Abstract
Context and objective
We examined whether a prevalent caveolin-1 gene (CAV1) variant, previously related to insulin resistance, is associated with metabolic syndrome (MetS).
Patients and methods
We included subjects genotyped for the CAV1 variant rs926198 from two cohorts: 735 Caucasians from the HyperPATH multicenter study, and 810 Hispanic participants from the HTN-IR cohort.
Results
Minor allele carriers from HyperPATH cohort (57% of subjects) had higher Framingham risk scores, higher odds of diabetes (10.7% vs 5.7%, p=0.016), insulin resistance (44.3% vs 35.1%, p=0.022), low HDL (49.3% vs 39.6%, p=0.018) and MetS (33% vs 20.5%, p<0.001) but similar BMI. Consistently, minor allele carriers exhibited higher odds of MetS, even when adjusted for confounders and relatedness (OR 2.83 (1.73–4.63), p<0.001). The association with MetS was replicated in the Hispanic cohort HTN-IR (OR 1.61, [1.06–2.44], p=0.025). Exploratory analyses suggest that MetS risk is modified by a CAV1 variant - BMI status interaction, whereby the minor allele carrier status strongly predicted MetS (OR 3.86 [2.05–7.27], p<0.001) and diabetes (OR 2.27 [1.07–4.78], p=0.03) in non-obese, but not in obese subjects. In addition, we observed a familial aggregation for MetS diagnosis in minor allele carriers.
Conclusion
The prevalent CAV1 gene variant rs926198 is associated with MetS in separate Caucasian and Hispanic cohorts. These findings appear to be driven by an interaction between the genetic marker and obesity status, suggesting that the CAV1 variant may improve risk profiling in non-obese subjects. Additional studies are needed to confirm the clinical implications of our results.
Keywords: caveolin 1, metabolic syndrome, dyslipidemia, insulin resistance, diabetes, cardiovascular risk
1. INTRODUCTION
The metabolic syndrome (MetS) is a composite of central obesity, hyperglycemia/insulin resistance (IR), dyslipidemia and hypertension that is highly prevalent worldwide [2]. MetS is also associated with all-cause mortality, myocardial infarction, and stroke in subjects with and without diabetes [3]. Interestingly, current evidence shows that metabolically unhealthy normal-weight individuals have similar mortality and cardiovascular disease risk compared with metabolically unhealthy overweight or obese persons [4, 5]. These prospective observations suggest that individual cardiometabolic risk should involve more in-depth considerations than body-mass index (BMI) alone.
Our group has previously described that variations in the CAV1 gene were associated with IR in hypertensive humans, consistent with our bench studies that demonstrated that CAV1 deficiency in mice leads to abnormal glucose metabolism and hyperinsulinemia [6–8]. CAV1, the main component of plasma membrane caveolae, has been widely studied in cardiovascular and kidney tissues for its critical role in signal transduction and trafficking, and also for its interplay with steroid receptors and ion channel activation [6]. In adipose tissue, caveolae are typically abundant, occupying up to 30% of the surface area, and have been shown to regulate adipocyte differentiation, transport and lipid droplet formation [9]. Of note, CAV1 knockout mice display abnormal glucose tolerance, hypertension and dyslipidemia despite a lean body habitus. Also, humans with undetectable CAV1 expression due to severe CAV1 gene (CAV1) mutations display a lipodystrophic phenotype associated with insulin resistance, acanthosis nigricans, diabetes mellitus, and hypertriglyceridemia, suggesting that CAV1-related metabolic disorders may be secondary to adipocyte dysfunction rather than to increased fat mass [9–11].
In the present study, we tested the hypothesis that a prevalent CAV1 variant is associated with MetS. We investigated this hypothesis in a large Caucasian cohort with subsequent replication in a Hispanic cohort; in contrast to our previous report [6], the current analysis included not only hypertensives, but also normotensives and diabetics. We further performed exploratory analyses to investigate the role of this CAV1 variant in (1) predicting MetS in non-obese individuals and (2) in familial aggregation for MetS diagnosis.
2. METHODS
2.1 HyperPATH protocol (Caucasian cohort)
Participants were selected from the HyperPATH Cohort, a protocol that controls for factors that influence the renin-angiotensin-aldosterone-system (RAAS). A total of 735 Caucasian adults with available genotype were included in the analysis. All participants were studied under a common protocol in Clinical Research Centers located at Brigham and Women’s Hospital (Boston, MA), University of Utah Medical Center (Salt Lake City, UT), Vanderbilt University Medical Center (Nashville, TN), Hospital Broussais (Paris, France), and San Salvatore Hospital (Rome, Italy). HTN was defined as described previously [12]. Type 2 diabetes mellitus and prediabetes were defined as per American Diabetes Association criteria [13]. IR estimation was calculated by HOMA-IR and IR as a categorical variable considering the upper quartile of HOMA-IR in the HyperPATH Cohort (HOMA-IR ≥2.8). As previously published by our group and following World Health Organization suggestions, to be diagnosed with MetS a participant needed to have diabetes, prediabetes or IR plus ≥2 of the following characteristics: 1) HTN, 2) dyslipidemia (triglyceride measurement ≥150 mg/dL or high-density lipoprotein <35 mg/dL in men and <39 mg/dL in women), or 3) BMI ≥30 kg/m2 and/or waist: hip ratio >0.9 in men, >0.85 in women [12, 14]. Sensitivity analysis using the harmonized MetS criteria was also performed [15]. Participants were also classified as “metabolically unhealthy normal-weight” if they presented MetS with a BMI in the 20–27 kg/m2 range as proposed in non-Asian subjects [16, 17].
Framingham risk score and laboratory analyses were performed using standardized and validated methods, as previously described [12].
2.2 The HTN-IR Cohort and Study Protocol (Hispanic cohort)
Hispanic subjects with available genotype (810 participants) were analyzed from the Mexican-American Hypertension-Insulin Resistance (HTN-IR) cohort and recruited through the Hypertension Clinic at Los Angeles County, University of Southern California Medical Center or the Clinical Research Center at the University of California [18]. The criteria for MetS diagnosis were the same as outlined above for Caucasian subjects. For the definition of IR as a categorical variable, we used the upper HOMA quartile in this cohort (HOMA-IR ≥4.9).
2.3 Genotype, quality control, and expression profile analysis
In the current study, we used rs926198 as the only candidate variant to be explored using a dominant model. This SNP was chosen based on our previous work where we analyzed eleven HapMap variants of the CAV1 gene, in relation to IR [6]. The dominant model analysis (grouping heterozygous and minor allele homozygous as minor allele carriers) was selected based on the frequency of the minor allele in our population, and on the similarity of the phenotype displayed by the minor allele carriers subgroups with the trait of interest. DNA was genotyped using the Sequenom platform, with a completion rate of greater than 95% and adequate quality control. The genotype and allele frequencies were in Hardy-Weinberg equilibrium (HWE) in both cohorts: HyperPATH 65.8% for major allele T and 34.2% for minor allele C, p=0.62; HTN-IR: 81.4% for major allele T and 18.6% for minor allele C, p=0.42.
In the case where rs926198 was not available in a specific subject, as a proxy we used the CAV1 variant rs917664 within the HyperPATH cohort, which is in perfect linkage disequilibrium (r2=1) in both HapMap and 1000 Genomes datasets.
We evaluated rs926198 as an expression quantitative trait locus (eQTL) for CAV1 gene expression, by using two shared resources that provide information on specific gene expression by variant: the GENEVAR (GENe Expression VARiation) and the Genotype-Tissue Expression (GTEx) databases [19]. For this specific analysis we assessed rs926198-associated CAV1 expression in lymphoblastoid cell lines; in addition, the analysis in GENEVAR was performed in HapMap subjects by race of interest [6, 20]
2.4 Statistical analysis
Data and results are presented as mean ± SD and percentages for categorical variables. Univariate analyses were performed using a Chi-square test and an unpaired Student’s t test. Observed and expected values for allele frequencies to evaluate HWE were compared with a Chi-square test. The odds of MetS were compared by genotype, using a mixed effects logistic regression model. Covariates in the model were used as fixed effects and were chosen based on their clinical importance. Age, gender, site of study and BMI were included. We used a random effects model to account for participant relatedness. Secondary analyses were performed and included the analyses for the different components that cluster in MetS to test the robustness of our primary outcome and to evaluate potential differences of each trait. Furthermore, because of the described effect of low CAV1 on metabolic disorders despite a lean phenotype we evaluated the interaction of the CAV1 variant with the BMI status. Sensitivity analyses also included evaluating the outcome with a second definition for MetS (Harmonized criteria) and incorporating HOMA-IR as a covariate in the model, thus assessing if the association between the CAV1 variant and MetS was beyond the described effect on IR. Bootstrapping with 1000 iterations was selected to corroborate the performance of regression models and possible influence of non-normality and outliers.
We explored whether familial aggregation exceeded chance aggregation in minor allele carriers compared to homozygous for the major allele using 348 subjects (142 sibships) from the HyperPATH cohort using a Chi-square analysis as previously described [21]. Next, the ratio of the observed-to-expected concordance of MetS by genotype was adjusted for confounders and relatedness in a mixed model logistic regression. We replicated our findings using the HTN-IR cohort; associations between MetS and the CAV1 variant were analyzed with the same covariates and family aggregation used in HyperPATH. Analyses were performed using STATA 11 and SAS 9.2. A p-value <0.05 was considered significant.
3. RESULTS
3.1 HyperPATH study population (Caucasian cohort)
Cohort characteristics of the 735 subjects analyzed are: age 45.1 ± 10.4 years, BMI 27.2 ± 4.2 kg/m2, 45% female, 67% HTN, 9% diabetes, and 30% MetS. The rs926198 minor allele carrier status (CT/CC) was observed in 57% of participants versus 43% who were homozygous for the major allele (TT), consistent with this cohort being in HWE. The observed allele and genotype frequencies are similar to those reported in HapMap for Caucasians [6].
3.2 Clinical and biochemical characteristics by CAV1 genotype in HyperPATH
Compared to major allele homozygotes, minor allele carriers of the CAV1 variant had similar age, gender and HTN prevalence, but increased IR. Moreover, CAV1 minor allele carriers also had a higher 10-year CV risk by Framingham score, and higher prevalence of diabetes, dyslipidemia and MetS (Table 1). Of note, the higher cardiometabolic risk profile of the CAV1 minor allele carriers was not explained by differences in BMI between groups (p=0.247).
TABLE 1.
Clinical and biochemical characteristics categorized by the CAV1 variant rs926198 in Caucasian subjects
| Variable | Major Allele Homozygotes [n=315] TT |
Minor Allele Carriers [n=420] CT/CC |
p-value |
|---|---|---|---|
| Gender (female) | 131/315 (42%) | 198/420 (47%) | 0.134 |
| Age (y) | 44.2 ± 11.1 | 45.7 ± 9.8 | 0.052 |
| Body Mass Index (kg/m2) | 27.0 ± 4.2 | 27.3 ± 4.2 | 0.247 |
| Hypertension | 208/315 (66%) | 291/420 (69%) | 0.350 |
| Type 2 Diabetes | 18/315 (5.7%) | 45/420 (10.7%) | 0.016 |
| Type 2 Diabetes/Pre-Diabetes | 55/315 (17.4%) | 99/420 (24%) | 0.035 |
| IR | 91/259 (35.1%) | 161/363 (44.3%) | 0.021 |
| Triglycerides > 150 mg/dL | 91/272 (33.5%) | 153/378 (40.5%) | 0.060 |
| Low HDL (< 35 or 39 mg/dl) | 102/257 (39.6%) | 177/359 (49.3%) | 0.018 |
| Metabolic Syndrome* | 64/312 (20.5%) | 149/409 (33%) | <0.001 |
| Framingham risk score | 7.5 ± 7.0 | 8.9 ± 6.3 | 0.011 |
MetS if they had type 2 diabetes, IFG, IGT or HOMA>2.8 plus ≥2 criteria for dyslipidemia (triglycerides or HDL), hypertension or Obesity
3.3 Role of CAV1 genotype in MetS status and related disorders in HyperPATH
The association between MetS status, our primary outcome, and the presence of at least one minor allele was highly significant, even when adjusting for participant relatedness and known risk factors for MetS such as male gender, older age, and increased BMI (OR 2.83 [1.73–4.73], p<0.001, Table 2). Sensitivity analysis, including HOMA-IR as a covariate, did not modify the association between the CAV1 variant and MetS (adjusted OR 2.70 [1.59–4.61], p<0.001), suggesting that the effect extends beyond IR. Moreover, we confirmed that the CAV1 minor allele carrier status predicted higher odds for MetS using the new harmonized criteria definition in 582 subjects (OR 1.92 [1.25–2.91], p=0.003)
TABLE 2.
Crude and adjusted odds ratios for metabolic syndrome (primary outcome) and related disorders categorized by the CAV1 genotype rs926198 in Caucasians
| Cardiometabolic Outcome | rs926198 CAV1 variant Minor allele carriers versus major allele homozygotes |
|
|---|---|---|
| Crude OR (95% CI), p value | Adjusted OR* (95% CI), p value | |
| Metabolic syndrome† | 2.35 (1.60 – 3.462), p <0.001 | 2.83 (1.73 – 4.63), p <0.001 |
| Type 2 diabetes | 3.01 (1.14 – 7.98), p =0.027 | 2.38 (1.07 – 5.32), p =0.034 |
| Insulin resistance‡ | 1.52 (1.04 – 2.20), p =0.029 | 1.68 (1.15 – 2.44), p =0.007 |
| Hypertension | 1.48 (0.65 – 3.36), p =0.345 | 1.13 (0.54 – 2.40), p =0.744 |
| Low HDL Cholesterol | 1.56 (1.07 – 2.29), p =0.022 | 1.73 (1.12 – 2.68), p =0.014 |
| High Triglycerides | 1.59 (0.93 – 2.73), p =0.093 | 1.67 (0.95 – 2.94), p =0.078 |
| Obesity | 1.12 (0.66 – 1.89), p =0.671 | 0.70(0.29 – 1.69), p =0.425 |
Adjusted by age, gender, body mass index and site of the study as fixed effects and relatedness as random effect.
MetS if they had type 2 diabetes, IFG, IGT or HOMA upper quartile ≥2 criteria for Dyslipidemia (Triglycerides > 150 mg/dl or HDL<35 mg/dL in men, <39 mg/dL in women), Hypertension or Obesity
Criteria for IR: upper quartile of HOMA-IR in the HyperPATH Cohort (HOMA-IR ≥2.8)
We also determined whether the presence of the rs926198 minor allele was associated with specific traits that are used to define MetS. We observed that minor allele carriers of the CAV1 variant rs926198 also had higher odds for diabetes, IR and low HDL in both crude and adjusted models (Table 2). Notably, we observed no association of the CAV1 genotype with HTN or obesity, with a non-significant trend for triglyceride levels (Table 2).
3.4 Replication Cohort (Hispanic cohort)
We then tried to replicate our finding in the HTN-IR Cohort of 810 Hispanic adults, with MetS status as the primary outcome and adjusted for the same covariates. The HTN-IR cohort has the following characteristics: age 39.3 ± 15.1 years, BMI 29.1 ± 5.6 kg/m2, 59% female, 24% HTN, 15% diabetes, and 24% had MetS. Prevalence of minor allele carriers was 33%, consistent with the HapMap genotype frequency for Mexican ancestry in Los Angeles. Consistent with our Caucasian cohort, minor allele carriers of rs926198 CAV1 displayed higher odds for MetS in the adjusted model (OR 1.61, [1.06–2.44], p=0.025). We again observed no differences in BMI by genotype suggesting that the described metabolic abnormalities are not related to body mass. Baseline characteristics of the subjects are reported in Supplementary Table 1.
Fisher’s combined P value for the two ethnically different cohorts showed that the presence of a minor allele in the rs926198 variant was strongly associated with an increased risk of MetS (p<0.0001).
3.5 Expression profile by CAV1 genotype
We assessed whether the variant rs926198 can be explained by altered CAV1 gene expression. Available expression profile (additive model) for the CAV1 gene in lymphoblastoid cell lines showed a significant negative association with the number of rs926198 minor alleles in HapMap Caucasians (CEU) (rho=−0.194, p=0.04), i.e. the lowest expression level was in minor allele homozygotes (CC) and the highest - in major allele homozygotes (TT). Consistent with the above-shown association the CC genotype was also associated with lower CAV1 expression in the HapMap Hispanic (MEX) population (rho=−0.307, p=0.04). These results were confirmed in lymphocytes from the GTEx database, where the minor rs926198 allele was associated with lower CAV1 expression levels (p=0.03).
3.6 Exploratory analyses in Caucasians for the role of CAV1 genotype in non-obese subjects and familial aggregation of MetS
Since we observed a significantly higher prevalence of MetS in minor allele carriers versus major allele homozygotes despite similar BMI (Table 1), and since minor allele carrier status did not predict higher risk of obesity, we explored the effect of obesity status on MetS by genotype. This analysis confirmed a significant interaction between genotype and BMI when categorized by obesity status (p=0.03 for interaction). Further we evaluated the ability of the CAV1 genotype to predict MetS in obese (BMI≥30 kg/m2, 200 subjects, MetS prevalence 62%) versus non-obese individuals (BMI<30 kg/m2, 535 subjects, MetS prevalence 18%). In the non-obese group, the minor allele carrier status improved the predictive capacity of the variant, showing a 4-fold odds ratio for MetS even when controlling for confounders (Table 3). The association between the CAV1 variant and MetS remained significant in non-obese subjects even when including HOMA-IR as a covariate, thus supporting the notion that the association of rs926198 with MetS cannot be explained by IR alone. Moreover, subjects with the risk allele had 4-fold higher odds to be classified as metabolically unhealthy normal-weight (adjusted OR 4.30 [1.34–13.84], p=0.01). Consistently, the adjusted prevalence of diabetes among the non-obese was two-fold higher in minor allele carriers of rs926198 when compared to major allele homozygotes (OR 2.27 [1.07–4.78], p=0.03, Table 3).
TABLE 3.
Adjusted odds ratio for metabolic syndrome by the CAV1 genotype rs926198 sorted by obesity status in Caucasians.
| Category* | rs926198 CAV1 variant Minor allele carriers versus major allele homozygous |
|
|---|---|---|
| Adjusted OR† (95% CI), p value | ||
| Metabolic syndrome | Non Obese | 3.86 (2.05 – 7.27), p <0.001 |
| Obese | 1.78(0.73 – 4.33), p =0.202 |
Participants were categorized as non-obese (BMI <30 kg/m2, n=535) or obese (≥30 kg/m2, n=200). Both groups differ in their relation to MetS by genotype (p for interaction=0.03).
Adjusted by age, gender, body mass index and site of the study as fixed effects and relatedness as random effect
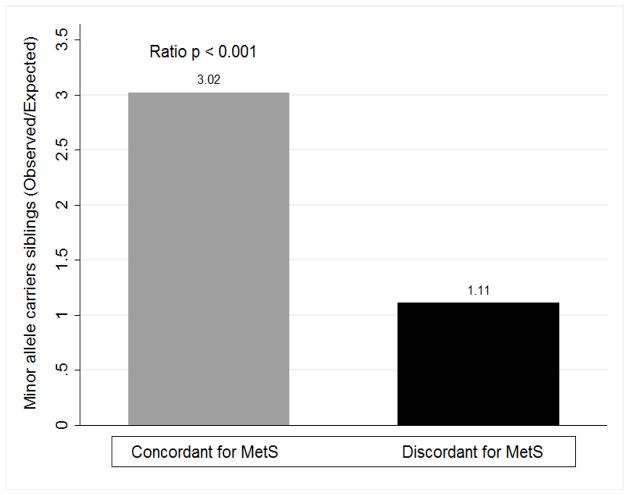
The observed concordance for MetS diagnosis was 3-fold higher in minor allele carriers indicating familial aggregation for the association of the trait of interest and the risk allele herein reported (Figure 1). The observed discordance for MetS status by genotype was similar to that expected randomly, as observed in Figure 1. Further, in an adjusted mixed model weighted by number of members in the family, concordant siblings with MetS diagnosis had 5-fold higher odds of carrying the CAV1 minor allele compared to discordant siblings (OR 5.57 [1.20–25.82], p=0.028).
FIGURE 1.
FAMILIAL AGGREGATION OF METABOLIC SYNDROME BY CAVEOLIN 1 VARIANT STATUS
Ratio of observed to expected proportions of Caucasian siblings concordant for the diagnosis of MetS was 3-fold higher in minor allele carriers compared to major allele homozygous indicative of familial aggregation of the trait of interest and rs926198 CAV1 variant status. Observed concordance of MetS by genotype differed significantly from expected (p<0.001). Observed discordance of siblings for MetS by genotype was similar to that expected suggesting it is randomly distributed.
4. DISCUSSION
Herein we confirm our primary hypothesis that the CAV1 variant rs926198 is associated with MetS in both Caucasians and Hispanics. Moreover, we show that CAV1 genetic variation is associated with diabetes, low HDL, cardiovascular risk as assessed by the Framingham score, and lower CAV1 expression. Further analyses suggest that non-obese minor allele carriers of the rs926198 variant display a particularly increased risk of developing MetS compared to their major allele homozygous counterparts. Thus, the rs926198 minor allele carrier status may be a valuable marker for several cardiometabolic risk factors for non-obese individuals–a group that, up to this point, has been extremely difficult to identify early as “at risk” for MetS.
Our group previously reported that CAV1 variants were associated with IR in two hypertensive, non-diabetic, ethnically different cohorts [6]. Herein we took the next logical step–to evaluate if the CAV1 polymorphism was associated with MetS in a phenotypically diverse and larger Caucasian cohort (including not only hypertensive but also normotensive and diabetic subjects), as well as in a much larger replication Hispanic cohort. Our hypothesis has important clinical implications; it is well known that MetS is a stronger predictor of CV disorders and mortality, as compared to IR or obesity status alone [22, 23]. In this study we report the novel finding that rs926198 strongly predicts MetS in both Caucasian and Hispanic adult subjects. Analyses of the individual components of MetS confirmed that rs926198 variant is not only related to IR, as previously described by us [6], but also to diabetes and dyslipidemia, particularly low HDL cholesterol. We first analyzed Caucasian subjects and showed that minor allele carriers had roughly 3-fold higher odds for MetS even when adjusted for confounders and relatedness. Next, we confirmed increased odds for MetS by genotype in a Hispanic cohort. From a clinical perspective, MetS has been validated as a single entity in all age groups, races and both genders, thus becoming an accessible tool to estimate cardiometabolic risk [24]. To date, information concerning the CAV1 gene and MetS is scarce, and is mainly based on animal studies [10, 25]. Of note, a particular region in chromosome 7 close to the CAV1 gene has been linked to both glucose and lipid factors but not increased adiposity in Asians, being the most associated region to both metabolic traits [26]. Also, linkage analysis showed that a region close to 7q.31 where CAV1 is located was related to metabolic syndrome [27]. Mechanistically, plasma membrane caveolae are active participants in signal transduction and interplay with several receptors and ion channels, thus highlighting the key role of CAV1 in several metabolic pathways [28, 29]. A potential role for CAV1 dysfunction in cardiometabolic disorders is supported by the described phenotypes of the CAV1-deficient mice (which exhibit IR, dyslipidemia and hypertension), as well as by those observed in humans with severe, non-sense CAV1 mutations (diabetes, IR and dyslipidemia) [10, 30, 31].
The observed increased risk for diabetes in Caucasian minor allele carriers could be attributed to the crucial role played by caveolae not only in insulin signaling (direct binding to the insulin receptor), but also in glucose homeostasis given that CAV1 has been reported to regulate glucose transporter-4 levels and mitochondrial function in adipose tissue [28, 32]. The relationship between diabetes and CAV1 deficiency has been studied mainly in animal models; thus, our novel finding in humans requires future validation [30, 33]. The reasons underlying the apparent higher odds for MetS and diabetes in Caucasian vs. Hispanic carriers of the CAV1 variant are unclear, and future studies are needed to explore whether this difference is due to variations in allele frequency/race/ethnicity or to dissimilarities in the cohort characteristics [34].
The association between the CAV1 variant and dyslipidemia is supported by genome-wide studies showing association of CAV1 gene proximal regions to low HDL and high triglycerides in two ethnically different cohorts [26, 35, 36]. Additionally, CAV1 KO mice display alterations in lipid composition (triglycerides, LDL and HDL), lending further support to a role for CAV1 in the complex regulation of lipoprotein metabolism and cholesterol efflux [8]. Despite preliminary findings in rodents, we did not find an association between the CAV1 variant and HTN in humans. The reason for this discrepancy is not clear; however, the elevated blood pressure and vascular dysfunction phenotypes described in CAV1 deficient rodent models were only present during a short term high sodium diet intervention, and thus they may reflect a salt-sensitive response [6, 37, 38].
Furthermore, we showed that rs926198 is an eQTLfor lower CAV1 expression in both Caucasian and Hispanic HapMap populations. These findings, replicated in a second eQTL database, lend strong support to an association between lower CAV1 levels and MetS risk. Furthermore, our in silico analyses support a translational link with rodent KO models with metabolic disorders, previously described by our group.
Interestingly, the CAV1 variant was related to Mets despite no difference in BMI between genotypes. Of note, both the CAV1 KO animal models as well as null autosomal recessive mutations of the CAV1 gene in humans associate with metabolic dysfunction despite a lean phenotype with decreased adiposity [10, 31]. Therefore, we then explored the predictive value of rs926198 for MetS in non-obese subjects. Our secondary analyses revealed that rs926198 associated with (1) higher odds for MetS in non-obese (more than 3-fold) and in metabolically unhealthy normal-weight subjects (roughly 4-fold) and with (2) higher odds for diabetes in non-obese subjects (2-fold), despite a lower prevalence of metabolic disorders in these subjects. The underlying mechanism of the abnormal metabolic phenotype in non-obese subjects is multifactorial and not fully elucidated, contributing to difficulties in screening the non-obese for MetS in clinical practice [39, 40]. Analysis of the Framingham Offspring study has shown that MetS diagnosis predicts long-term future CV events regardless of BMI category of the participants [22]. This prospective study and others confirm that there is no linear relationship of cardiovascular risk with BMI, thus highlighting the importance of individual risk profiling before severe clinical events [5, 23, 41]. Finally, the observed concordance for MetS diagnosis in siblings was 3-fold higher in minor allele carriers, indicative of familial aggregation for the trait of interest. The clustering in families supports our results relating CAV1 as the potential mechanism for the selected metabolic trait. Although studies have shown a familial aggregation for MetS [35], to our knowledge this is the first study to perform such an analysis for a CAV1 variant.
From a clinical perspective, since current practice lacks accurate predictive tools for MetS, the identification of genetic markers for MetS may help provide early diagnosis and would allow the implementation of simpler, preventive measures. Thus, the potential use of rs926198 as biomarker for MetS, especially in non-obese individuals, warrants further investigation.
Although we describe several novel findings linking a specific CAV1 genotype to cardiometabolic risk factors, our report has several potential limitations. First, both human protocols lack longitudinal follow up. Also, despite the wide use of MetS as a cardiovascular risk factor, a number of overlapping definitions are being used in the current literature; thus in our study we included the analysis of two definitions and each of the individual traits clustered in the definition of MetS. While we used two available eQTL datasets to determine the relationship between CAV1 expression and the variant of interest, we could not measure CAV1 levels in the recruited subjects to confirm our in silico findings. Furthermore, the mechanisms that underlie the association between CAV1 gene variants and the metabolic syndrome have yet to be studied
4.1 CONCLUSIONS
We describe a prevalent CAV1 variant that is associated with higher odds for MetS and lower CAV1 expression in both Caucasian and Hispanic subjects. Notably, our exploratory analyses suggest that the tested CAV1 variant improves MetS and diabetes diagnosis in non-obese subjects and with family aggregation. Therefore, prospective studies are needed to test if this particular genotype may improve individual risk profiling and personalized treatment.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support of the dietary, nursing, administrative and laboratory staff of all the Clinical Research Centers in which these studies were performed. Parts of the results of this study were shown as an oral presentation at the ICE/ENDO 2014 meeting.
FUNDING SOURCES:
This work was supported by grants from the National Institutes of Health: HL104032 (LHP), HL67974 (JIR), P50-HL55005 (JIR), K24 HL103845 (GKA), K23HL111771 (AV), F32NR013318 (PCU), NCATS UL1TR000124 (Cedars-Sinai Medical Center), M01-RR000425 (Cedars-Sinai Medical Center), M01-RR000043 (University of Southern California), P30-DK063491 (Southern California Diabetes Endocrinology Research Center), from the American Heart Association 14GRNT20500000 (LHP), from the Doris Duke Charitable Foundation (AV) and from the Chilean National Science and Technology Research Fund (FONDECYT) 1130427(RB), 1150437 (RB), 1150327(RB) and CORFO 13CTI-21526-P1 (RB).
ABBREVIATIONS
- MetS
Metabolic syndrome
- CAV1
caveolin-1
- HOMA
homeostasis model assessment
- IR
insulin resistance
- eQTL
expression quantitative trait loci
- KO
knockout
Footnotes
DISCLOSURES:
No potential conflicts of interest relevant to this article were reported.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pohl J, Ring A, Ehehalt R, Schulze-Bergkamen H, Schad A, Verkade P, et al. Long-chain fatty acid uptake into adipocytes depends on lipid raft function. Biochemistry. 2004;43:4179–87. doi: 10.1021/bi035743m. [DOI] [PubMed] [Google Scholar]
- 2.Ford ES. The metabolic syndrome and mortality from cardiovascular disease and all-causes: findings from the National Health and Nutrition Examination Survey II Mortality Study. Atherosclerosis. 2004;173:309–14. doi: 10.1016/j.atherosclerosis.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Mottillo S, Filion KB, Genest J, Joseph L, Pilote L, Poirier P, et al. The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. Journal of the American College of Cardiology. 2010;56:1113–32. doi: 10.1016/j.jacc.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 4.Kramer CK, Zinman B, Retnakaran R. Are metabolically healthy overweight and obesity benign conditions?: A systematic review and meta-analysis. Ann Intern Med. 2013;159:758–69. doi: 10.7326/0003-4819-159-11-201312030-00008. [DOI] [PubMed] [Google Scholar]
- 5.Romero-Corral A, Somers VK, Sierra-Johnson J, Korenfeld Y, Boarin S, Korinek J, et al. Normal weight obesity: a risk factor for cardiometabolic dysregulation and cardiovascular mortality. Eur Heart J. 2010;31:737–46. doi: 10.1093/eurheartj/ehp487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pojoga LH, Underwood PC, Goodarzi MO, Williams JS, Adler GK, Jeunemaitre X, et al. Variants of the caveolin-1 gene: a translational investigation linking insulin resistance and hypertension. J Clin Endocrinol Metab. 2011;96:E1288–92. doi: 10.1210/jc.2010-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grilo A, Fernandez ML, Beltran M, Ramirez-Lorca R, Gonzalez MA, Royo JL, et al. Genetic analysis of CAV1 gene in hypertension and metabolic syndrome. Thromb Haemost. 2006;95:696–701. [PubMed] [Google Scholar]
- 8.Frank PG, Pavlides S, Cheung MW, Daumer K, Lisanti MP. Role of caveolin-1 in the regulation of lipoprotein metabolism. Am J Physiol Cell Physiol. 2008;295:C242–8. doi: 10.1152/ajpcell.00185.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garg A, Agarwal AK. Caveolin-1: a new locus for human lipodystrophy. J Clin Endocrinol Metab. 2008;93:1183–5. doi: 10.1210/jc.2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuengsamarn S, Garza AE, Krug AW, Romero JR, Adler GK, Williams GH, et al. Direct renin inhibition modulates insulin resistance in caveolin-1-deficient mice. Metabolism. 2013;62:275–81. doi: 10.1016/j.metabol.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao H, Alston L, Ruschman J, Hegele RA. Heterozygous CAV1 frameshift mutations (MIM 601047) in patients with atypical partial lipodystrophy and hypertriglyceridemia. Lipids Health Dis. 2008;7:3. doi: 10.1186/1476-511X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaidya A, Underwood PC, Hopkins PN, Jeunemaitre X, Ferri C, Williams GH, et al. Abnormal aldosterone physiology and cardiometabolic risk factors. Hypertension. 2013;61:886–93. doi: 10.1161/HYPERTENSIONAHA.111.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36(Suppl 1):S67–74. doi: 10.2337/dc13-S067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetic medicine: a journal of the British Diabetic Association. 1998;15:539–53. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 15.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–5. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 16.Ruderman N, Chisholm D, Pi-Sunyer X, Schneider S. The metabolically obese, normal-weight individual revisited. Diabetes. 1998;47:699–713. doi: 10.2337/diabetes.47.5.699. [DOI] [PubMed] [Google Scholar]
- 17.Karelis AD, St-Pierre DH, Conus F, Rabasa-Lhoret R, Poehlman ET. Metabolic and body composition factors in subgroups of obesity: what do we know? J Clin Endocrinol Metab. 2004;89:2569–75. doi: 10.1210/jc.2004-0165. [DOI] [PubMed] [Google Scholar]
- 18.Xiang AH, Azen SP, Buchanan TA, Raffel LJ, Tan S, Cheng LS, et al. Heritability of subclinical atherosclerosis in Latino families ascertained through a hypertensive parent. Arterioscler Thromb Vasc Biol. 2002;22:843–8. doi: 10.1161/01.atv.0000015329.15481.e8. [DOI] [PubMed] [Google Scholar]
- 19.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS genetics. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamarthi B, Kolatkar NS, Hunt SC, Williams JS, Seely EW, Brown NJ, et al. Urinary free cortisol: an intermediate phenotype and a potential genetic marker for a salt-resistant subset of essential hypertension. J Clin Endocrinol Metab. 2007;92:1340–6. doi: 10.1210/jc.2006-2093. [DOI] [PubMed] [Google Scholar]
- 22.Meigs JB, Wilson PW, Fox CS, Vasan RS, Nathan DM, Sullivan LM, et al. Body mass index, metabolic syndrome, and risk of type 2 diabetes or cardiovascular disease. J Clin Endocrinol Metab. 2006;91:2906–12. doi: 10.1210/jc.2006-0594. [DOI] [PubMed] [Google Scholar]
- 23.Fan J, Song Y, Chen Y, Hui R, Zhang W. Combined effect of obesity and cardio-metabolic abnormality on the risk of cardiovascular disease: a meta-analysis of prospective cohort studies. Int J Cardiol. 2013;168:4761–8. doi: 10.1016/j.ijcard.2013.07.230. [DOI] [PubMed] [Google Scholar]
- 24.Viitasalo A, Lakka TA, Laaksonen DE, Savonen K, Lakka HM, Hassinen M, et al. Validation of metabolic syndrome score by confirmatory factor analysis in children and adults and prediction of cardiometabolic outcomes in adults. Diabetologia. 2014 doi: 10.1007/s00125-014-3172-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhang WZ. An association of metabolic syndrome constellation with cellular membrane caveolae. Pathobiol Aging Age Relat Dis. 2014:4. doi: 10.3402/pba.v4.23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam CH, Lam VK, So WY, Ma RC, Chan JC, Ng MC. Genome-wide linkage scan for factors of metabolic syndrome in a Chinese population. BMC Genet. 2010;11:14. doi: 10.1186/1471-2156-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W, Miller MB, Rich SS, North KE, Pankow JS, Borecki IB, et al. Linkage analysis of a composite factor for the multiple metabolic syndrome: the National Heart, Lung, and Blood Institute Family Heart Study. Diabetes. 2003;52:2840–7. doi: 10.2337/diabetes.52.11.2840. [DOI] [PubMed] [Google Scholar]
- 28.Fruhbeck G, Lopez M, Dieguez C. Role of caveolins in body weight and insulin resistance regulation. Trends Endocrinol Metab. 2007;18:177–82. doi: 10.1016/j.tem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Baudrand R, Pojoga LH, Romero JR, Williams GH. Aldosterone’s mechanism of action: roles of lysine-specific demethylase 1, caveolin and striatin. Current opinion in nephrology and hypertension. 2014;23:32–7. doi: 10.1097/01.mnh.0000436543.48391.e0. [DOI] [PubMed] [Google Scholar]
- 30.Pojoga LH, Yao TM, Opsasnick LA, Garza AE, Reslan OM, Adler GK, et al. Dissociation of Hyperglycemia from Altered Vascular Contraction and Relaxation Mechanisms in Caveolin-1 Null Mice. The Journal of pharmacology and experimental therapeutics. 2013 doi: 10.1124/jpet.113.209189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim CA, Delepine M, Boutet E, El Mourabit H, Le Lay S, Meier M, et al. Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy. J Clin Endocrinol Metab. 2008;93:1129–34. doi: 10.1210/jc.2007-1328. [DOI] [PubMed] [Google Scholar]
- 32.Yuan T, Hong S, Yao Y, Liao K. Glut-4 is translocated to both caveolae and non-caveolar lipid rafts, but is partially internalized through caveolae in insulin-stimulated adipocytes. Cell Res. 2007;17:772–82. doi: 10.1038/cr.2007.73. [DOI] [PubMed] [Google Scholar]
- 33.Oh YS, Lee TS, Cheon GJ, Jang IS, Jun HS, Park SC. Modulation of insulin sensitivity and caveolin-1 expression by orchidectomy in a nonobese type 2 diabetes animal model. Mol Med. 2011;17:4–11. doi: 10.2119/molmed.2009.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassy JL, Shrader P, Yang Q, Liu T, Yesupriya A, Chang MH, et al. Genetic associations with metabolic syndrome and its quantitative traits by race/ethnicity in the United States. Metab Syndr Relat Disord. 2011;9:475–82. doi: 10.1089/met.2011.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loos RJ, Katzmarzyk PT, Rao DC, Rice T, Leon AS, Skinner JS, et al. Genome-wide linkage scan for the metabolic syndrome in the HERITAGE Family Study. J Clin Endocrinol Metab. 2003;88:5935–43. doi: 10.1210/jc.2003-030553. [DOI] [PubMed] [Google Scholar]
- 36.Shearman AM, Ordovas JM, Cupples LA, Schaefer EJ, Harmon MD, Shao Y, et al. Evidence for a gene influencing the TG/HDL-C ratio on chromosome 7q32.3-qter: a genome-wide scan in the Framingham study. Hum Mol Genet. 2000;9:1315–20. doi: 10.1093/hmg/9.9.1315. [DOI] [PubMed] [Google Scholar]
- 37.Pojoga LH, Yao TM, Sinha S, Ross RL, Lin JC, Raffetto JD, et al. Effect of dietary sodium on vasoconstriction and eNOS-mediated vascular relaxation in caveolin-1-deficient mice. Am J Physiol Heart Circ Physiol. 2008;294:H1258–65. doi: 10.1152/ajpheart.01014.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gildea JJ, Kemp BA, Howell NL, Van Sciver RE, Carey RM, Felder RA. Inhibition of renal caveolin-1 reduces natriuresis and produces hypertension in sodium-loaded rats. Am J Physiol Renal Physiol. 2011;300:F914–20. doi: 10.1152/ajprenal.00380.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wildman RP, Muntner P, Reynolds K, McGinn AP, Rajpathak S, Wylie-Rosett J, et al. The obese without cardiometabolic risk factor clustering and the normal weight with cardiometabolic risk factor clustering: prevalence and correlates of 2 phenotypes among the US population (NHANES 1999–2004) Arch Intern Med. 2008;168:1617–24. doi: 10.1001/archinte.168.15.1617. [DOI] [PubMed] [Google Scholar]
- 40.Conus F, Rabasa-Lhoret R, Peronnet F. Characteristics of metabolically obese normal-weight (MONW) subjects. Appl Physiol Nutr Metab. 2007;32:4–12. doi: 10.1139/h06-092. [DOI] [PubMed] [Google Scholar]
- 41.Choi KM, Cho HJ, Choi HY, Yang SJ, Yoo HJ, Seo JA, et al. Higher mortality in metabolically obese normal-weight people than in metabolically healthy obese subjects in elderly Koreans. Clin Endocrinol (Oxf) 2013 doi: 10.1111/cen.12154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.