB. Retinoic acid induces multiple hallmarks of the prospermatogonia-to-spermatogonia transition in the neonatal mouse. Biol Reprod. 2014;90:64. doi: 10.1095/biolreprod.113.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction. 2015;149:329–338. doi: 10.1530/REP-14-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder EM, Small C, Griswold MD. Retinoic acid availability drives the asynchronous initiation of spermatogonial differentiation in the mouse. Biol Reprod. 2010;83:783–790. doi: 10.1095/biolreprod.110.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod. 2008;79:35–42. doi: 10.1095/biolreprod.107.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogarth CA, Griswold MD. Retinoic acid regulation of male meiosis. Curr Opin Endocrinol Diabetes Obes. 2013 doi: 10.1097/MED.0b013e32836067cf. [DOI] [PubMed] [Google Scholar]
- 8.Kluin PM, Kramer MF, de Rooij DG. Spermatogenesis in the immature mouse proceeds faster than in the adult. Int J Androl. 1982;5:282–294. doi: 10.1111/j.1365-2605.1982.tb00257.x. [DOI] [PubMed] [Google Scholar]
- 9.de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- 10.de Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J Androl. 2012;33:1085–1095. doi: 10.2164/jandrol.112.016832. [DOI] [PubMed] [Google Scholar]
- 11.Kluin PM, Kramer MF, de Rooij DG. Proliferation of spermatogonia and Sertoli cells in maturing mice. Anat Embryol (Berl) 1984;169:73–78. doi: 10.1007/BF00300588. [DOI] [PubMed] [Google Scholar]
- 12.Endo T, Romer KA, Anderson EL, Baltus AE, de Rooij DG, Page DC. Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci U S A. 2015;112:E2347–2356. doi: 10.1073/pnas.1505683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan F, Oatley MJ, Kaucher AV, Yang QE, Bieberich CJ, Shashikant CS, Oatley JM. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28:1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod. 2008;78:537–545. doi: 10.1095/biolreprod.107.064337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JJ, Bickel PJ, Biggin MD. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ. 2014;2:e270. doi: 10.7717/peerj.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 18.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol. 2015;397:140–149. doi: 10.1016/j.ydbio.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chappell VA, Busada JT, Keiper BD, Geyer CB. Translational activation of developmental messenger RNAs during neonatal mouse testis development. Biol Reprod. 2013;89:61. doi: 10.1095/biolreprod.113.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, Geyer CB, McCarrey JR. Transcriptional and Translational Heterogeneity among Neonatal Mouse Spermatogonia. Biol Reprod. 2015 doi: 10.1095/biolreprod.114.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 23.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 2005;25:100–113. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson MA, Cronland E, Dunkelbarger S, Contreras V, Strome S, Keiper BD. A germ line-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J. Cell Science. 2009;122:1529–1539. doi: 10.1242/jcs.046771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 30.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 31.Song A, Labella S, Korneeva NL, Keiper BD, Aamodt EJ, Zetka M, Rhoads RE. A C. elegans eIF4E-family member upregulates translation at elevated temperatures of mRNAs encoding MSH-5 and other meiotic crossover proteins. J. Cell Sci. 2010;123:2228–2237. doi: 10.1242/jcs.063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochem Soc Trans. 2006;34:12–16. doi: 10.1042/BST20060012. [DOI] [PubMed] [Google Scholar]
- 33.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 35.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 36.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 37.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Rosenfeld SV, Blagosklonny MV. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10:4230–4236. doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- 40.Feng LX, Ravindranath N, Dym M. Stem cell factor/c-kit up-regulates cyclin D3 and promotes cell cycle progression via the phosphoinositide 3-kinase/p70 S6 kinase pathway in spermatogonia. J Biol Chem. 2000;275:25572–25576. doi: 10.1074/jbc.M002218200. [DOI] [PubMed] [Google Scholar]
- 41.Hobbs RM, La HM, Makela JA, Kobayashi T, Noda T, Pandolfi PP. Distinct germline progenitor subsets defined through Tsc2-mTORC1 signaling. EMBO Rep. 2015 doi: 10.15252/embr.201439379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142:468–479. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kofman AE, Huszar JM, Payne CJ. Transcriptional analysis of histone deacetylase family members reveal similarities between differentiating and aging spermatogonial stem cells. Stem Cell Rev. 2013;9:59–64. doi: 10.1007/s12015-012-9392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashworth RE, Wu J. Mammalian target of rapamycin inhibition in hepatocellular carcinoma. World J Hepatol. 2014;6:776–782. doi: 10.4254/wjh.v6.i11.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Pablo A, Santos F, Sole A, Borro JM, Cifrian JM, Laporta R, Monforte V, Roman A, de la Torre M, Ussetti P, Zurbano F. Recommendations on the use of everolimus in lung transplantation. Transplant Rev (Orlando) 2013;27:9–16. doi: 10.1016/j.trre.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Eyre TA, Collins GP, Goldstone AH, Cwynarski K. Time now to TORC the TORC? New developments in mTOR pathway inhibition in lymphoid malignancies. Br J Haematol. 2014;166:336–351. doi: 10.1111/bjh.12945. [DOI] [PubMed] [Google Scholar]
- 47.Ganschow R, Pollok JM, Jankofsky M, Junge G. The role of everolimus in liver transplantation. Clin Exp Gastroenterol. 2014;7:329–343. doi: 10.2147/CEG.S41780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng VC, Johnson JJ, Cuellar S. Targeting the mammalian target of rapamycin pathway with everolimus: Implications for the management of metastatic breast cancer. J Oncol Pharm Pract. 2014 doi: 10.1177/1078155214540732. [DOI] [PubMed] [Google Scholar]
- 49.Peddi VR, Wiseman A, Chavin K, Slakey D. Review of combination therapy with mTOR inhibitors and tacrolimus minimization after transplantation. Transplant Rev (Orlando) 2013;27:97–107. doi: 10.1016/j.trre.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Boobes Y, Bernieh B, Saadi H, Raafat Al Hakim M, Abouchacra S. Gonadal dysfunction and infertility in kidney transplant patients receiving sirolimus. Int Urol Nephrol. 2010;42:493–498. doi: 10.1007/s11255-009-9644-8. [DOI] [PubMed] [Google Scholar]
- 51.Framarino-dei-Malatesta M, Derme M, Manzia TM, Iaria G, De Luca L, Fazzolari L, Napoli A, Berloco P, Patel T, Orlando G, Tisone G. Impact of mTOR-I on fertility and pregnancy: state of the art and review of the literature. Expert Rev Clin Immunol. 2013;9:781–789. doi: 10.1586/1744666X.2013.824243. [DOI] [PubMed] [Google Scholar]
- 52.Huyghe E, Zairi A, Nohra J, Kamar N, Plante P, Rostaing L. Gonadal impact of target of rapamycin inhibitors (sirolimus and everolimus) in male patients: an overview. Transpl Int. 2007;20:305–311. doi: 10.1111/j.1432-2277.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- 53.Zaza G, Tomei P, Ria P, Granata S, Boschiero L, Lupo A. Systemic and nonrenal adverse effects occurring in renal transplant patients treated with mTOR inhibitors. Clin Dev Immunol. 2013;2013:403280. doi: 10.1155/2013/403280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuber J, Anglicheau D, Elie C, Bererhi L, Timsit MO, Mamzer-Bruneel MF, Ciroldi M, Martinez F, Snanoudj R, Hiesse C, Kreis H, Eustache F, et al. Sirolimus may reduce fertility in male renal transplant recipients. Am J Transplant. 2008;8:1471–1479. doi: 10.1111/j.1600-6143.2008.02267.x. [DOI] [PubMed] [Google Scholar]
- 55.Deutsch MA, Kaczmarek I, Huber S, Schmauss D, Beiras-Fernandez A, Schmoeckel M, Ochsenkuehn R, Meiser B, Mueller-Hoecker J, Reichart B. Sirolimus-associated infertility: case report and literature review of possible mechanisms. Am J Transplant. 2007;7:2414–2421. doi: 10.1111/j.1600-6143.2007.01929.x. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Zhang Z, Lin Y, Lin H, Li M, Nie P, Chen L, Qiu J, Lu Y, Chen L, Xu B, Lin W, et al. Long-term impact of immunosuppressants at therapeutic doses on male reproductive system in unilateral nephrectomized rats: a comparative study. Biomed Res Int. 2013;2013:690382. doi: 10.1155/2013/690382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker WH. Testosterone signaling and the regulation of spermatogenesis. Spermatogenesis. 2011;1:116–120. doi: 10.4161/spmg.1.2.16956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sahin P, Sahin Z, Gungor-Ordueri NE, Donmez BO, Celik-Ozenci C. Inhibition of mammalian target of rapamycin signaling pathway decreases retinoic acid stimulated gene 8 expression in adult mouse testis. Fertil Steril. 2014;102:1482–1490.e1483. doi: 10.1016/j.fertnstert.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 59.Drumond AL, Meistrich ML, Chiarini-Garcia H. Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction. 2011;142:145–155. doi: 10.1530/REP-10-0431. [DOI] [PubMed] [Google Scholar]
- 60.Kluin PM, de Rooij DG. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int J Androl. 1981;4:475–493. doi: 10.1111/j.1365-2605.1981.tb00732.x. [DOI] [PubMed] [Google Scholar]
- 61.de Rooij DG. Stem cells in the testis. Int J Exp Pathol. 1998;79:67–80. doi: 10.1046/j.1365-2613.1998.t01-1-00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Rooij DG, Lok D. Regulation of the density of spermatogonia in the seminiferous epithelium of the Chinese hamster: II. Differentiating spermatogonia. Anat Rec. 1987;217:131–136. doi: 10.1002/ar.1092170204. [DOI] [PubMed] [Google Scholar]
- 63.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 64.Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 66.Lopez F, Belloc F, Lacombe F, Dumain P, Reiffers J, Bernard P, Boisseau MR. Modalities of synthesis of Ki67 antigen during the stimulation of lymphocytes. Cytometry. 1991;12:42–49. doi: 10.1002/cyto.990120107. [DOI] [PubMed] [Google Scholar]
- 67.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 69.Masek T, Valasek L, Pospisek M. Polysome analysis and RNA purification from sucrose gradients. Methods Mol Biol. 2011;703:293–309. doi: 10.1007/978-1-59745-248-9_20. [DOI] [PubMed] [Google Scholar]
- 70.Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Griswold MD, Hogarth CA, Bowles J, Koopman P. Initiating meiosis: the case for retinoic acid. Biol Reprod. 2012;86:35. doi: 10.1095/biolreprod.111.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Feret B, Vernet N, Codreanu CA, Chambon P, Ghyselinck NB. STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J Cell Sci. 2008;121:3233–3242. doi: 10.1242/jcs.035071. [DOI] [PubMed] [Google Scholar]
- 73.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 74.Hay N. Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta. 2011;1813:1965–1970. doi: 10.1016/j.bbamcr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koubova J, Hu YC, Bhattacharyya T, Soh YQ, Gill ME, Goodheart ML, Hogarth CA, Griswold MD, Page DC. Retinoic acid activates two pathways required for meiosis in mice. PLoS Genet. 2014;10:e1004541. doi: 10.1371/journal.pgen.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 77.Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 78.Ballow DJ, Xin Y, Choi Y, Pangas SA, Rajkovic A. Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expr Patterns. 2006;6:1014–1018. doi: 10.1016/j.modgep.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Grasso M, Fuso A, Dovere L, de Rooij DG, Stefanini M, Boitani C, Vicini E. Distribution of GFRA1-expressing spermatogonia in adult mouse testis. Reproduction. 2012;143:325–332. doi: 10.1530/REP-11-0385. [DOI] [PubMed] [Google Scholar]
- 80.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen N, Napoli JL. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARalpha. Faseb j. 2008;22:236–245. doi: 10.1096/fj.07-8739com. [DOI] [PubMed] [Google Scholar]
- 82.Chen N, Onisko B, Napoli JL. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J Biol Chem. 2008;283:20841–20847. doi: 10.1074/jbc.M802314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maghsoodi B, Poon MM, Nam CI, Aoto J, Ting P, Chen L. Retinoic acid regulates RARalpha-mediated control of translation in dendritic RNA granules during homeostatic synaptic plasticity. Proc Natl Acad Sci U S A. 2008;105:16015–16020. doi: 10.1073/pnas.0804801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Masia S, Alvarez S, de Lera AR, Barettino D. Rapid, nongenomic actions of retinoic acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic acid receptor. Mol Endocrinol. 2007;21:2391–2402. doi: 10.1210/me.2007-0062. [DOI] [PubMed] [Google Scholar]
- 85.Lopez-Carballo G, Moreno L, Masia S, Perez P, Barettino D. Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J Biol Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- 86.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 88.Hogarth CA, Arnold S, Kent T, Mitchell D, Isoherranen N, Griswold MD. Processive pulses of retinoic acid propel asynchronous and continuous murine sperm production. Biol Reprod. 2015;92:37. doi: 10.1095/biolreprod.114.126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bowles J, Koopman P. Retinoic acid, meiosis and germ cell fate in mammals. Development. 2007;134:3401–3411. doi: 10.1242/dev.001107. [DOI] [PubMed] [Google Scholar]
- 90.O'Donnell L, Nicholls PK, O'Bryan MK, McLachlan RI, Stanton PG. Spermiation: The process of sperm release. Spermatogenesis. 2011;1:14–35. doi: 10.4161/spmg.1.1.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, Soares V, Angeles M, Whitlow SR, Manova K, Besmer P. Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. EMBO J. 2000;19:1312–1326. doi: 10.1093/emboj/19.6.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Notini AJ, McClive PJ, Meachem SJ, van den Bergen JA, Western PS, Gustin SE, Harley VR, Koopman P, Sinclair AH. Redd1 is a novel marker of testis development but is not required for normal male reproduction. Sex Dev. 2012;6:223–230. doi: 10.1159/000339723. [DOI] [PubMed] [Google Scholar]
- 93.Puighermanal E, Marsicano G, Busquets-Garcia A, Lutz B, Maldonado R, Ozaita A. Cannabinoid modulation of hippocampal long-term memory is mediated by mTOR signaling. Nat Neurosci. 2009;12:1152–1158. doi: 10.1038/nn.2369. [DOI] [PubMed] [Google Scholar]
- 94.Busada JT, Kaye EP, Renegar RH, Geyer CB. Retinoic Acid Induces Multiple Hallmarks of the Prospermatogonia-to-Spermatogonia Transition in the Neonatal Mouse. Biol Reprod. 2014 doi: 10.1095/biolreprod.113.114645. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A. 2006;103:8090–8095. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.