Synopsis
Hearing loss (HL) is one of the most common birth defects in developed countries and is a diverse pathology with different classifications. One of these is based on the association with other clinical features, defined as syndromic hearing loss (SHL). Determining the etiology of the HL in these patients is extremely beneficial as it enables a personalized approach to caring for the individual. Early screening can further aid in optimal rehabilitation for a child’s development and growth. Pathogenic variants in forty-five genes, encoding proteins functioning as ion channels, transcription factors, molecular motors and more, are known to lead to eleven forms of SHL. The development of high-throughput sequencing technology is facilitating rapid and low-cost diagnostics for patients with SHL.
Keywords: Deafness, Hearing loss, Genetics, Genome, Sequencing
INTRODUCTION
Hearing loss (HL) is the most prevalent sensory impairment in both childhood and adulthood1, 2. According to the last update of the World Health Organization (WHO), approximately 360 million people worldwide, equaling 5% of the world’s population, suffer from a disabling HL (Table 1). The majority of these people live in low and middle-income countries, where treatments for HL are more difficult to obtain and consanguinity increases the risk of recessive disease. HL is an etiologically heterogeneous pathology caused by different genetic and environmental factors, with half of the cases estimated to be genetic3. HL can also be a result of infections, injuries and exposure to excessive noise.
Table 1.
Informative websites for SHL
| WHO | http://www.who.int/topics/deafness/en/ |
| ASHA | http://www.asha.org/public/hearing/Degree-of-Hearing-Loss/) |
| NIDCD | http://www.nidcd.nih.gov/health/statistics/Pages/quick.aspx |
| Hereditary Hearing Loss Homepage | http://hereditaryhearingloss.org/ |
| Deafness Variation Database | http://deafnessvariationdatabase.org/ |
| Leiden Open Variation Database | http://grenada.lumc.nl/LOVD2/WS/ |
| Genetics Home Reference | http://ghr.nlm.nih.gov/ |
| Online Mendelian Inheritance in Man | http://www.ncbi.nlm.nih.gov/omim/ |
| UCSC Genome Browser | https://genome.ucsc.edu/ |
| Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/ |
| GATK | https://www.broadinstitute.org/gatk/ |
| GenVec | http://www.genvec.com/product-pipeline/cgf-166-hearing-loss |
HEARING LOSS
Our ability to hear is orchestrated by the auditory system. The vestibular system is responsible for balance, three-dimensional orientation and gravity perception. The ear is a three-chambered organ divided into the external, the middle and the inner ear, which are all essential for the intact activity of the auditory and the vestibular systems. The external and middle ear are responsible for collecting and conducting the sound wave’s energy to the inner ear4,5. The sensorineural end organ of hearing is the snail-shaped organ of Corti that resides in the inner ear. It is composed of a single row of inner hair cell (IHC), three rows of outer hair cells (OHC) and supporting cells. The IHC act as sensory transducers, capturing stimulus energy, interpreting it as electrical responses and sending the impulses to the brain through the auditory nerve. The OHC are responsible for enhancing the signal6.
According to the American Speech-Language-Hearing Association (ASHA) (Table 1), normal hearing occurs in the range of −10 to 15 dB, with a slight HL if the range of loss is within 16 to 25 dB. Mild HL occurs when the hearing loss ranges between 26 to 40 dB, moderate HL is when the hearing loss ranges between 41 to 55 dB and moderate to severe HL ranges between 56 to 70 dB. Individuals with HL in these ranges are considered to be ‘hard of hearing’ and can benefit from hearing aids and assistive listening devices. With severe or profound HL, one is considered to be ‘deaf,’ when hearing loss ranges between 71 to 90 dB in the former case, or profound, when the hearing loss range is above 91 dB. Individuals with this kind of HL may benefit only from cochlear implants7.
HL is the most common neurosensory disorder in humans. It can be a congenital pathology, caused by genetic factors or by complications during pregnancy and childbirth. It can also be acquired later in life, at any age. Acquired HL can be caused by infectious diseases, physical injuries, the use of ototoxic drugs and genetic pathogenic variants or mutations. Congenital HL is the most commonly occurring condition for which newborns are screened for, with about 1 out of 1,000 infants born affected. Nearly one in five individuals aged 12 and older suffer from unilateral or bilateral hearing loss in the United States alone8. Age related hearing loss (ARHL) is the most prevalent sensory deficit in the elderly9, with nearly 25 percent of those aged 65 to 74 and 50 percent of those who are 75 and older suffering from a disabling HL in the United States alone, according to the National Institute on Deafness and Other Communication Disorders (NIDCD; Table 1). Half of the HL cases are estimated to be genetically related3 and account for about 50 to 60 percent of childhood HL cases in developed countries2. Over the years more than 100 deafness-related loci and their associated genes have been identified and studied, revealing the genetic basis of different deafness-related pathologies10.
The diversity of ear disease pathologies is classified according to the etiology of the case, which can be genetically related or due to environmental causes. If the pathology is genetically related, it is further classified according to the pattern of inheritance (dominant, recessive, X-linked or Y-linked). HL is also classified according to the onset of the pathology, the type, the severity, uni- or bilateral and the association with other disorders as syndromic hearing loss (SHL) versus non-syndromic hearing loss (NSHL)11.
For NSHL, HL loci are classified and named according to their mode of inheritance, with a prefix of DFN (for DeaFNess) (Hereditary Hearing Loss Homepage; Table 1). DFNA refers to autosomal dominant inheritance, DFNB refers to autosomal recessive inheritance and DFNX refers to an X- linked mode of inheritance. Furthermore, Y-chromosome linked genes and maternal inheritance linked to mitochondria have also been identified12. Each locus name also contains a number that represents the order in which these loci were identified in association with deafness. In many cases, the genes for the DFN loci have subsequently been identified13 (Hereditary Hearing Loss Homepage, Deafness Variation Database; Table 1; Chapter this book)
As HL is one of the most common birth defects in developed countries, newborn screening for hearing defects has an important role in treatment and rehabilitation strategies, such as cochlear implantation14. The early diagnosis of the etiology of a child’s HL can allow the monitoring of possible complications and can indicate which therapy is the most suitable and effective one. It also allows for more accurate genetic counseling for parents who want to have more children10.
SYNDROMIC HEARING LOSS
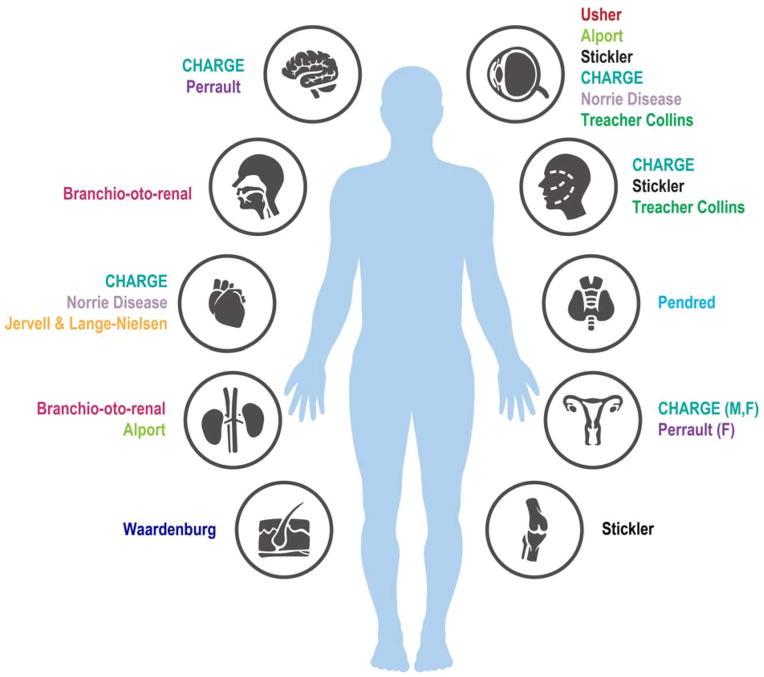
SHL is a form of HL accompanied by additional clinical features. Approximately 30% of the genetic cases of HL are considered to be syndromic11. SHL consists of HL that presents with anomalies of the eye, kidney, the musculoskeletal and the nervous systems, as well as pigmentary disorders and others15 (Figure 1). Among the well-known syndromes are Usher, Waardenburg and Pendred. Of these syndromes, Pendred and Usher syndromes are the most common10. Several genes associated with SHL (Figure 2) are also involved with NSHL, as in the case of SLC26A4 mutations leading to Pendred syndrome and DFNB410. The genes associated with SHL are represented in Table 1 and Figure 1.
Fig. 1.
Different organs are involved in the clinical symptoms of patients with SHL, in addition to the phenotype in the inner ear. The organs affected in each syndrome are indicated. F, female genitals; M, male genitals.
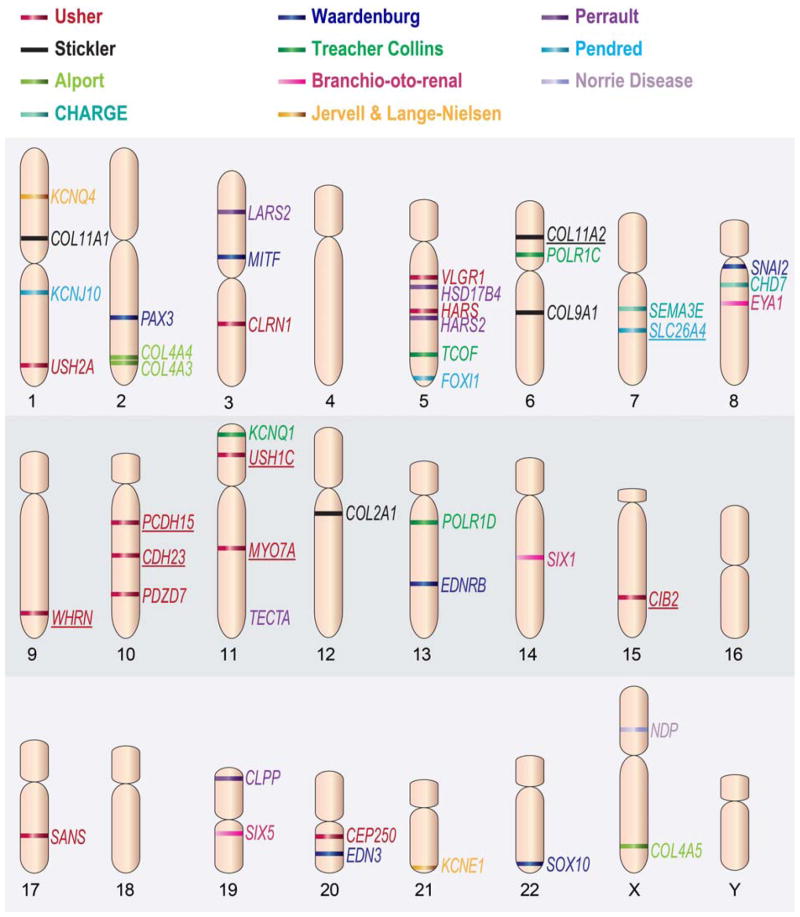
Fig. 2.
Schematic representation of the chromosomal location of genes associated with SHL. The genes are color coded according to the syndrome they are associated with. The genes associated with both syndromic and non-syndromic hearing loss are underlined. Adapted from Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010;68:293–308; with permission.
FORMS OF SYNDROMIC HEARING LOSS
Usher Syndrome
The eye and the ear are the sensory organs responsible for vision, balance and hearing in mammals. These organs are essential for both communication and environmental perception16, 17. Diseases affecting the inner ear and retina of the eye can cause major impairments for human communication systems. Syndromes that include symptoms of both blindness and hearing impairment are widely known. In humans there are approximately 40 syndromes that include both impairments, and about half of the affected cases are caused by mutations attributed to Usher syndrome18. Usher syndrome is an autosomal recessive genetic disease with clinically and genetically heterogeneous characteristics. In humans it is defined by congenital, bilateral deafness and a later onset of vision impairment, caused by retinitis pigmentosa16. Epidemiological studies have estimated that the prevalence of Usher syndrome ranges from 1/6,000 to 1/10,00019, 20. Usher syndrome is subclassified into three clinical types, USH1, USH2, and USH3, based on the severity of the sensorineural hearing loss (SNHL), the presence or absence of vestibular dysfunction and the age at onset of retinitis pigmentosa. USH1 patients suffer from severe to profound congenital bilateral HL accompanied by congenital vestibular dysfunction. In terms of retinitis pigmentosa symptoms, night blindness may be detected during childhood, followed by a narrowing of the visual field, which progresses to severe blindness16, 21. USH2 patients suffer from moderate to severe congenital HL with no vestibular abnormalities. Retinitis pigmentosa is usually diagnosed between the ages of 10 to 4022. In USH3 patients, hearing impairment begins before the 3rd decade of life and is characterized by variable progression. In most cases the patient eventually becomes profoundly deaf. Vestibular defects are variable and retinitis pigmentosa usually begins from the age of 2022. Early symptoms of retinitis pigmentosa are night blindness and loss of peripheral vision. This form of Usher syndrome is the least common in the general population23, but is more prevalent in the Finnish and Ashkenazi Jewish populations24, 25.
To date, 16 independent loci on different chromosomes are known to be associated with Usher syndrome. These loci are further divided into USH1A-G, USH2A-C and USH3A. Moreover, 13 genes have been identified. The USH1 genes are MYO7A for the USH1B locus, encoding the motor protein myosin VIIA26. USH1C encodes harmonin27, 28 and USH1G encodes SANS29, both of which are scaffold proteins. CDH23 mutations are responsible for USH1D, which encodes cadherin 2330, PCDH15 mutations lead to USH1F and encodes protocadherin 1531, both of which are cell adhesion molecules. CIB2 mutations are the cause of USH1J, which encodes calcium and integrin binding protein 232. The USH2 genes are USH2A, encoding usherin33 and ADGRV1 for USH2C, encoding adhesion G protein-coupled receptor VI, also referred to as GPCR98 or VLGR134, 35. Both genes are transmembrane proteins that are involved in signaling. Another gene associated with USH2 is WHRN, encoding whirlin for USH2D36. The USH3 genes are CLRN1 for USH3A, encoding Clarin 1, and HARS (Histidyl tRNA synthetase)24, 37–39. Moreover, another two USH genes have recently been identified. PDZD7 encodes the protein PDZ domain containing 7 and CEP250 encodes centrosome associated protein 250. Usher syndrome has recently been proposed to be an oligogenic disease, due to digenic inheritance of PDZD7 and USH2A or ADGRV1 in patients with Usher syndrome. Two genes have been proposed to lead to variable phenotypes of Usher syndrome, depending on the dosage. CEP250 is associated with early onset HL and severe retinitis pigmentosa in conjunction with two mutant alleles of C2orf71. The patients have mild HL and retinal degeneration with one mutant allele of C2or7140, 41. C2orf71 mutations are associated with retinitis pigmentosa and proposed to encode a ciliary protein42. Importantly, many of the genes listed above have been reported to cause NSHL (Figure 1). For example, different mutations in MYO7A are known to cause recessive deafness DFNB2 and dominant deafness DFNA1143, 44.
Waardenburg Syndrome
Waardenburg syndrome was considered to be an autosomal dominant inherited disease of the neural crest cells, but this syndrome is more clinically and genetically heterogeneous than originally known45. Waardenburg syndrome is characterized mostly by SNHL and pigmentation abnormalities that can occur in the eyes, hair, skin and the cochlear stria vascularis. Other features can be found in a subset of patients. These features are used for clinical classification of the syndrome. Waardenburg syndrome is estimated to have a prevalence of 1/42,000 and is responsible for 1–3% of all congenital HL cases45. During embryonic development, the pluripotent neural crest cells migrate from the neural tube and give rise to different cell types, among them, melanocytes of the skin and inner ear, glia, neurons of the peripheral and enteric nervous systems and some of the skeletal tissue. The symptoms associated with Waardenburg syndrome results from an abnormal proliferation, survival, migration or differentiation of neural crest-derived melanocytes45. Waardenburg syndrome is subdivided into four subtypes, WS1, WS2, WS3 and WS4, on the basis of the presence or absence of additional symptoms46. WS1 is further characterized by dystopia canthourm, an appearance of wide-set eyes due to a prominent broad nasal root, while WS2 has no further significant features. WS1 and WS2 are the most frequent among the four subtypes. WS3 is further characterized by dystopia canthorum and musculoskeletal abnormalities of the upper limbs. WS4 is associated with Hirschsprung disease, characterized by a blockage of the large intestine due to improper muscular bowel movement and neurological defects. Neurological features were also observed in a subset of WS2 patients47. Among the symptoms of this syndrome, the SNHL is the most frequent one with 60% in WS1 to 90% in WS248. Six genes are associated with this syndrome: PAX3 (Paired box 3)49, MITF (Microphthalmia-associated transcription factor)50, EDNBR (Endothelin receptor type B)51, EDN3 (Endothelin 3)52, SOX10 (SRY box10)53, and SNAI2 (Snail homolog 2)54. These genes are known to be involved in the regulation of melanocyte differentiation45. A database for the Waardenburg syndrome-associated genes can be found at the Leiden Open Variation Database (Table 1).
Pendred syndrome
Pendred syndrome is one of the most common autosomal recessive syndromic causes of HL. The audiological phenotype is quite broad, ranging from mild to profound and can be congenital or with a later onset and be progressive55. A common feature among patients is an enlarged vestibular aqueduct (EVA), a common radiological malformation of the inner ear. In addition to SNHL, patients who suffer from this syndrome also show features of congenital and severe to profound temporal bone abnormalities, in addition to goiter partial iodine organification defects resulting in a positive perchlorate discharge test from the goiter, usually in late childhood to early adulthood. Pendred syndrome also features thyroid dysfunction, ranging from euthyroid to hypothyroidism56, 57, and vestibular dysfunction, demonstrated in approximately 65% of affected individuals. The vestibular dysfunction can range from mild unilateral canal paresis to gross bilateral absence of function58.
The estimated prevalence of Pendred syndrome is 7.5 per 100,000 newborns and it accounts for approximately 1 to 8% of the cases of congenital deafness59. Approximately half of the Pendred syndrome cases are caused by a mutation in one of three genes. SLC26A4, encoding the protein pendrin, is an iodide-chloride transporter. Mutations in this gene are responsible for both Pendred syndrome and DFNB4, a form of NSHL. Pendrin is expressed in the kidneys, the inner ear and thyroid59. Approximately 50% of Pendred syndrome affected individuals have a mutation in this gene (Genetics Home Reference, Table 1). Less than 2% of the rest of the affected individuals have mutations in FOXI1 encoding Forkhead box protein I1 or KCNJ10 encoding the ATP-sensitive inward rectifier potassium channel 1058. More than 280 SLC26A4-Pendred syndrome and DFNB4-causing mutations have been identified, but in different ethnic groups, unique pathogenic alleles are found more frequently than others, reflecting a few prevalent founder mutations58.
Additional syndromes
In addition to the above-mentioned syndromes, there are over 700 genetic syndromes that have been described with features of hearing impairment15. Alport syndrome is characterized by renal defects, SNHL and ocular abnormalities60 with a prevalence of 1 in 50,000 (Genetics Home Reference).Three genes are associated with this syndrome: COL4A3 encoding collagen, type IV, alpha 3, and COL4A4 encoding collagen, type IV, alpha 4 for the autosomal inherited types61, 62 and COL4A5 encoding collagen, type IV, alpha 5 for the X-linked type63.
Branchio-oto-renal (BOR) syndrome is an autosomal dominant disease, characterized by defects in the development of the tissues in the neck and malformations of the ear and kidney64. It is estimated that the prevalence of this syndrome is 1 in 40,000 (Genetics Home Reference). Approximately 40% of the individuals affected test positive for mutations in the EYA1 gene encoding the eyes absent homolog 1. An additional 5% and 4% of affected individuals have a mutation in SIX5 encoding the homeobox protein SIX5 and SIX1 encoding the homeobox protein SIX1, respectively64.
CHARGE syndrome is an autosomal dominant syndrome that features coloboma, heart defects, choanal atresia, retardation in growth and development, genital abnormalities, hearing loss and vestibular dysfunction. It is estimated that the prevalence of CHARGE syndrome is 1 in 8,500 to 10,000 individuals (Genetics Home Reference). This syndrome is mostly caused by mutations in the CHD7 gene encoding Chromodomain-helicase-DNA-binding protein 7, an ATP- dependent chromatin remodeling protein65.
Jervell and Lange-Nielsen syndrome is an autosomal recessive disease with features of arrhythmia, SNHL and a significantly higher risk of fainting and sudden death as a result of prolongation of the QTc interval66. This syndrome is estimated to affect 1.6 to 6 per 1,000,000 people worldwide, with a higher frequency in Denmark (Genetics Home Reference; Table 1). The genes associated with this syndrome are KCNQ1 encoding the potassium channel, voltage gated KQT-like subfamily Q, member 1 and KCNE1 encoding potassium channel, voltage gated subfamily E regulatory beta subunit 166–68. Approximately 90% of the cases are caused by a mutation in the KCNQ1 gene, with the rest of the cases caused by mutations in KCNE169, 70.
Norrie disease is characterized by a spectrum of fibrous vascular changes of the retina at birth that progresses to visual impairment with age. About 30 to 50 percent of males with Norrie disease have developmental delays or other forms of intellectual disability, behavioral abnormalities or psychotic-like features. Moreover, the majority of the males also develop HL. This syndrome is X-linked, recessively inherited and caused by mutations in the NDP gene encoding the norrin protein. Mutations in this gene are responsible for about 95% of the affected individuals71. The prevalence of this syndrome is unknown and it is not associated with any racial or ethnic group (Genetics Home Reference, Table 1).
Stickler syndrome can be both dominant and recessive and is characterized by ocular, skeletal, orofacial and auditory abnormalities72, 73. The prevalence of this syndrome is about 1 in 7,500 to 1 in 9,000 newborns (Genetics Home Reference; Table 1). Stickler syndrome is subdivided into five subtypes based on its underlying genetic collagen defect. For the autosomal dominant form of Stickler syndrome, three genes have been identified. Type I Stickler syndrome (STL1) is associated with mutations in COL2A1 encoding collagen, type II, alpha-174. Moreover, mutations in COL11A1 encoding collagen, type XI, alpha-1 are associated with type II (STL2)75 and mutations in COL11A2 encoding collagen, type XI, alpha-2 are associated with type III (STL3)76. The autosomal recessive forms of Stickler syndrome are STL4 and STL5, and their identified related genes are COL9A1 encoding collagen, type IX, alpha-1 and COL9A2 encoding collagen, type IX, alpha-2, respectively77, 78. There is a degree of variability in HL frequency and severity in the different types of this syndrome, even within the same family79.
Treacher-Collins syndrome is usually an autosomal dominant syndrome that affects the development of the bones and other tissues of the face. These abnormalities contribute to speech and language difficulties, visual impairment, conductive HL and breathing difficulties. The symptoms of this syndrome can range from undetectable to severe. Half of the affected individuals suffer from HL caused by defects of the three bones of the middle ear or defects in the development of the ear canal. One in 50,000 people will suffer from this syndrome (Genetics Home Reference; Table 1), caused by mutations in three genes: TCOF1 encoding the treacle protein, POLR1C encoding polymerase I polypeptide C and POLR1D encoding polymerase I polypeptide D. The majority of patients have a mutation in TCOF1, with 1% of the cases caused by a recessive form of this syndrome, with mutations in POLR1C80.
Perrault syndrome is an autosomal recessive disease characterized by SNHL in both sexes and ovarian dysfunction in females. The HL symptoms are bilateral and ranges from moderate with early childhood onset to profound at birth. Moreover, the early childhood form can be progressive. The ovarian dysfunction symptoms can also vary and affected females also show, in some cases, neurological features, such as developmental delay and cerebellar ataxia. Less than 100 affected individuals have been documented (Genetics Home Reference; Table 1), most probably due to difficulties in diagnosis. Four genes have been associated with this syndrome: HARS2 encoding Histidyl-tRNA Synthetase 2, HSD17B4 encoding hydroxysteroid (17-beta) dehydrogenase 4, LARS2 encoding leucyl-tRNA synthetase2 and CLPP encoding the caseinolytic mitochondrial matrix peptidase proteolytic subunit. This syndrome is subdivided into 4 types: type I, II, III and IV, also called PRLTS1, 2, 3, and 4, respectively. The classification of the subtypes is determined according to the neurological involvement and its state, progressive or non-progressive. This classification is now being reconsidered as mutations in CLPP were found to include both types of cases, with or without neurological symptoms81. The clinical features and the molecular genetics information of these syndromes are comprehensively described in OMIM, the Online Mendelian Inheritance in Man database (Table 1).
GENETICS OF HEARING LOSS
Single-gene disorders may be inherited in an autosomal recessive or dominant mode, be carried on the X-chromosome or inherited through the mitochondria. Each form of inheritance may have implications for the number of children to manifest the disease in each case, or whether one or both sexes will be affected. As a result, an accurate and thorough family history is essential upon examining the patient.
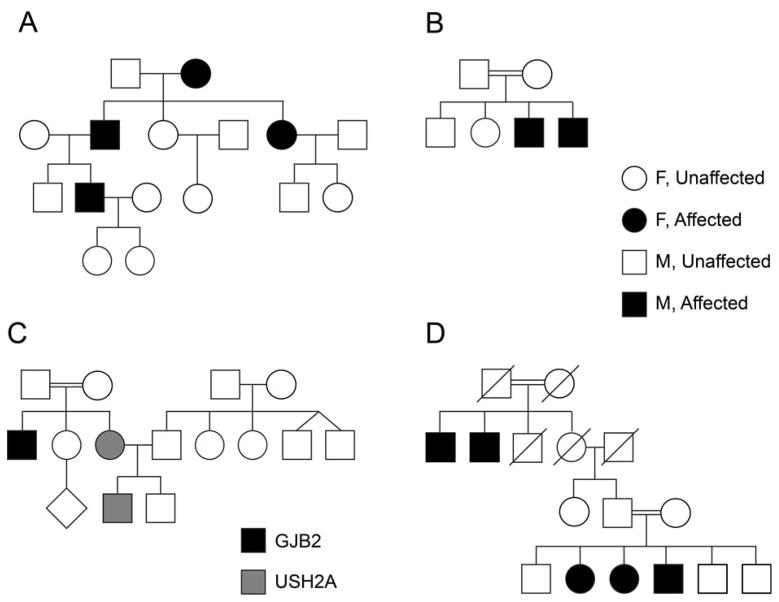
Diseases inherited in a recessive manner are often the most severe in nature and are expressed at birth (congenital) or soon thereafter. As recessive inheritance ‘skips’ a generation, and depends on two carriers of the pathogenic variant, there is often no family history in patients with a recessively inherited disease (Figure 3A). In other cases, multiply affected patients in every other generation are very clear signs of recessive diseases (Figure 3B). Consanguinity, where parents are related, may increase the incidence of recessive disease and is more prevalent in regions of the world such as the Middle East, India and Pakistan (Figure 3A, B). Both females and males are affected in equal proportions. Many forms of SHL are inherited in a recessive manner, including Jervell and Lange syndrome, Usher syndrome, Pendred syndrome, Perrault syndrome and some forms of Alport and Waardenburg syndrome (Table 2).
Fig. 3.
Representative pedigrees of families with HL. (A) Recessive inheritance with no previous family history. After biallelic variants were found in the child, the parents were each found to be carriers of the variant, validating the recessive pattern of inheritance. (B) Recessive inheritance with family history. The family represented in the pedigree presented with symptoms of USH1, with five affected individuals. The chromosomal critical region was defined by the use of microsatellite DNA markers, pinpointing the locus to chromosome 15q2286. Several years later, CIB2 was found to be the pathogenic variant responsible for this new form of Usher syndrome, USH1J32. (C) Dominant inheritance, with an affected individual in three out of four generations, which may be observed for Waardenburg syndrome with a PAX3, SNAI2 or MITF mutation. (D) A family with both SHL and NSHL. Two patients have Usher syndrome type II, with pathogenic variants in the USH2A gene. One patient has NSHL, due to biallelic pathogenic variants in the GJB2 gene111.
Table 2.
Syndromic hearing loss-associated genes
| Syndrome | Gene | Protein | OMIM1 entry |
|---|---|---|---|
| Alport Syndrome | COL4A5 | Collagen, type IV, alpha 5 | 303630 |
| COL4A3 | Collagen, type IV, alpha 3 | 120070 | |
| COL4A4 | Collagen, type IV, alpha 4 | 120131 | |
|
| |||
| Branchio-oto-renal Syndrome | EYA1 | Eyes absent homolog 1 | 601653 |
| SIX5 | Homeobox protein SIX5 | 600963 | |
| SIX1 | Homeobox protein SIX1 | 601205 | |
|
| |||
| CHARGE Syndrome | SEMA3E | Semaphorin 3E | 608166 |
| CHD7 | Chromodomain-helicase-DNA-binding protein 7 | 608892 | |
|
| |||
| Jervell & Lange-Nielsen Syndrome | KCNQ1 | Potassium channel, voltage gated KQT-like subfamily Q, member 1 | 607542 |
| KCNE1 | Potassium channel, voltage gated subfamily E regulatory beta subunit 1 | 176261 | |
|
| |||
| Norrie Disease | NDP | Norrie disease protein | 300658 |
|
| |||
| Pendred Syndrome | SLC26A4 | Pendrin | 605646 |
| FOXI1 | Forkhead box protein I1 ATP-sensitive inward rectifier | 601093 | |
| KCNJ10 | potassium channel 10 | 602208 | |
|
| |||
| Stickler Syndrome | COL2A1 | Collagen, type II, alpha-1 | 120140 |
| COL11A1 | Collagen, type XI, alpha-1 | 120280 | |
| COL11A2 | Collagen, type XI, alpha-2 | 120290 | |
| COL9A1 | Collagen, type IX, alpha-1 | 120210 | |
| COL9A2 | Collagen, type IX, alpha-2 | 120260 | |
|
| |||
| Treacher Collins Syndrome | TCOF1 | Treacher Collins-Franceschetti syndrome 1 | 606847 |
| POLR1D | Polymerase I Polypeptide D | 613715 | |
| POLR1C | Polymerase I Polypeptide C | 610060 | |
|
| |||
| Usher Syndrome | MYO7A | Myosin VIIA | 276903 |
| USH1C | Harmonin | 605242 | |
| CDH23 | Cadherin 23 | 605516 | |
| PCDH15 | Protocadherin 15 | 605514 | |
| SANS | Scaffold protein containing ankyrin repeats and sam domain | 607696 | |
| CIB2 | Calcium and integrin binding protein 2 | 605564 | |
| USH2A | Usherin | 608400 | |
| VLGR1 | Very large G-coupled protein receptor isoform b | 602851 | |
| WHRN | Whirlin | 607928 | |
| CLRN1 | Clarin 1 | 606397 | |
| HARS | Histidyl tRNA synthetase | 142810 | |
| PZDZ7 | PDZ domain containing 7 | NA2 | |
| CEP250 | 250 | NA | |
|
| |||
| Waardenburg Syndrome | PAX3 | Paired box 3 | 606567 |
| SNAI2 | Snail homolog 2 | 602150 | |
| EDN3 | Endothelin 3 | 131242 | |
| EDNRB | Endothelin receptor type B | 131244 | |
| MITF | Microphthalmia-associated transcription factor | 156845 | |
| SOX10 | SRY box10 | 602229 | |
|
| |||
| Perrault Syndrome | HSD17B4 | Hydroxysteroid (17-beta) dehydrogenase 4 | 601860 |
| HARS2 | Histidyl-TRNA Synthetase 2 | 600783 | |
| CLPP | Caseinolytic mitochondrial matrix peptidase proteolytic subunit | 601119 | |
| LARS2 | Leucyl-tRNA synthetase2 | 604544 | |
OMIM: Online Mendelian Inheritance in Man
NA: Not available
Dominant inheritance tends to involve onset of the disease phenotype later in life and may be less severe than recessively inherited diseases (Figure 3C). Only one parent need to carry the pathogenic variant and is affected by the disease as well. Complications in diagnosis may arise due to reduced penetrance, whereby a patient harbors the genotype but not the phenotype of the disease.
It is important to note that several syndromes may be inherited in either a recessive or dominant fashion, depending on the gene and variant involved. Furthermore, families may harbor mutations in more than one gene, leading to the presence of more than one disease in an extended family (Figure 3D).
A more complex scenario arises due to the oligogenic nature of some diseases. It has become clearer in recent years that the phenotype of patients, even with what appears to be a single-gene disorder, may be due to multiple variants. An example is described above for Usher syndrome40, 41.
DIAGNOSIS OF HEARING LOSS
Screening and identifying the etiology of HL is extremely important to provide the best opportunities for care and rehabilitation in children. The introduction of newborn screening for HL in developed countries has led to earlier diagnosis and improvement in ascertainment and potential outcomes82, as treatment strategies are examined and applied sooner. Determining the etiology of HL is even more crucial in cases of SHL, as the associated clinical features are usually more severe83. Early diagnosis can help predict the progression of the HL in the patient and the prescribed course of action in treating the patient, as well as provide warnings for future potentially life threatening abnormalities.
A continuing challenge in medical genetics has been to determine the etiology of each disease. For example, the presence of SLC26A4 mutations in a child may help predict whether the child will develop goiter and hypothyroidism after puberty, or be more susceptible to acute HL following head trauma83. Patients with Jervell and Lange-Nielsen syndrome will be more sensitive to syncope and are at risk for sudden death83. As a result, being aware that a child has a KCNQ1 mutation can be extremely informative for his or her health. Today, mutations in at least 45 genes are known to be associated with SHL, providing an opportunity for patients with these mutations to benefit from this knowledge. Most of these discoveries have only been made in the last two decades, and some even more recently. Despite this progress, the underlying genetic cause of hundreds of inherited syndromes is still unknown and the continuing challenge is to uncover the etiology for the affected children and adults.
Identifying disease genes through linkage mapping with genetic markers
The large size of the genome and the high number of genes it contains made the identification of disease-associated genes a tedious task from a historical perspective. Before the Human Genome Project (HGP) was completed in 200184, researchers used various techniques for identifying and mapping genes associated with Mendelian and single-gene inherited disorders. Linkage mapping, the most common technique used, was aimed at finding the approximate location of the disease gene relative to a DNA segment with a known chromosomal location, a genetic marker85. The process included scanning the genomes of members of an affected family and using highly polymorphic DNA markers to identify regions linked with the disease. The affected family members shared specific variants of the DNA markers more frequently than in the general population, suggesting linkage with the presence of a nearby disease gene. To focus on the disease gene, multiple markers in the linked region were further genotyped to define the critical region. DNA markers were generally in the form of microsatellites, repetitive DNA elements present every 1000 bp in the human genome. Their polymorphic nature and presence of multiple alleles in the population rendered these markers to be extremely useful. The most commonly used ones were CA repeats, which could be purchased commercially and could be used for automated genotyping as they were labeled with fluorescent markers. The next generation of DNA markers involved single nucleotide polymorphisms (SNPs). Although they are less polymorphic, having only two alleles per SNP, they are more frequent in the genome, at every 100–300 bp, and facilitate linkage analysis. As most of the human genes are annotated, examination of the region using genome browsers, such as the UCSC Genome Browser (Table 1), can provide a list of the genes in the region. The challenge is then to identify the pathogenic variant that leads to the disease under study. Oftentimes there was a gap of years from identification of the chromosomal location of the putative gene, to identification and validation of the pathogenic variant. For example, a number of families with USH1 from Pakistan were studied by linkage analysis with microsatellite markers86. Years later, one of the families in this group was found to harbor a mutation in CIB2, defining a new form of Usher syndrome, USH1J (pedigree in Figure 3B). The majority of genes for SHL were identified in this way.
Validation of the pathogenic variant is subsequently performed using capillary Sanger sequencing, which allows for sequencing of regions of approximately 800–1000 bp. A comparison of the patient’s sequence to unaffected family members can help define the critical variant. However, additional criteria are required for the variant to be defined as the disease causing change in the DNA. Functional analysis, by replicating the variant in cell culture, may help define the pathogenicity further. For example, the SLC26A4 variant c.1458_1459insT, when replicated in COS7 cells, was mislocalized, providing further evidence that this is a pathogenic variant87. Finally, reduction of a particular gene in an animal model can provide the most compelling circumstantial evidence of pathogenicity, as well as demonstrate the mechanisms involved. CIB2, a candidate for Usher syndrome, was shown to be essential for mechanosensory hair cell function in zebrafish32. Gene-targeted mutagenesis of Slc26a4 in the mouse revealed that Pendrin, the protein encoded by this gene, is important for the development of the inner ear and provided an understanding of the cause for deafness in Pendred syndrome patients88. An additional Slc26a4 mutation in mice, Slc26a4 loop/loop, had symptoms of deafness but without the enlarged thyroid gland symptoms that characterizes Pendred syndrome89. However, histological analysis of the thyroid tissue showed morphological defects and the inner ear showed molecular and morphological defects consistent with the symptoms seen in patients with disrupted thyroid hormone activity. This study led to the proposal that thyroid hormone deprivation, caused by this Slc26a4 mutation, contributes to the deafness in this mouse model.
Identifying disease genes through high-throughput sequencing
The development of high-throughput sequencing technology has revolutionized both discovery and diagnosis of pathogenic variants for disease. Massive parallel sequencing (MPS), also known as Next Generation Sequencing (NGS), enables a more rapid and low-cost method of DNA sequencing. Compared to the traditional capillary or Sanger sequencing, these methods perform large-scale sequencing, generating a billion bases within a single run90. The MPS output is aligned to the human reference genome, a consensus representation of the genome (Genome Reference Consortium; Table 1), enabling the identification of differences between the reads of the current genome being sequenced and the reference genome. Bioinformatics tools, such as the Genome Analysis Toolkit (GATK) software package (Table 1), are next used for detecting the variants, examining whether the variant is unique, the frequency of this variant, and the potential damage of this variant to the protein structure and function. Moreover, the sequence conservation is also examined as well as the mode of inheritance, which should match the one of the disease analyzed. The potential pathogenic variants are then screened for segregation in the affected family. This analysis is less complicated if the variant detected is a known deafness mutation, while variants in genes not previously associated with deafness are more challenging to prove91.
The development of MPS changed the field of gene sequencing, enabling multiple genomes to be sequenced simultaneously. However, the complexity of genomic analysis for the large number of variants that arise, in particular from non-coding regions of the genome, remains challenging. To circumvent this problem, researchers studying highly heterogeneous diseases such as Usher syndrome92, 93, inherited eye diseases94, or deafness95, 96, have screened only for genes already known to be associated with the disease under study. In cases where pathogenic variants are not found using a select number of genes, Whole Exome Sequencing (WES) becomes the next option. As the costs are decreasing, WES is sometimes the first approach97. In this method, only coding exons of genes are sequenced, alleviating the need to analyze non-coding regions, with a substantial savings of cost. Whole Genome Sequencing (WGS) has been used for identification of human disease genes in severe cases of neonatal disease98. Though considerably more costly, WGS is considered to be the most comprehensive method for detecting all variants, in particular copy number variants (CNV) that may be involved in disease98. Furthermore, linkage analysis, described above, may be used in conjunction with WGS to facilitate localization of the pathogenic variant98. Overall, currently, NGS technology has a molecular diagnostic success rate of 25%99 and is predicted to solve the genetic basis of 60% of Mendelian inherited disorders in the near future100.
The importance of genetic research in understanding the processes involved in hearing and deafness
To date there are a number of different treatments available for hearing impairment. These include amplification of sound using hearing aids and cochlear or auditory brainstem implants that stimulate the cochlear nerve or the nuclei, respectively11. Although the physical structure and activity of the ear are well understood, the specific part of the ear that each deafness-associated gene functions in is not fully characterized. Defining the genes and pathways responsible for normal hearing are paving the way towards developing new treatments for hearing impairments. Among the different approaches for newly developed hearing impairment treatments are the regeneration of inner ear sensory cells101, 102 and the use of viral vectors for gene therapy103. These potential treatments will be helpful for both genetically and environmentally-related HL11.
PROSPECTS OF FUTURE TREATMENT MODALITIES FOR HL
Although hearing aids and cochlear implants are far from being ideal, HL treatments today rely mostly on these approaches. With the aim of providing more optimal treatments for HL patients, the research for alternative treatments has progressed toward different fields, including the use of gene therapy. Understanding the genetic basis of the HL and advances in DNA delivery methods are both essential for achieving the goal of gene therapy development for HL and for others disorders as well104. One approach for HL therapy is the use of antisense oligonucleotides. These were successfully used in a mouse model with a loss of function mutation in USH1C, which correlates with the human Acadian USH1C mutation105. This specific mutation leads to defects in splicing of the USH1C messenger RNA (mRNA), encoding the harmonin protein. Antisense oligonucleotides can modulate posttranscriptional regulation by gene silencing or alteration of RNA metabolism104. In this case the oligonucleotides were used to restore the correct splicing of the gene, enabling expression of the protein. The most effective results were achieved when 3 to 5 day old mutant mice were injected intraperitoneally with the antisense oligonucleotides. Although high-frequency hearing was not developed in these mice at this stage, low and mid- frequency hearing levels were the same as those measured in non-mutant mice. These results were maintained for six months105. Implementing this treatment for humans is more complex. Human newborns can hear, which might require gene therapy intervention during gestation. Moreover, the delayed onset of hearing development in mice makes comparisons to humans difficult. Nevertheless, these initial results are promising, providing hope for the future104,106.
Another possible approach to rescue HL is the use of hair cell regenerative treatments, as most HL cases are caused by irreversible damage to hair cells106. As mammalian cochlear hair cells do not regenerate naturally, different strategies are being employed to regenerate or transplant hair cells. The basis of regenerative research relies on the observation that hair cell regeneration is possible in birds, fish and amphibians101. Regenerative research is aimed at discovering the exact formula of reagents that will trigger the regeneration of mammalian cochlear hair cells from precursor cells. The field of transplantation focuses on coaxing either inner-ear stem cells or even pluripotent stem cells into hair cell lineage. These cells may subsequently be transplanted into the inner ear. Although very promising, both strategies are in very early stages for HL treatment106, 107. Novartis and Genvec are currently implementing the first clinical trial using the Atoh1 gene, which can induce the differentiation of sensory cells in the inner ear (Table 1).
Efforts are also being made in the pharmacological field with the aim of discovering novel drugs for HL. This field is not limited to orally delivered pharmacological compounds. The use of non-invasive intra-tympanic or invasive intra-cochlear routes are options being examined as well108. Animal models, including zebrafish and mice, are widely used as preclinical models for drug use, both in vivo and in vitro with cochlear cultures109, 110. While there is potential in HL drug treatments, to date, there are no Food and Drug Administration (FDA) approved drugs on the market106.
Summary
Determining the etiology of deafness in a patient with SHL is a key goal in order to provide optimal rehabilitation options. In the future, understanding the mechanisms of HL due to each genetic mutation will pave the way for therapeutic delivery. Alleviating the isolation caused by deafness will greatly improve the quality of life of these patients. The recent development and implementation of new high-throughput sequencing technology will facilitate a significant improvement in identification of novel syndromic and non-syndromic HL-associated pathogenic variants and genes.
Key Points.
Syndromic hearing loss (SHL) is a form of hearing loss (HL) accompanied by additional clinical features in the visual, nervous system, endocrine and other systems. The most prevalent syndromes are Usher, Waardenburg and Pendred.
Genetic diagnostics can detect pathogenic variants and provide an answer regarding the cause of the HL, as well as the associated clinical symptoms of the SHL, to care for the patient.
Linkage analysis with DNA markers and PCR diagnostics is often used to detect these variants in clinical settings. High-throughput sequencing methods, focusing on either specific genes, the exons of genes or the entire genome of a patient, are moving into the clinic to provide more cost effective and efficient methods for diagnostics.
Acknowledgments
Research in the Karen Avraham laboratory is supported by the National Institutes of Health (NIDCD) R01DC011835, I-CORE Gene Regulation in Complex Human Disease Center No. 41/11, Israel Science Foundation 1320/11, Human Frontier Science Program RGP0012/2012, and United States-Israel Binational Science Foundation (BSF) 2013027.
Abbreviations
- ARHL
age related hearing loss
- ASHA
American Speech-Language-Hearing Association
- ATP
adenosine triphosphate
- BOR
Branchio-oto-renal
- bp
base pairs
- CHARGE
Coloboma, Heart defect, Atresia choanae, Retarded growth and development, Genital hypoplasia, Ear anomalies/deafness syndrome
- CNV
copy number variants
- COS7
CV-1 in Origin with SV40 genes
- dB
decibel
- DFN
DeaFNess
- DFNA
nonsyndromic deafness, autosomal dominant
- DFNB
nonsyndromic deafness autosomal recessive
- DFNX
nonsyndromic deafness, X- linked
- DNA
deoxyribonucleic acid
- EVA
Enlarged vestibular aqueduct
- FDA
Food and Drug Administration
- HARS
histidyl tRNA synthetase
- HGP
Human Genome Project
- HL
hearing loss
- GATK
Genome Analysis Toolkit
- IHC
inner hair cell
- JLNS
Jervell and Lange-Nielsen syndrome
- MPS
massive parallel sequencing
- mRNA
messenger RNA
- NGS
next generation sequencing
- NIDCD
National Institute on Deafness and Other Communication Disorders
- NSHL
non-syndromic hearing loss
- OHC
outer hair cell
- OMIM
Online Mendelian Inheritance in Man
- PCR
polymerase chain reaction
- PRLTS1
Perrault syndrome 1
- QTc
Corrected QT Interval
- SHL
syndromic hearing loss
- SNHL
sensorineural hearing loss
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
- STL1
Type I Stickler syndrome
- UCSC
University of California, Santa Cruz
- USH1
2, 3, Usher syndrome 1, 2, 3
- WES
whole exome sequencing
- WGS
whole genome sequencing
- WHO
World Health Organization
- WS1
2, 3, 4, Waardenburg syndrome 1, 2, 3, 4
Footnotes
Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Tal Koffler, Email: talkoffler@post.tau.ac.il.
Kathy Ushakov, Email: kathyushakov@post.tau.ac.il.
Karen B. Avraham, Email: karena@post.tau.ac.il.
References
- 1.Quaranta N, Coppola F, Casulli M, et al. Epidemiology of age related hearing loss: A review. Hearing, Balance and Communication. 2015:1–5. [Google Scholar]
- 2.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 3.Nance WE. The genetics of deafness. Ment Retard Dev Disabil Res Rev. 2003;9:109–119. doi: 10.1002/mrdd.10067. [DOI] [PubMed] [Google Scholar]
- 4.Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010;68:293–308. doi: 10.1016/j.neuron.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Ross M, Pawlina W. Histology: A Text and Atlas, with Correlated Cell and Molecular Biology. 6. 2010. Paperback. [Google Scholar]
- 6.Raphael Y, Altschuler RA. Structure and innervation of the cochlea. Brain Res Bull. 2003;60:397–422. doi: 10.1016/s0361-9230(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 7.Clark JG. Uses and abuses of hearing loss classification. ASHA. 1981;23:493–500. [PubMed] [Google Scholar]
- 8.Lin FR, Thorpe R, Gordon-Salant S, Ferrucci L. Hearing loss prevalence and risk factors among older adults in the United States. J Gerontol A Biol Sci Med Sci. 2011;66:582–590. doi: 10.1093/gerona/glr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Q, Tang J. Age-related hearing loss or presbycusis. Eur Arch Otorhinolaryngol. 2010;267:1179–1191. doi: 10.1007/s00405-010-1270-7. [DOI] [PubMed] [Google Scholar]
- 10.Parker M, Bitner-Glindzicz M. Genetic investigations in childhood deafness. Arch Dis Child. 2014 doi: 10.1136/archdischild-2014-306099. [DOI] [PubMed] [Google Scholar]
- 11.Kalatzis V, Petit C. The fundamental and medical impacts of recent progress in research on hereditary hearing loss. Hum Mol Genet. 1998;7:1589–1597. doi: 10.1093/hmg/7.10.1589. [DOI] [PubMed] [Google Scholar]
- 12.Petit C, Levilliers J, Hardelin JP. Molecular genetics of hearing loss. Annu Rev Genet. 2001;35:589–646. doi: 10.1146/annurev.genet.35.102401.091224. [DOI] [PubMed] [Google Scholar]
- 13.Vona B, Nanda I, Hofrichter MA, et al. Non-syndromic hearing loss gene identification: a brief history and glimpse into the future. Mol Cell Probes. 2015 doi: 10.1016/j.mcp.2015.03.008. [DOI] [PubMed]
- 14.Pimperton H, Kennedy CR. The impact of early identification of permanent childhood hearing impairment on speech and language outcomes. Arch Dis Child. 2012;97:648–653. doi: 10.1136/archdischild-2011-301501. [DOI] [PubMed] [Google Scholar]
- 15.Toriello HV, Reardon W, Gorlin RJ. Hereditary hearing loss and its syndromes. Oxford university press; Oxford: 2004. [Google Scholar]
- 16.Reiners J, Nagel-Wolfrum K, Jurgens K, Marker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Mathur P, Yang J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. Biochim Biophys Acta. 2015;1852:406–420. doi: 10.1016/j.bbadis.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernon M. Usher's syndrome--deafness and progressive blindness. Clinical cases, prevention, theory and literature survey. J Chronic Dis. 1969;22:133–151. doi: 10.1016/0021-9681(69)90055-1. [DOI] [PubMed] [Google Scholar]
- 19.Kimberling WJ, Hildebrand MS, Shearer AE, et al. Frequency of Usher syndrome in two pediatric populations: Implications for genetic screening of deaf and hard of hearing children. Genet Med. 2010;12:512–516. doi: 10.1097/GIM.0b013e3181e5afb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope CI, Bundey S, Proops D, Fielder AR. Usher syndrome in the city of Birmingham--prevalence and clinical classification. Br J Ophthalmol. 1997;81:46–53. doi: 10.1136/bjo.81.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Soest S, Westerveld A, de Jong PT, Bleeker-Wagemakers EM, Bergen AA. Retinitis pigmentosa: defined from a molecular point of view. Surv Ophthalmol. 1999;43:321–334. doi: 10.1016/s0039-6257(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 22.El-Amraoui A, Petit C. The retinal phenotype of Usher syndrome: pathophysiological insights from animal models. C R Biol. 2014;337:167–177. doi: 10.1016/j.crvi.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Millan JM, Aller E, Jaijo T, Blanco-Kelly F, Gimenez-Pardo A, Ayuso C. An update on the genetics of usher syndrome. J Ophthalmol. 2011;2011:417217. doi: 10.1155/2011/417217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joensuu T, Hamalainen R, Yuan B, et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet. 2001;69:673–684. doi: 10.1086/323610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ness SL, Ben-Yosef T, Bar-Lev A, et al. Genetic homogeneity and phenotypic variability among Ashkenazi Jews with Usher syndrome type III. J Med Genet. 2003;40:767–772. doi: 10.1136/jmg.40.10.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weil D, Blanchard S, Kaplan J, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 27.Verpy E, Leibovici M, Zwaenepoel I, et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 28.Bitner-Glindzicz M, Lindley KJ, Rutland P, et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet. 2000;26:56–60. doi: 10.1038/79178. [DOI] [PubMed] [Google Scholar]
- 29.Weil D, El-Amraoui A, Masmoudi S, et al. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet. 2003;12:463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- 30.Bolz H, von Brederlow B, Ramirez A, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 31.Ahmed ZM, Riazuddin S, Ahmad J, et al. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12:3215–3223. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- 32.Riazuddin S, Belyantseva IA, Giese AP, et al. Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat Genet. 2012;44:1265–1271. doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eudy JD, Weston MD, Yao S, et al. Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science. 1998;280:1753–1757. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- 34.Kremer H, van Wijk E, Marker T, Wolfrum U, Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum Mol Genet. 2006;15(Spec No 2):R262–270. doi: 10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- 35.Weston MD, Luijendijk MW, Humphrey KD, Moller C, Kimberling WJ. Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am J Hum Genet. 2004;74:357–366. doi: 10.1086/381685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebermann I, Scholl HP, Charbel Issa P, et al. A novel gene for Usher syndrome type 2: mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet. 2007;121:203–211. doi: 10.1007/s00439-006-0304-0. [DOI] [PubMed] [Google Scholar]
- 37.Puffenberger EG, Jinks RN, Sougnez C, et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One. 2012;7:e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adato A, Vreugde S, Joensuu T, et al. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur J Hum Genet. 2002;10:339–350. doi: 10.1038/sj.ejhg.5200831. [DOI] [PubMed] [Google Scholar]
- 39.Fields RR, Zhou G, Huang D, et al. Usher syndrome type III: revised genomic structure of the USH3 gene and identification of novel mutations. Am J Hum Genet. 2002;71:607–617. doi: 10.1086/342098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebermann I, Phillips JB, Liebau MC, et al. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J Clin Invest. 2010;120:1812–1823. doi: 10.1172/JCI39715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khateb S, Zelinger L, Mizrahi-Meissonnier L, et al. A homozygous nonsense CEP250 mutation combined with a heterozygous nonsense C2orf71 mutation is associated with atypical Usher syndrome. J Med Genet. 2014;51:460–469. doi: 10.1136/jmedgenet-2014-102287. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura DY, Baye LM, Perveen R, et al. Discovery and functional analysis of a retinitis pigmentosa gene, C2ORF71. Am J Hum Genet. 2010;86:686–695. doi: 10.1016/j.ajhg.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu XZ, Walsh J, Mburu P, et al. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet. 1997;16:188–190. doi: 10.1038/ng0697-188. [DOI] [PubMed] [Google Scholar]
- 44.Liu XZ, Walsh J, Tamagawa Y, et al. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet. 1997;17:268–269. doi: 10.1038/ng1197-268. [DOI] [PubMed] [Google Scholar]
- 45.Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 46.Read AP, Newton VE. Waardenburg syndrome. J Med Genet. 1997;34:656–665. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bondurand N, Dastot-Le Moal F, Stanchina L, et al. Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am J Hum Genet. 2007;81:1169–1185. doi: 10.1086/522090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton V. Hearing loss and Waardenburg's syndrome: implications for genetic counselling. J Laryngol Otol. 1990;104:97–103. doi: 10.1017/s002221510011196x. [DOI] [PubMed] [Google Scholar]
- 49.Tassabehji M, Read AP, Newton VE, et al. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature. 1992;355:635–636. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- 50.Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 51.Attie T, Till M, Pelet A, et al. Mutation of the endothelin-receptor B gene in Waardenburg-Hirschsprung disease. Hum Mol Genet. 1995;4:2407–2409. doi: 10.1093/hmg/4.12.2407. [DOI] [PubMed] [Google Scholar]
- 52.Edery P, Attie T, Amiel J, et al. Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome) Nat Genet. 1996;12:442–444. doi: 10.1038/ng0496-442. [DOI] [PubMed] [Google Scholar]
- 53.Pingault V, Bondurand N, Kuhlbrodt K, et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I. SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet. 2002;11:3231–3236. doi: 10.1093/hmg/11.25.3231. [DOI] [PubMed] [Google Scholar]
- 55.King KA, Choi BY, Zalewski C, et al. SLC26A4 genotype, but not cochlear radiologic structure, is correlated with hearing loss in ears with an enlarged vestibular aqueduct. Laryngoscope. 2010;120:384–389. doi: 10.1002/lary.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reardon W, Trembath RC. Pendred syndrome. J Med Genet. 1996;33:1037–1040. doi: 10.1136/jmg.33.12.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Masindova I, Varga L, Stanik J, et al. Molecular and hereditary mechanisms of sensorineural hearing loss with focus on selected endocrinopathies. Endocr Regul. 2012;46:167–186. doi: 10.4149/endo_2012_03_167. [DOI] [PubMed] [Google Scholar]
- 58.Alasti F, Van Camp G, Smith RJH. Pendred Syndrome/DFNB4. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 59.Hone SW, Smith RJ. Genetic screening for hearing loss. Clin Otolaryngol Allied Sci. 2003;28:285–290. doi: 10.1046/j.1365-2273.2003.00700.x. [DOI] [PubMed] [Google Scholar]
- 60.Alport AC. Hereditary familial congenital haemorrhagic nephritis. Br Med J. 1927;1:504–506. doi: 10.1136/bmj.1.3454.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemmink HH, Mochizuki T, van den Heuvel LP, et al. Mutations in the type IV collagen alpha 3 (COL4A3) gene in autosomal recessive Alport syndrome. Hum Mol Genet. 1994;3:1269–1273. doi: 10.1093/hmg/3.8.1269. [DOI] [PubMed] [Google Scholar]
- 62.Mochizuki T, Lemmink HH, Mariyama M, et al. Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat Genet. 1994;8:77–81. doi: 10.1038/ng0994-77. [DOI] [PubMed] [Google Scholar]
- 63.Barker DF, Hostikka SL, Zhou J, et al. Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science. 1990;248:1224–1227. doi: 10.1126/science.2349482. [DOI] [PubMed] [Google Scholar]
- 64.Smith RJH. Branchiootorenal Spectrum Disorders. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 65.Martin DM. Epigenetic Developmental Disorders: CHARGE syndrome, a case study. Curr Genet Med Rep. 2015;3:1–7. doi: 10.1007/s40142-014-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neyroud N, Tesson F, Denjoy I, et al. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- 67.Tyson J, Tranebjaerg L, Bellman S, et al. IsK and KvLQT1: mutation in either of the two subunits of the slow component of the delayed rectifier potassium channel can cause Jervell and Lange-Nielsen syndrome. Hum Mol Genet. 1997;6:2179–2185. doi: 10.1093/hmg/6.12.2179. [DOI] [PubMed] [Google Scholar]
- 68.Schulze-Bahr E, Wang Q, Wedekind H, et al. KCNE1 mutations cause Jervell and Lange-Nielsen syndrome. Nat Genet. 1997;17:267–268. doi: 10.1038/ng1197-267. [DOI] [PubMed] [Google Scholar]
- 69.Tyson J, Tranebjaerg L, McEntagart M, et al. Mutational spectrum in the cardioauditory syndrome of Jervell and Lange-Nielsen. Hum Genet. 2000;107:499–503. doi: 10.1007/s004390000402. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz PJ, Spazzolini C, Crotti L, et al. The Jervell and Lange-Nielsen syndrome: natural history, molecular basis, and clinical outcome. Circulation. 2006;113:783–790. doi: 10.1161/CIRCULATIONAHA.105.592899. [DOI] [PubMed] [Google Scholar]
- 71.Sims KB. NDP-Related Retinopathies. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 72.Rose PS, Levy HP, Liberfarb RM, et al. Stickler syndrome: clinical characteristics and diagnostic criteria. Am J Med Genet A. 2005;138A:199–207. doi: 10.1002/ajmg.a.30955. [DOI] [PubMed] [Google Scholar]
- 73.Temple IK. Stickler's syndrome. J Med Genet. 1989;26:119–126. doi: 10.1136/jmg.26.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahmad NN, Ala-Kokko L, Knowlton RG, et al. Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy) Proc Natl Acad Sci U S A. 1991;88:6624–6627. doi: 10.1073/pnas.88.15.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richards AJ, Yates JR, Williams R, et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum Mol Genet. 1996;5:1339–1343. doi: 10.1093/hmg/5.9.1339. [DOI] [PubMed] [Google Scholar]
- 76.Vikkula M, Mariman EC, Lui VC, et al. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell. 1995;80:431–437. doi: 10.1016/0092-8674(95)90493-x. [DOI] [PubMed] [Google Scholar]
- 77.Van Camp G, Snoeckx RL, Hilgert N, et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am J Hum Genet. 2006;79:449–457. doi: 10.1086/506478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mayne R, Brewton RG, Mayne PM, Baker JR. Isolation and characterization of the chains of type V/type XI collagen present in bovine vitreous. J Biol Chem. 1993;268:9381–9386. [PubMed] [Google Scholar]
- 79.Acke FR, Dhooge IJ, Malfait F, De Leenheer EM. Hearing impairment in Stickler syndrome: a systematic review. Orphanet J Rare Dis. 2012;7:84. doi: 10.1186/1750-1172-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kadakia S, Helman SN, Badhey AK, Saman M, Ducic Y. Treacher Collins Syndrome: the genetics of a craniofacial disease. Int J Pediatr Otorhinolaryngol. 2014;78:893–898. doi: 10.1016/j.ijporl.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 81.Jenkinson EM, Rehman AU, Walsh T, et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am J Hum Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choo D, Meinzen-Derr J. Universal newborn hearing screening in 2010. Curr Opin Otolaryngol Head Neck Surg. 2010;18:399–404. doi: 10.1097/MOO.0b013e32833d475d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parker M, Bitner-Glindzicz M. Genetic investigations in childhood deafness. Arch Dis Child. 2015;100:271–278. doi: 10.1136/archdischild-2014-306099. [DOI] [PubMed] [Google Scholar]
- 84.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 85.Weber JL, May PE. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 86.Ahmed ZM, Riazuddin S, Khan SN, Friedman PL, Friedman TB. USH1H, a novel locus for type I Usher syndrome, maps to chromosome 15q22–23. Clin Genet. 2009;75:86–91. doi: 10.1111/j.1399-0004.2008.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brownstein ZN, Dror AA, Gilony D, Migirov L, Hirschberg K, Avraham KB. A novel SLC26A4 (PDS) deafness mutation retained in the endoplasmic reticulum. Arch Otolaryngol Head Neck Surg. 2008;134:403–407. doi: 10.1001/archotol.134.4.403. [DOI] [PubMed] [Google Scholar]
- 88.Everett LA, Belyantseva IA, Noben-Trauth K, et al. Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum Mol Genet. 2001;10:153–161. doi: 10.1093/hmg/10.2.153. [DOI] [PubMed] [Google Scholar]
- 89.Dror AA, Lenz DR, Shivatzki S, Cohen K, Ashur-Fabian O, Avraham KB. Atrophic thyroid follicles and inner ear defects reminiscent of cochlear hypothyroidism in Slc26a4-related deafness. Mamm Genome. 2014;25:304–316. doi: 10.1007/s00335-014-9515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 91.Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol. 2012;13:245. doi: 10.1186/gb-2012-13-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bujakowska KM, Consugar M, Place E, et al. Targeted exon sequencing in Usher syndrome type I. Invest Ophthalmol Vis Sci. 2014;55:8488–8496. doi: 10.1167/iovs.14-15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aparisi MJ, Aller E, Fuster-Garcia C, et al. Targeted next generation sequencing for molecular diagnosis of Usher syndrome. Orphanet J Rare Dis. 2014;9:168. doi: 10.1186/s13023-014-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Consugar MB, Navarro-Gomez D, Place EM, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2015;17:253–261. doi: 10.1038/gim.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brownstein Z, Friedman LM, Shahin H, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Riahi Z, Bonnet C, Zainine R, et al. Whole exome sequencing identifies mutations in usher syndrome genes in profoundly deaf tunisian patients. PLoS One. 2015;10:e0120584. doi: 10.1371/journal.pone.0120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur J Hum Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rubel EW, Furrer SA, Stone JS. A brief history of hair cell regeneration research and speculations on the future. Hear Res. 2013;297:42–51. doi: 10.1016/j.heares.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Q, Chen P, Wang J. Molecular mechanisms and potentials for differentiating inner ear stem cells into sensory hair cells. Dev Biol. 2014;390:93–101. doi: 10.1016/j.ydbio.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 103.Chien WW, Monzack EL, McDougald DS, Cunningham LL. Gene therapy for sensorineural hearing loss. Ear Hear. 2015;36:1–7. doi: 10.1097/AUD.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 104.Avraham KB. Rescue from hearing loss in Usher's syndrome. N Engl J Med. 2013;369:1758–1760. doi: 10.1056/NEJMcibr1311048. [DOI] [PubMed] [Google Scholar]
- 105.Lentz JJ, Jodelka FM, Hinrich AJ, et al. Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat Med. 2013;19:345–350. doi: 10.1038/nm.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Muller U, Barr-Gillespie PG. New treatment options for hearing loss. Nat Rev Drug Discov. 2015 doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- 107.Hu Z, Ulfendahl M. The potential of stem cells for the restoration of auditory function in humans. Regen Med. 2013;8:309–318. doi: 10.2217/rme.13.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rivera T, Sanz L, Camarero G, Varela-Nieto I. Drug delivery to the inner ear: strategies and their therapeutic implications for sensorineural hearing loss. Curr Drug Deliv. 2012;9:231–242. doi: 10.2174/156720112800389098. [DOI] [PubMed] [Google Scholar]
- 109.Stawicki TM, Esterberg R, Hailey DW, Raible DW, Rubel EW. Using the zebrafish lateral line to uncover novel mechanisms of action and prevention in drug-induced hair cell death. Front Cell Neurosci. 2015;9:46. doi: 10.3389/fncel.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiong W, Wagner T, Yan L, Grillet N, Muller U. Using injectoporation to deliver genes to mechanosensory hair cells. Nat Protoc. 2014;9:2438–2449. doi: 10.1038/nprot.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Behar DM, Davidov B, Brownstein Z, Ben-Yosef T, Avraham KB, Shohat M. The many faces of sensorineural hearing loss: one founder and two novel mutations affecting one family of mixed Jewish ancestry. Genet Test Mol Biomarkers. 2014;18:123–126. doi: 10.1089/gtmb.2013.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]