Summary
Salmonellae survive and propagate in macrophages to cause serious systemic disease. Periplasmic superoxide dismutase plays a critical role in this survival by combating phagocytic superoxide. Salmonella Typhimurium strain 14028 produces two periplasmic superoxide dismutases, SodCI and SodCII. Although both proteins are produced during infection, only SodCI is functional in the macrophage phagosome. We have previously shown that SodCI, relative to SodCII, is both protease resistant and tethered within the periplasm, and that either of these properties is sufficient to allow a SodC to protect against phagocytic superoxide. Tethering is defined as remaining cell-associated after osmotic shock or treatment with cationic antimicrobial peptides. Here we show that SodCI non-covalently binds peptidoglycan. SodCI binds to Salmonella and Bacillus peptidoglycan, but not peptidoglycan from Staphylococcus. Moreover, binding can be inhibited by a diaminopimelic acid containing tripeptide, but not a lysine containing tripeptide, showing that the protein recognizes the peptide portion of the peptidoglycan. Replacing nine amino acids in SodCII with the corresponding residues from SodCI confers tethering, partially delineating an apparently novel peptidoglycan binding domain. These changes in sequence increase the affinity of SodCII for peptidoglycan fragments to match that of SodCI, and allow the now tethered SodCII to function during infection.
Keywords: Salmonella, superoxide, peptidoglycan, SodC
Introduction
Salmonellae are food-borne pathogens that cause morbidity and mortality throughout the world (Buckle et al., 2012; Feasey et al., 2012; Scallan et al., 2011). Serious and sometimes lethal infection results from the ability of Salmonella to survive and propagate in macrophages. The bacteria circumvent the normal killing mechanisms of these phagocytic cells using a variety of virulence factors, including periplasmic copper/zinc co-factored superoxide dismutase (SodC), which is critical for resistance to the antimicrobial phagocytic superoxide (Slauch, 2011; Burton et al., 2014)
Salmonella enterica serovar Typhimurium strain 14028 produces two periplasmic superoxide dismutases, SodCI and SodCII (Fang et al., 1999). The SodCII enzyme is encoded in the chromosome, whereas the SodCI enzyme is encoded on the fully functional lambdoid phage Gifsy-2, which is found in many strains of Salmonella (Figueroa-Bossi and Bossi, 1999). Although both enzymes are produced during Salmonella infection of animals and during growth in macrophages in tissue culture, only SodCI contributes to infection; SodCII plays no role in the ability of Salmonella to cause disease, even in the absence of SodCI (Uzzau et al., 2002; Krishnakumar et al., 2004; Krishnakumar et al., 2007; Ammendola et al., 2008).
The properties of the two SodC enzymes, which are ~60% identical at the amino acid level, have been characterized in some detail. SodCI is a 32 kDa dimeric protein that is protease resistant (Pesce et al., 2000; Gabbianelli et al., 2004; Krishnakumar et al., 2007). SodCII is a 16 kDa monomeric protein that is protease sensitive (Gabbianelli et al., 2004; Krishnakumar et al., 2007; Mori et al., 2008). SodCII behaves like a classic periplasmic protein in that it is efficiently released from the cell upon osmotic shock (Krishnakumar et al., 2004; Krishnakumar et al., 2007). Treatment of Salmonella with a sublethal concentration of antimicrobial peptide mimics an osmotic shock, causing the release of periplasmic contents, including SodCII (Vaara and Vaara, 1981; Kim et al., 2010). In contrast, SodCI has the unusual property of being “tethered” within the periplasm, defined as remaining cell associated upon either osmotic shock or treatment with antimicrobial peptides (Krishnakumar et al., 2004; Krishnakumar et al., 2007; Kim et al., 2010). SodCI and SodCII are enzymatically very similar, although SodCI has a slightly higher specific activity and a slightly higher affinity for, and ability to acquire, copper and zinc in its active site (Ammendola et al., 2008; Krishnakumar et al., 2004; Kim et al., 2010). Despite these various differences, we have shown clearly that protease resistance and tethering are the key properties that independently allow SodCI to function during infection (Krishnakumar et al., 2007; Kim et al., 2010; Rushing and Slauch, 2011). By exchanging various sequences between the homologous SodCI and SodCII proteins, we could show that either a protease resistant, but non-tethered SodCII, or a protease sensitive, but tethered SodCII enzyme were fully capable of complementing SodCI function during infection (Rushing and Slauch, 2011).
The nature of tethering has not been elucidated, but there are indirect data that suggest a tethering target (Krishnakumar et al., 2007; Krishnakumar et al., 2004). First, SodCI remains tethered even when ~10-fold overexpressed. This suggests that SodCI binds to something abundant. Second, SodCI is tethered in both E. coli and Salmonella, so SodCI binds something that is common between these two species. Third, SodCI can be released by a high salt wash after osmotic shock. This indicates that SodCI tethering within the periplasm is mediated by non-covalent bonding, likely an ionic interaction. Finally, SodCI is released when lysozyme is used to generate spheroplasts. Based on these results, we hypothesized that SodCI might bind to the peptidoglycan (PGN). In this study we show that SodCI binds to the peptide part of PGN and the affinity of SodCI is ~3 fold higher than SodCII. We also show that SodCI tethering is dependent on nine amino acids within the protein. Replacing the corresponding sequences in SodCII with these nine amino acids creates a tethered SodCII that complements SodCI activity during infection.
Results
SodCI binds to Salmonella peptidoglycan
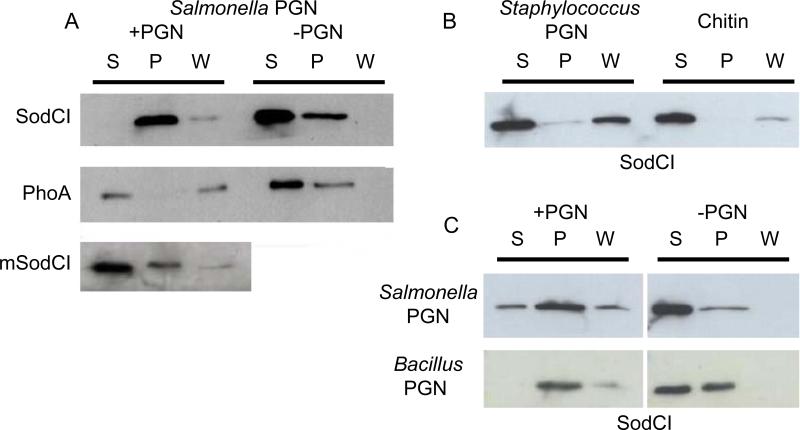
Based on the accumulated data described above, we hypothesized that SodCI binds to peptidoglycan to remain tethered within the periplasm. To test this hypothesis, we purified SodCI containing a carboxy-terminal His-tag. This His-tag has no effect on SodCI activity in vitro or during an animal infection (Kim et al., 2010). As a control, we also purified His-tagged monomeric SodCI (mSodCI). This mutant protein contains two amino acid changes, Y87E and Y109E, that render SodCI enzymatically active, but monomeric and released by osmotic shock (Krishnakumar et al., 2007). As a further control, we purified His-tagged PhoA, the well-characterized periplasmic alkaline phosphatase from E. coli; the 100 kDa PhoA homodimer is released by osmotic shock (Neu and Heppel, 1965). Salmonella peptidoglycan (PGN) was also purified (Young, 1996). The purified proteins and PGN were incubated together for a short time and then the PGN was pelleted by ultra-centrifugation. Control experiments were performed without added PGN. Figure 1A shows that SodCI, in the absence of PGN, was found primarily in the supernatant, although we routinely recovered some protein in the pellet. However, in the presence of PGN, most of the SodCI was found in the pellet. This suggests that SodCI binds to Salmonella PGN. In contrast, both PhoA and mSodCI were found primarily in the supernatant in the presence of PGN. These data suggest that binding to Salmonella PGN is specific to SodCI and that there is a correlation between binding to PGN and release by osmotic shock. Given the fact that both PGN and SodCI were purified, binding is also apparently direct and does not require any additional protein.
Figure 1. SodCI binds to diaminopimelic acid-containing murein sacculus.
A) Purified SodCI, PhoA or monomeric SodCI (mSodCI) was incubated with or without purified Salmonella peptidoglycan sacculus (PGN). Supernatant (S) after 1st centrifugation (100,000×g) was saved. The remaining pellet was washed gently with buffer and another supernatant (W) and pellet (P) were obtained after 2nd centrifugation (100,000×g). All fractions were brought to the same volume and proteins were separated via PAGE. His-tagged proteins were detected by Western blot. B) SodCI binding to purified PGN from Staphylococcus or Chitin were tested as above. C) SodCI binding to Salmonella and Bacillus subtilis PGN.
To further investigate the nature of PGN recognition by SodCI, we asked if the protein would bind to PGN from other organisms. The PGN of Salmonella and the Gram-positive bacterium Staphylococcus have the same carbohydrate structure, but differ in the structure of the peptide and crosslink (Vollmer et al., 2008). On the other hand, Bacillus PGN has the same carbohydrate and peptide structure as Salmonella PGN, including the meso-diaminopimelic acid at the third position of the peptide (Vollmer et al., 2008). When Staphylococcus PGN was used in our assay, SodCI was found mostly in the supernatant and wash fractions, but not in the pellet (Figure 1B). In contrast, the data suggested that SodCI binds Bacillus PGN in a manner similar to Salmonella PGN (Figure 1C). Thus, SodCI binds some structure common to Salmonella and Bacillus PGN, but not found in Staphylococcus PGN. Chitin is a polymer of N-acetylglucosamine, one of the two sugars found in all PGN. SodCI did not pellet with chitin in our assay. Taken together, these data suggest that SodCI recognition of Salmonella PGN involves the peptide portion of the molecule.
SodCI binding to PGN is blocked by muramyl-peptides
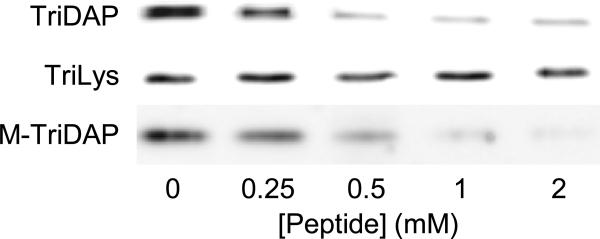
The results above show that SodCI binds directly to PGN and suggest that recognition involves the peptide portion of PGN. If true, then specific binding could be blocked by fragments of the PGN muropeptide structure. We used a competitive protein-binding assay with L-Ala-γ-D-Glu-mDAP (Tri-DAP) and MurNAc-L-Ala-γ-D-Glu-mDAP (M-TriDAP), which correspond to the Salmonella peptide structure, and L-Ala-γ-D-Glu-Lys (Tri-Lys), which is equivalent to part of the Staphylococcus peptide structure. Purified SodCI was first mixed with increasing concentrations of each of the muramyl peptides. Purified PGN was then added and the mixture was incubated, after which the PGN was pelleted by ultra-centrifugation. SodCI present in the pellets was detected by Western blot analysis. As shown in figure 2, increasing concentrations of either TriDAP or M-TriDAP efficiently competed for SodCI, apparently blocking SodCI binding to PGN. TriLys, on the other hand, had no effect. These data support the hypothesis that SodCI specifically binds to the peptide region of Salmonella PGN. Moreover, the recognition apparently involves the diaminopimelic acid residue, in that TriLys did not compete for PGN binding. TriDAP and TriLys differ by a single carboxylic acid group.
Figure 2. SodCI binding to PGN is competitively inhibited by muramyl-peptides.
Binding of purified SodCI to purified Salmonella sacculus was determined by ultracentrifugation and Western blot analysis of the pellet fractions. Binding was determined in the absence or presence of increasing concentrations of the designated muramyl-peptide.
Identifying amino acid residues in SodCI essential for tethering
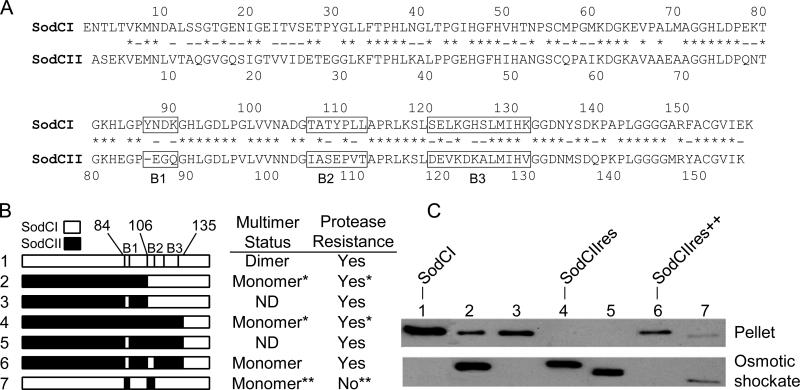
In a previous study (Rushing and Slauch, 2011), we showed that periplasmic tethering of SodCII is attained if amino acids 84 to 135 of SodCI replace the corresponding sequence of SodCII. Therefore, the sequences that account for tethering should include this region. Alignment of the SodCI and SodCII sequences shows three short stretches of amino acid sequences that differ in this region, designated boxes 1, 2 and 3 (Figure 3A). In order to identify the amino acid sequences necessary for tethering, we generated a series of SodC hybrid proteins, where the distinctive sequences of SodCI and SodCII were switched. Biochemical characterization of these hybrids allowed us to correlate tethering properties with the corresponding sequence. For technical reasons, most of these hybrid proteins contain the C-terminal end of SodCI, which confers protease resistance, and a carboxy terminal His-tag. The C-terminal sequences of SodCI and SodCII do not affect tethering (Rushing and Slauch, 2011).
Figure 3. Properties of SodCI-SodCII hybrid proteins.
A) Sequence alignment of mature SodCI and SodCII. Amino acid identity is indicated by asterisks, conserved substitutions are indicated by hyphens. The boxes indicating differential sequences of interest (B1, B2, and B3) are marked. B) Hybrid SodC genes were generated by PCR as described in Materials and Methods. SodCI sequences are indicated in white, while SodCII sequences are designated in black. The point of hybrid cross-over is indicated, numbered according to the mature wild-type SodCI protein. The boxes of differential sequence are indicated. Protease resistance was defined as a whole-cell extract retaining > 50% of SOD activity after 2 h incubation in 1 mg ml-1 Proteinase K. Multimerization status was determined by gel filtration chromatography. *Data from Rushing and Slauch, 2011. **Based on results from Y87E Y109E mutant, Krishnakumar et al., 2007. ND, not determined. C) Release of SodC protein by osmotic shock was determined via Western blot analysis with anti-His antibodies. (All proteins had a C-terminal His tag.) The lane numbers correspond to the indicated hybrid SodC proteins. Lysates of the osmotic shockate and pelleted cells (suspended in equal concentration) were used to determine if the protein is released or tethered, respectively.
We initially suspected, based on extrapolation from several hybrid constructs, that amino acids from 121 to 131, corresponding to box 3, were most likely responsible for tethering (Rushing and Slauch, 2011). Therefore, we initially changed several amino acids of SodCI in box 3 to the corresponding SodCII amino acids (L122V, G124D, H125K, S126A, K131V). This mutant protein had no change in its tethering properties, as measured by SOD activity following osmotic shock. (Nine percent of total mutant SodCI activity was released by osmotic shock compared to 12% of the wild type SodCI activity.) When additional amino acids in box 3 were mutated (E121A or K123A) in the context of the above mutant there was still no significant change in the percent of activity released by osmotic shock. We therefore concluded that the amino acids in box 3 do not account for SodCI tethering and focused on the amino acids in boxes 1 and 2.
As monitored by Western blot analysis, SodCI was found in the cell (pellet) fraction after osmotic shock (Figure 3C, lane 1), whereas the protease resistant SodCII1-133::SodCI136-157 (termed SodCIIres) was found in the supernatant (osmotic shockate; lane 4). The hybrid protein SodCII1-103::SodCI106-157, which includes the SodCI amino acids of boxes 2 and 3, was mostly released by osmotic shock (lane 2). When the box 1 sequence of this hybrid protein was replaced by SodCI sequence, the protein become fully tethered (lane 3). To determine if the tethering properties of the protein depended exclusively on the amino acids in box 1, we examined tethering of a SodCIIres hybrid in which the sequence of SodCI box 1 replaced the SodCII sequence. The box 1 amino acids were not sufficient to change the tethering properties of the protein (lane 5). However, replacing both SodCII box 1 and box 2 sequences with the corresponding sequences of SodCI in the SodCIIres hybrid protein conferred full tethering to the protein (lane 6). These results show that the tethering properties of SodC are dependent on both box 1 and 2 sequences, which constitute changes in 9 amino acids in the SodCII sequence (E86YN, G87D, Q88K, I104T, S106T, E107Y V109L, and T110L). When the SodCI protein sequences in boxes 1 and 2 were replaced by SodCII sequences, the protein became mostly released by osmotic shock. However, this modified protein is expected to be monomeric and protease sensitive based on our previous results showing that dimerization of SodCI is dependent on the two tyrosines at positions 87 and 109 that partially define the dimer interface (Krishnakumar et al., 2007; Rushing and Slauch, 2011; Bordo et al., 1999; Forest et al., 2000; Pesce et al., 2000). Exposing this dimer interface confers protease sensitivity, which is distinct from the protease sensitivity conferred by the carboxy-terminus of SodCII (Rushing and Slauch, 2011). Given these results, we examined the dimerization status (as determined by size exclusion chromatography) and protease sensitivity of the SodCIIres hybrid protein with SodCI box 1 and box 2 sequences. This tethered protein (lane 6) remained monomeric and protease resistant, as expected (data not shown).
SodCI and SodCII with box 1 and 2 sequences bind PGN muropeptides with equal affinity
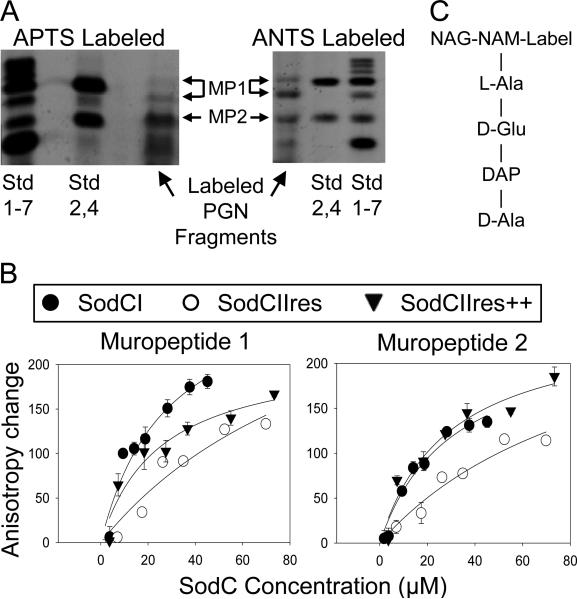
The results above suggest that the SodCI protein specifically binds the peptide portion of PGN and that this recognition is dependent on specific amino acid sequences in the protein. We wished to further determine the portions of PGN recognized by SodCI and attempt to determine relative affinities for binding. To this end, we digested S. Typhimurium PGN with mutanolysin, which cleaves the β-N-acetylmuramyl-(1→4)-N-acetylglucosamine bond, leaving a reducing end. We then labeled the resulting muropeptides with a fluorescent molecule, ran the samples on native polyacrylamide gels, and isolated various small molecular weight fragments. We used two different fluorescent labels: ANTS is superior for separation and visualization on gels (so-called fluorophore-assisted carbohydrate electrophoresis or FACE) (Jackson, 1996; Young, 1996), while APTS labeled fragments were ideal for fluorescence anisotropy analysis (Chen et al., 1996), described below. We also labeled the reducing ends of glucose and glucose polymers to maltoheptaose as size markers (Jackson, 1996). The digested and labeled PGN yielded four main products (Figure 4). The larger muropeptides were extracted from gels, dried, and suspended in phosphate buffer.
Figure 4. Fluorescence anisotropy analysis of SodC-muropeptide interactions.
A) Muropeptides were prepared from digested Salmonella PGN, labeled with the fluorescent dye APTS (left) or ANTS (right) and separated by electrophoresis through a 30% polyacrylamide gel. Standards (Std) are fluorescently labeled glucose (1), maltose (2), and additional glucose polymers to maltoheptaose (3-7). “Std 1-7” is comprised of all the labeled sugars, whereas “Std 2,4” contains labeled maltose and maltotetraose. Two different APTS labeled muropeptide derivatives were isolated (MP-1 and MP-2) and used in a fluorescence anisotropy assay. B) The isolated APTS-muropeptides were incubated with increasing amounts of purified SodCI, SodCIIres or SodCIIres++. The latter contains SodCI sequences in boxes 1 and 2. “Anisotropy Change” represents the difference between anisotropy values measured at each concentration of SodCI and the anisotropy measured with no added SodC. C) Schematic structure of labeled monomeric muropeptide.
We attempted to specifically define these various molecules using several mass spectrometry methods, but we were unsuccessful. However, our PAGE results are comparable with previous FACE studies performed on E. coli PGN (Li et al., 2004; Young, 1996). Based on these comparisons, we conclude that the fragments labeled MP-1 (a mixture of two species) are crosslinked species while those labeled MP-2 represent uncrossinked PGN subunits. The purified muropeptides were used in binding assays with three different SodC species: SodCI, SodCIIres and SodCIIres with SodCI box1 and 2 sequences (labeled SodCIIres++). We used an ultra-low-volume microplate rapid fluorescence anisotropy assay (FAMA). Fluorescently-labeled muropeptides and the indicated amounts of the three proteins were mixed and anisotropy values were measured (Figure 4). In those cases where binding reached saturation, plotting anisotropy change (muropeptide bound) versus SodC concentration yielded approximate Kd (dissociation constants). The Kd of the SodCI and SodCIIres++ proteins with muropeptide 1 or 2 was ~30 ± 10 μM while SodCIIres showed a Kd that was ~3.5 fold higher. Thus, SodCI apparently binds both crosslinked and un-crosslinked muropeptides with similar affinities. Altering the sequence of SodCII in boxes 1 and 2 confers the ability to bind the muropeptides with affinities essentially identical to SodCI. Surprisingly, SodCII does bind the muropeptides, albeit with an affinity 3-4 fold lower than SodCI.
SodCII containing SodCI box 1 and 2 sequences complements loss of SodCI during animal infection
In a previous study we demonstrated that periplasmic tethering is sufficient for virulence (Rushing and Slauch, 2011). We wanted to further test this concept by asking if SodCII containing the SodCI sequences in boxes 1 and 2 could function during infection. Therefore, we recombined the different sodCI and sodCII alleles with various tethering profiles into the normal sodCI or sodCII loci and tested the resulting strains for virulence using a mouse competition assay. Note that the SodCII constructs used in the animal experiments have the normal carboxy-terminus and are, thus, protease sensitive (Rushing and Slauch, 2011). As shown in Table 1, a strain producing only wild type SodCII was ~10 fold attenuated compared to a strain producing SodCI. This is consistent with all previous results showing that SodCII does not function during Salmonella infection in the animal (Uzzau et al., 2002; Krishnakumar et al., 2004; Krishnakumar et al., 2007; Ammendola et al., 2008). A strain producing SodCI (SodCII box 1 and 2) was also attenuated ~16 fold compared to a strain producing SodCI. This suggests that this SodCI mutant is a complete loss of function, but these results are difficult to interpret given that these mutations cause the protein to be monomeric and protease sensitive in addition to affecting tethering. The strain producing the SodCII (SodCI box 1 and 2) allele was still attenuated compared to the strain producing SodCI, although it appeared to be less attenuated than the wild type SodCII-producing strain. The most accurate measure of this was to compete these two strains directly. The SodCII (SodCI box 1 and 2) producing strain was 2.6 fold more virulent than the strain producing only wild type SodCII. These data show that the SodCII (SodCI box 1 and 2) protein, containing 9 amino acids from SodCI, is partially functional during animal infection, albeit not equal to SodCI. Thus, as we have previously shown (Rushing and Slauch, 2011), conferring tethering to SodCII allows it to function during infection, independent of other properties of the protein, such as protease sensitivity, regulation, or metal acquisition.
Table 1.
Competition assays to measure the contribution of hybrid SodCs during infection.
| Strain Aa | Strain Ba | Median CIb | No. of mice | P c |
|---|---|---|---|---|
| SodCII | SodCI | 0.11 | 5 | 0.001 |
| SodCI (SodCII Box 1+2) | SodCI | 0.06 | 5 | 0.042 |
| SodCII (SodCI Box 1+2) | SodCI | 0.21 | 5 | 0.0009 |
| SodCII (SodCI Box 1+2) | SodCII | 2.57 | 10 | 0.046 |
Strains produce only the indicated SodC from their normal loci. Strains used were JS2040, JS2041, JS2042, JS2043, and JS2044.
Competitive Index (CI) = (percent strain A recovered/percent strain B recovered)/(percent strain A inoculated/percent strain B inoculated).
Student's t-test was used to compare the output and the inoculums.
Discussion
Salmonella Typhimurium strain 14028 produces two periplasmic superoxide dismutases, SodCI and SodCII. Despite being 60% identical at the amino acid level with very similar enzymatic properties, only SodCI functions during infection to protect the bacterium from phagocytic superoxide (Uzzau et al., 2002; Krishnakumar et al., 2004; Krishnakumar et al., 2007; Ammendola et al., 2008). Our previous results (Krishnakumar et al., 2004; Krishnakumar et al., 2007; Kim et al., 2010; Rushing and Slauch, 2011) show that SodCI is both protease resistant and “tethered” within the periplasm. Tethering is defined as remaining cell-associated after osmotic shock or treatment with sublethal concentrations of antimicrobial peptide. Protease resistance and tethering independently allow SodCI to function during infection. Our combined results (Krishnakumar et al., 2004; Krishnakumar et al., 2007; Kim et al., 2010; Rushing and Slauch, 2011; Golubeva and Slauch, 2006) lead to the following model. Salmonella within the macrophage phagosome is exposed to antimicrobial peptides, proteases, and superoxide. The antimicrobial peptides partially disrupt the outer membrane of Salmonella in the phagosome. This allows periplasmic proteins, including SodCII, to leak out into the phagosome and/or proteases access to the periplasm. SodCII, and likely other periplasmic proteins, are degraded. SodCI is induced in this environment, is tethered within the periplasm, and is protease resistant, thereby allowing it to survive and combat the superoxide. Support for this model includes the fact SodCII can partially or fully complement SodCI function during infection if 1) the outer membrane is protected against antimicrobial peptides, 2) SodCII is made protease resistant, or 3) SodCII is tethered within the periplasm (Kim et al., 2010; Rushing and Slauch, 2011).
Although tethering is important for SodCI function, the nature of tethering was not previously defined. Here we show that SodCI binds the peptidoglycan (PGN) cell wall, specifically recognizing the peptide stem. Evidence for recognition of the peptide includes the fact that SodCI binds Salmonella and Bacillus PGN, but not Staphylococcus PGN, and that SodCI binding to Salmonella PGN could be competed with the tripeptide L-Ala-γ-D-Glu-mDAP (Tri-DAP), but not the corresponding tripeptide L-Ala-γ-D-Glu-Lys (Tri-Lys). Although overall binding could be to a larger portion of the PGN that includes even the polysaccharide, these data strongly suggest that binding involves specific recognition of the diaminopimelic acid moiety, which differs from lysine by a single carboxylic acid group. Interestingly, our data suggest that SodCI binds equally well to both crosslinked and uncrosslinked muropeptides. However, the crosslinked species retains a free diaminopimelic acid moiety (Vollmer et al., 2008), so it is not clear if SodCI can recognize the diaminopimelic acid side chain involved in the crosslink.
Replacing amino acids in the non-tethered SodCII with the corresponding amino acids in SodCI showed that changing nine amino acids in the sequences, designated boxes 1 and 2 (Figure 3), was sufficient to confer tethering to SodCII. Making the reciprocal changes in SodCI caused the mutant protein to be mostly released by osmotic shock. Thus, these amino acid sequences are necessary for binding to peptidoglycan. It is doubtful that they are sufficient for binding and the overall protein region required for binding likely encompasses sequences that are identical in the two proteins. The concept that sequences conserved between SodCI and SodCII are important for PGN binding is also supported by the fact that SodCII does have significant affinity for PGN fragments, only 3-4 fold less than SodCI, as determined by fluorescence anisotropy assays. Thus, SodCII, and likely other closely related bacterial SodCs, do apparently associate with PGN, but the binding is not sufficiently strong to withstand an osmotic shock or, in the case of SodCII, survive release and proteolytic degradation in the macrophage phagosome (Kim et al., 2010). SodC proteins are found throughout the three domains of life. Although the region of SodCI required for tethering has homology in all SodC proteins, one presumes that peptidoglycan binding is relevant to only bacterial enzymes, and likely only a subset of those. The implication is that this region in close SodCI homologs, an inherent and relatively conserved part of all SodC proteins, subsequently evolved to bind peptidoglycan.
The fact that the amino acids implicated in PGN binding are at the dimer interface of SodCI complicates our understanding of the interaction. Figure 5 shows the SodCI crystal structure (Pesce et al., 2000) with the identified amino acids in yellow. The residues are in a cleft along the both sides of the dimer and one could imagine a strand of PGN fitting nicely into this groove. However, dimerization is not required for binding (e.g. SodCIIres++ is monomeric, yet tethered), and labeling these same amino acid residues in one of the polypeptides (structurally equivalent to the SodCII monomer) gives a very different impression of the apparent binding surface. Structures of SodCI, and perhaps mutant SodCII, bound to PGN fragments will be required to fully understand the interaction. Given that the monomer binds, it is possible that SodCI has two independent binding sites that could cooperate in their interaction with PGN. However, none of our data suggest such cooperatively, nor is any such cooperatively required for tethering in vitro or function during infection.
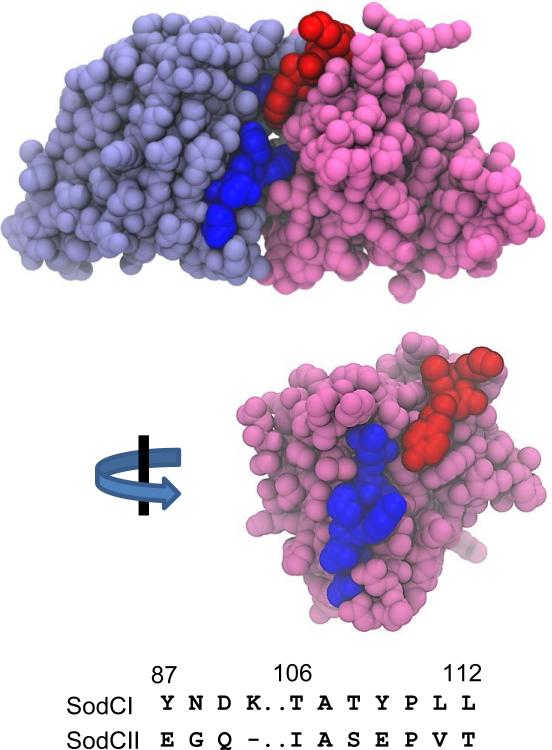
Figure 5. Amino acids required for tethering on SodCI structure.
Top structure shows dimeric SodCI (1EQW; Pesce et al., 2000). The box 1 amino acids are colored red on the pink polypeptide while the box 2 residues are colored dark blue on the light blue polypeptide. The bottom structure shows just the pink polypeptide. This view is facing the dimeric interface with the box 2 residues colored blue.
Peptidoglycan is a hallmark of the bacteria and is essential for cell structure and viability. Not surprisingly, more than 13 different protein families and motifs have evolved to specifically interact with various elements of peptidoglycan. These binding motifs are found in proteins involved in peptidoglycan synthesis and cell division (e.g. AMIN and SPOR domains; (Rocaboy et al., 2013; Williams et al., 2013)), cell envelope structure (e.g. OmpA and Pal; (Parsons et al., 2006)), antibacterial cell lysis (e.g. lysozymes and bacteriophage murein hydrolases), as well as immune recognition (e.g. PGRPs; (Royet and Dziarski, 2007)). The region of SodCI defined here as binding to peptidoglycan appears to be unique in both sequence and fold and does not match any of these previously defined peptidoglycan recognition motifs.
In summary, our results suggest that SodCI and other closely related bacterial SodCs have evolved to bind peptidoglycan. In the context of the macrophage phagosome, this is clearly an advantage, in that cationic antimicrobial peptides are capable of disrupting the outer member and causing apparent loss of freely diffusible periplasmic proteins (Kim et al., 2010). Indeed, increasing the binding affinity of SodCII to peptidoglycan confers the ability to contribute to resistance to phagocytic superoxide. For bacteria that do not propagate in phagocytic cells, the role of periplasmic superoxide is unknown, in that we do not know of any specific environment in which the bacteria experience significant extracellular superoxide. Nonetheless, SodC seems highly conserved and it is intriguing to suppose that for Gram-negative bacteria, such environments might also entail additional insults to the outer membrane, selecting for peptidoglycan binding of SodC, even in non-pathogens.
Experimental Procedures
Media and growth of strains
Bacteria were routinely cultured in Luria-Bertani (LB) broth at 37°C shaking for 16-18 h or grown on LB containing 1.5% agar (Silhavy et al., 1984). When required, media were supplemented with antibiotics at the following final concentrations: ampicillin, 50 mg ml−1; kanamycin, 50 mg ml−1 or chloramphenicol, 20 mg ml−1. When assaying for enzymatic activity, protease susceptibility, or release by osmotic shock, E. coli strains overexpressing a SodC from a plasmid were grown in LB/ampicillin supplemented with 0.25 mM CuSO4, 0.05 mM ZnCl2 and 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). For peptidoglycan purification, S. Typhimurium strain 14028 was cultured in M9 glucose minimal medium supplemented with Casamino acids (Li et al., 2004; Miller, 1992).
Strain construction
Bacterial strains and plasmids used in this study are listed in Table 2. Gene deletions/insertions were constructed by λ Red-mediated recombination (Datsenko and Wanner, 2000) as described (Ellermeier et al., 2002). All mutant alleles were confirmed by PCR analysis and sequence of the PCR amplification product and backcrossed into a wild-type background using phage P22 (HT105/1 int-201)-mediated transduction (Maloy, 1990). Hybrid sodC genes were generated by a PCR-mediated DNA stitching protocol. Briefly, sodC fragments corresponding to the N-terminal region of one sodC gene and a C-terminal region of a different sodC gene were generated in a standard PCR reaction using plasmid-encoded sodCI or sodCII as template. The primers were designed such that the resulting N-terminal and C-terminal fragments had 18-44 overlapping base pairs. For amino acid substitution, a primer with the desired coding sequence was used for the initial amplification. Amplified sodC fragments were used as templates in a subsequent reaction devoid of primers to allow fragments to anneal and be extended. Full-length hybrid sodC genes were selectively amplified using unpurified product from the extension reaction as template in a standard PCR reaction, with the N-terminal full-length primer encoding an EcoRI site and the sodCI ribosome binding site for cloning and expression, and the C-terminal full-length primer contained an XhoI site. Cloning of this fragment into the corresponding sites in pET21b resulted in a construct encoding the hybrid SodC with a C-terminal His-tagged tail including a linker of two amino acids (LEHHHHHH). All clones were confirmed by DNA sequence analysis and evaluated for protein expression following induction with 1 mm IPTG. To evaluate the tethering of hybrid SodC proteins, the coding sequence from each was moved to a low copy vector, pWKS30 (Wang and Kushner, 1991). Each clone was amplified by PCR with primers pWKS-up (CTCGAGGTCG ACGGTATCGA TAAGCTTACG AGGTAACTAA TGAAGCGATT AAGTTTAGC) and pWKS-down (CGGGGCGGCG CTCTAGATGC CTCAGCCGGA TCTCAGTGGT GGTG) and the products were digested with HindIII and XbaI and ligated into the corresponding sites in pWKS30, in which the hybrid gene is expressed from a lac promoter. These clones were confirmed by DNA sequence analysis and evaluated for protein expression following induction with 1 mM IPTG.
Table 2.
Bacterial strains and plasmids.
| Bacterial Strains | Genotypea | Source or referenceb |
|---|---|---|
| 14028 | Salmonella enterica serovar Typhimurium | ATCC |
| JS2040 | 14028 ΔsodCI::(sodCI-his-Kan) ΔsodCII-104::Cm | |
| JS2041 | 14028 ΔsodCI::(sodCI(Δ87Y N88E D89G K90Q T106I T108S Y109E L111V L112T)-his-Kan) ΔsodCII-104 | |
| JS2042 | 14028 ΔsodCI-105 ΔsodCII::(sodCII-his-Kan) | |
| JS2043 | 14028 ΔsodCI-105 ΔsodCII::(sodCII(E86YN G87D Q88K I104T S106T E107Y V109L T110L)-his-Kan) | |
| JS2044 | 14028 ΔsodCI-105::Cm ΔsodCII::(sodCII(E86YN G87D Q88K I104T S106T E107Y V109L T110L)-his-Kan) | |
| AS391 | E. coli AB1157(F- thr-1 leuB6 proA2 his-4 thi-1 argE2 lacY1 galK2 rpsL supE44 ara-14 xyl-15 mtl-1 tsx-33) (sodA::MudPR13)25 (sodB-kan)1-D2 sodC::Spec | Gort et al. (1999) |
| BL21(DE3) | E. coli B (F- ompT gal dcm lon hsdSB(rB- mB-) λ(DE3 [lacIq lacUV5-T7 gene 1 ind1 sam7 nin5]) | |
| Plasmids | ||
| pWKS30 | pSC101ori bla | (Wang and Kushner, 1991) |
| pET21b | pBR322ori lacI bla pT7 | Novagen |
| pBK104 | pWSK30 phoA-his | |
| pBK400 | pWKS30 sodCI(Y87E Y109E)-his | |
| pMR103 | pWKS30 sodCI | (Rushing and Slauch, 2011) |
| pMR317 | pWKS30 sodCI(L122V G124D H125K S126A K131V) | |
| pMR321 | pWKS30 sodCI(L122V G124D H125K S126A K131V E121A) | |
| pMR324 | pWKS30 sodCI(L122V G124D H125K S126A K131V K123A) | |
| pAT139 | pWKS30 sodCII(E86YN G87D Q88K I104T S106T E107Y V109L T110L)-his | |
| pAT101 | pET21b sodCI-his | |
| pAT108 | pET21b sodCII1-133sodCI136-157-his [called sodCIIres-his] | |
| pAT110 | pET21b sodCIIres(E86YN G87D Q88K I104T S106T E107Y V109L T110L)-his | |
| pAT123 | pWKS30 sodCI-his | |
| pAT130 | pWKS30 sodCIIres-his | |
| pAT131 | pWKS30 sodCIIres(E86YN G87D Q88K I104T S106T E107Y V109L T110L)-his | |
| pAT143 | pWKS30 sodCIIres(E86YN G87D Q88K)-his | |
| pAT128 | pWKS30 sodCII1-103sodCI106-157-his | |
| pAT129 | pWKS30 sodCII1-103SodCI106-157(E86YN, G87D, Q88K)-his | |
| pAT133 | pWKS30 sodCI(Δ87Y N88E D89G K90Q T106I T108S Y109E L111V L112T)-his |
All Salmonella strains are isogenic derivatives of serovar Typhimurium strain 14028. Subscript numbers in hybrid genes designate the amino acids of the mature protein encoded.
This study unless specified otherwise. ATCC, American Type Culture Collection.
To test the ability of mutant proteins to function in the animal, sodC constructs were integrated at the native sodC loci of strain 14028 as described previously (Rushing and Slauch, 2011). The resulting strains contain either a fully reconstructed sodCI or sodCII locus encoding His-tagged proteins and with a Kan resistance cassette inserted 120 bp downstream of the stop codon. The other sodC gene in each background was deleted.
Characterization of SodC proteins
Hybrid SodC proteins, produced from pET21b clones in BL21(DE3), were purified using a nickel affinity resin (HIS-Select Nickel Affinity Gel, Sigma). Cells from overnight cultures were pelleted, suspended in 1/10 volume of 50 mM potassium phosphate (KPi) buffer (pH 7.8), and then lysed using a French pressure cell. Extracts were clarified by centrifugation at 14,000 × g for 30 min at 4°C followed by filtration of the supernatant through a 0.22 μm filter. The resulting extract was loaded onto a nickel affinity column and the column was washed with KPi buffer, pH 7.8. His-tagged protein was eluted with 0.4 M imidazole, pH 8.0. The purified SodC hybrid proteins were dialyzed against the appropriate buffer for Sod activity assays (performed as described in McCord and Fridovich, 1969), Western blot analyses, size exclusion chromatography, fluorescenceanisotropy assays, and PGN binding assays. For size exclusion chromatography, supernatants were further clarified by centrifugation at 40,000 × g for 1 h at 4°C in a Beckman ultracentrifuge.
For osmotic shock, cells from overnight LB cultures (10 ml) were pelleted by centrifugation, washed three times in cold 50 mM potassium phosphate buffer (pH 7.8), and suspended in 5 ml of room temperature plasmolysis buffer (50 mM Tris, 2.5 mM EDTA, 20% (w/v) sucrose; pH 7.8). After incubation at room temp for 15 min, the cells were pelleted by centrifugation, resuspended in 2.5 ml of ice-cold deionized water, and allowed to stand for 15 min. Upon re-centrifugation, the collected supernatant was considered the osmotic shockate. The remaining pellet was suspended in 2.5 ml of 50 mM potassium phosphate buffer and the cells were lysed via French pressure cell. The resulting lysate was assayed for activity to determine the amount of SodC retained in the cells. SOD activity was determined by the xanthine oxidase - cytochrome c method (McCord and Fridovich, 1969). For most experiments, SodC proteins were overexpressed in an E. coli strain (AS391) devoid of any other SODs. In these cases, cyanide resistant SOD activity, which we routinely measure, was negligible. Total protein content of cellular extracts was determined by a Coomassie blue dye-based assay (Pierce). For Western blot analysis with anti-His antibodies, we used 25 μl (from the 2.5 ml sample).
To determine the protease susceptibility of SodC proteins, E. coli AS391 cells expressing the desired SodC were suspended in 20 mM Tris-HCl, pH 6.8, lysed in a French pressure cell, and the resulting extract was divided in half. One sample was incubated with Proteinase K (Sigma) at a final concentration of 1 mg ml−1. Samples with and without protease were incubated at 37°C and aliquots were removed at indicated time points and assayed for SOD activity. The percent residual activity at each time point is relative to the equivalent sample without added protease assayed at the same time. None of the samples without protease lost any significant activity over the time of the experiments.
Fast protein liquid chromatography (FPLC) was used to determine whether hybrid SodC proteins were monomeric or dimeric. Briefly, His-tagged proteins were purified as described above. These samples were dialysed against a solution of 20 mM Tris-HCl, 0.15 M NaCl, pH 7.0 at 4°C. This soluble protein fraction was then passed through a 0.22 μm syringe filter and loaded into a Superdex 75 10/300 GL gel filtration column. The column was eluted using degassed dialysis buffer at a flow rate of 0.4 ml min−1. Fractions (0.2 ml each) were collected and assayed for SodC activity. To approximate the molecular weight of the SodC proteins, elution profiles of protein standards (Albumin 67 kDa, Ovalbumin, 43 kDa, Chymotrypsin 25 kDa and Ribonuclease A 13.7 kDa) (Amersham Biosciences) were determined under identical conditions.
Peptidoglycan purification
A fresh overnight LB culture of S. Typhimurium strain 14028 was diluted 1:50 into 400 ml of M9 medium and grown at 37°C to an OD550 of 0.7-0.8. The culture was cooled rapidly to 4°C and cells were harvested by centrifugation at 10,000 × g for 15 min at 4°C. The wet weight of the pellet was determined. Cells were suspended to 0.2 g ml−1 in H2O and the mixture was added drop-wise with vigorous stirring to an equal volume (3-4 ml) of boiling 8% sodium dodecyl sulfate (SDS). The resulting mixture was boiled for an additional 30 min. The lysate was allowed to cool overnight to room temperature. Insoluble peptidoglycan was pelleted by ultracentrifugation at 100,000 × g for 45 min at room temperature. The pellet was washed and re-pelleted 10 times in distilled water until the SDS concentration was below 1 μg/ml, as determined by the methylene blue assay (Hayashi, 1975). The purified Salmonella PGN was resuspended in 1 ml deionized water. Bacillus subtilis PGN, Staphylococcus PGN and chitin were purchased from Sigma. PGN samples were sonicated briefly before each use.
Protein-PGN binding assay and Western blot
To measure the binding of purified SodC proteins to PGN, approximately 1 μg of a given protein was mixed with 30 μl of purified Salmonella PGN in 50 μl total KPi buffer, pH 7.4. After one hour incubation at 37°C, the sample was centrifuged at 100,000 × g for 20 mins to pellet the PGN. The supernatant was collected. The pellet was washed by adding 500 μl of 10 mM sodium phosphate buffer and mixing gently. The sample was centrifuged again for 20 mins at 100,000 × g. This supernatant (wash fraction) was collected and concentrated to 50 μl in a SpeedVac concentrator (Savant). The remaining pellet was suspended in 50 μl of the same buffer (pellet fraction). Control experiments without added PGN were performed identically to that described above except that an equivalent amount of buffer was added instead of PGN. To test SodCI binding to chitin, approximately 1 μg of SodCI was mixed with 150 μg of chitin in 10 mM sodium phosphate buffer. After incubation at 4°C for an hour, the sample was processed as described above. For binding assays with PGN from species other than Salmonella, 1 mg of PGN was used with the remaining steps as above. Each collected fraction was mixed with SDS loading buffer and boiled for 10 mins prior to SDS electrophoresis and Western blot analysis performed as describe previously (Kim et al., 2010).
SodC binding to PGN and competition assay
For the competitive protein-binding assay, 1 μg of SodCI was incubated with the indicated concentration of peptide (TriDAP, M-TriDAP, or TriLys; InvivoGen) in a total volume of 170 μl of 50 mM potassium phosphate buffer (pH 7.4) at room temperature for 30 minutes on a rotating mixer. A 30 μl aliquot of freshly prepared PGN was added to each sample and mixing was continued for one hour. After the incubation, the samples were centrifuged at 100,000 × g for 20 minutes to pellet the PGN. The pellet was washed by adding 500 μl of 10 mM sodium phosphate buffer to each tube and mixing gently. The samples were centrifuged again for 20 mins at 100,000 × g. The resulting pellet was suspended in 80 μl of the same buffer and 20 μl of the pellet solution was used for SDS electrophoresis and Western blot analysis.
Separation and visualization of labeled muropeptides by FACE
Fluorophore-assisted carbohydrate electrophoresis (FACE) was performed as previously described (Young KD, 1996). Muropeptide subunits were prepared by digesting 200 μl of purified Salmonella PGN with 200 units of mutanolysin (Sigma M9901) in a total volume of 250 μl for 16 hr at 37°C. The resulting samples were dried by SpeedVac and re-suspended in 16 μl of 0.2 M 8-Aminonaphthalene-1,3,6-trisulfonic acid (ANTS) solution or 0.2 M 9-Aminopyrene-1,4,6-trisulfonic acid (APTS) solution in 15% acetic acid with 16 μl of 1 M sodium cyanoborohydride (NaCNBH3 dissolved in dimethyl sulfoxide). The sample was incubate for 16 h at 37°C, then the volume of each sample was brought up to 300 μl with deionized water, filtered through 0.22 μM filter and dried by SpeedVac. The samples were re-suspended in 100 μl 62.5 mM Tris-HCl, 20% glycerol. A 25 μl aliquot of each fluorescently labeled muropeptide sample was loaded onto a 30% polyacrylamide gel (without SDS and β-mercaptoethanol) and the muropeptides separated by electrophoresis at 200 volts in a 4°C cold room. When using ANTS the run time was 2-2.5 hr; for APTS the run time was 4-5 hr. Labeled molecules were visualized by exposing the gel to a 365-nm wavelength light. Bands with muropeptide subunits were cut out from the polyacrylamide gels and placed in 5 ml of deionized water in 15 ml tubes. These samples were incubated at 37°C overnight shaking at 200 rpm. Gel fragments were removed by centrifugation and filtration of the resulting supernatant through a 0.45 μm syringe filter. The samples, designated MP1 and MP2, were then dried in a SpeedVac and suspended in 50 mM potassium phosphate buffer pH 7.8, 0.1 mM EDTA. The concentration of muropeptide subunits labeled with APTS were determined by absorbance at 475nm (ε = 1.75 × 104 M−1 cm−1) (Chen et al., 1996)
Calculation of Kd for SodC-muropeptide interactions
SodCI, SodCIIres and SodCIIres++ proteins were purified as described above, concentrated using a Microcon YM-10 filtration device (Millipore) and then suspended in 50 mM potassium phosphate buffer, pH 7.8, 0.1 mM EDTA to the desired final concentrations. Anisotropy assays were performed at room temperature in black 96-well low volume microplates (Molecular Devices) in a total volume of 10 μl, 50 mM potassium phosphate buffer, pH 7.8, 0.1 mM EDTA. Each well contained a 2.5 μl aliquot of 0.7 nM APTS-labeled muropeptide MP1 or MP2 and the indicated concentration of protein ranging from 0 to 70 μM. The plates were briefly centrifuged and incubated at room temperature for 10 min, and fluorescence anisotropy was measured using a Molecular Devices Analyst HT 96-384 microplate reader with excitation at 485 nm and emission at 530 nm. Fluorescence intensity was measured to ensure that both muropeptides were at the same concentration. Anisotropy values were automatically calculated using the Xfluor4 software. For binding affinity analysis, we plotted anisotropy change (measured value of sample – measured value of muropeptide without added protein) versus SodC concentration. Kd values were estimated using non-linear regression, least square fit to a single site binding model (SigmaPlot 11.0).
Mouse virulence assays
Mouse infection experiments were performed as described (Rushing and Slauch, 2011). Briefly, two isogenic Salmonella strains were injected in a 1:1 ratio (approximately 500–1000 total colony-forming units) into the peritoneum of female BALB/c mice (Harlan Sprague–Dawley). Infections proceeded approximately 4 days before mice were sacrificed and spleens were harvested. Extracted spleens were homogenized and dilutions were plated and the resulting colonies were replica plated to determine the output ratios. The competitive index was calculated as: (per cent strain A recovered/ per cent strain B recovered)/(per cent strain A inoculated/per cent strain B inoculated). The Student's t-test was employed to determine statistical significance of experiments. For most experiments, strains were rebuilt and retested to confirm the virulence phenotype. All animal experiments were reviewed and approved by the University of Illinois IACUC.
Acknowledgements
This study was supported by NIH Grants AI070805 and AI111455 to JMS. MDR was partially supported by NIH training Grant AI078876. We thank Dr. Chen Zhang, Director of the High-throughput Screening Facility at University of Illinois for the help in design and analysis of FAMA, Yekaterina Golubeva for technical assistance and helpful discussions, and Taras Pogorelov for the structure images.
References
- Ammendola S, Pasquali P, Pacello F, Rotilio G, Castor M, Libby SJ, et al. Regulatory and structural differences in the Cu,Zn-superoxide dismutases of Salmonella enterica and their significance for virulence. J Biol Chem. 2008;283:13688–13699. doi: 10.1074/jbc.M710499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo D, Matak D, Djinovic-Carugo K, Rosano C, Pesce A, Bolognesi M, et al. Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase. J Mol Biol. 1999;285:283–296. doi: 10.1006/jmbi.1998.2267. [DOI] [PubMed] [Google Scholar]
- Buckle GC, Walker CL, Black RE. Typhoid fever and paratyphoid fever: Systematic review to estimate global morbidity and mortality for 2010. J Glob Health. 2012;2:10401. doi: 10.7189/jogh.02.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NA, Schurmann N, Casse O, Steeb AK, Claudi B, Zankl J, et al. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe. 2014;15:72–83. doi: 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Chen FTA, Liu MS, Evangelista RA. Fluorescent labelled carbohydrates and their analysis. Vol. 29. Beckman Instruments, Inc.; Dec, 1996. US Patent US5569366 A. [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Janakiraman A, Slauch JM. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- Fang FC, DeGroote MA, Foster JW, Baumler AJ, Ochsner U, Testerman T, et al. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci USA. 1999;96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- Forest KT, Langford PR, Kroll JS, Getzoff ED. Cu,Zn superoxide dismutase structure from a microbial pathogen establishes a class with a conserved dimer interface. J Mol Biol. 2000;296:145–153. doi: 10.1006/jmbi.1999.3448. [DOI] [PubMed] [Google Scholar]
- Gabbianelli R, D'Orazio M, Pacello F, O'Neill P, Nicolini L, Rotilio G, Battistoni A. Distinctive functional features in prokaryotic and eukaryotic Cu,Zn superoxide dismutases. Biol Chem. 2004;385:749–754. doi: 10.1515/BC.2004.091. [DOI] [PubMed] [Google Scholar]
- Golubeva YA, Slauch JM. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutase SodCI is a member of the PhoPQ regulon and is induced in macrophages. J Bacteriol. 2006;188:7853–7861. doi: 10.1128/JB.00706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal Biochem. 1975;67:503–506. doi: 10.1016/0003-2697(75)90324-3. [DOI] [PubMed] [Google Scholar]
- Jackson P. The analysis of fluorophore-labeled carbohydrates by polyacrylamide gel electrophoresis. Mol Biotechnol. 1996;5:101–123. doi: 10.1007/BF02789060. [DOI] [PubMed] [Google Scholar]
- Kim B, Richards SM, Gunn JS, Slauch JM. Protecting from antimicrobial effectors in the phagosome allows SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192:2140–2149. doi: 10.1128/JB.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Craig M, Imlay JA, Slauch JM. Differences in enzymatic properties allow SodCI but not SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium strain 14028. J Bacteriol. 2004;186:5230–5238. doi: 10.1128/JB.186.16.5230-5238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kim B, Mollo EA, Imlay JA, Slauch JM. Structural properties of periplasmic SodCI that correlate with virulence in Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:4343–4352. doi: 10.1128/JB.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Holtje JV, Young KD. Comparison of high-performance liquid chromatography and fluorophore-assisted carbohydrate electrophoresis methods for analyzing peptidoglycan composition of Escherichia coli. Anal Biochem. 2004;326:1–12. doi: 10.1016/j.ab.2003.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR. Experimental techniques in bacterial genetics. Jones and Bartlett; Boston, MA: 1990. [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 1992. [Google Scholar]
- Mori M, Jimenez B, Piccioli M, Battistoni A, Sette M. The solution structure of the monomeric copper, zinc superoxide dismutase from Salmonella enterica: structural insights to understand the evolution toward the dimeric structure. Biochemistry. 2008;47:12954–12963. doi: 10.1021/bi801252e. [DOI] [PubMed] [Google Scholar]
- Neu HC, Heppel LA. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- Pesce A, Battistoni A, Stroppolo ME, Polizio F, Nardini M, Kroll JS, et al. Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene. J Mol Biol. 2000;302:465–478. doi: 10.1006/jmbi.2000.4074. [DOI] [PubMed] [Google Scholar]
- Rocaboy M, Herman R, Sauvage E, Remaut H, Moonens K, Terrak M, et al. The crystal structure of the cell division amidase AmiC reveals the fold of the AMIN domain, a new peptidoglycan binding domain. Mol Microbiol. 2013;90:267–277. doi: 10.1111/mmi.12361. [DOI] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- Rushing MD, Slauch JM. Either periplasmic tethering or protease resistance is sufficient to allow a SodC to protect Salmonella enterica serovar Typhimurium from phagocytic superoxide. Mol Microbiol. 2011;82:952–963. doi: 10.1111/j.1365-2958.2011.07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1984. [Google Scholar]
- Slauch JM. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol. 2011;80:580–583. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Bossi L, Figueroa-Bossi N. Differential accumulation of Salmonella [Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol Microbiol. 2002;46:147–156. doi: 10.1046/j.1365-2958.2002.03145.x. [DOI] [PubMed] [Google Scholar]
- Vaara M, Vaara T. Outer membrane permeability barrier disruption by polymyxin in polymyxin-susceptible and -resistant Salmonella typhimurium. Antimicrob Agents Chemother. 1981;19:578–583. doi: 10.1128/aac.19.4.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, De Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- Williams KB, Yahashiri A, Arends SJ, Popham DL, Fowler CA, Weiss DS. Nuclear magnetic resonance solution structure of the peptidoglycan-binding SPOR domain from Escherichia coli DamX: insights into septal localization. Biochemistry. 2013;52:627–639. doi: 10.1021/bi301609e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. A simple gel electrophoretic method for analyzing the muropeptide composition of bacterial peptidoglycan. J Bacteriol. 1996;178:3962–3966. doi: 10.1128/jb.178.13.3962-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]