Abstract
Interest in deep space exploration underlines the needs to investigate the effects of exposure to combined sources of space radiation. The lung is a target organ for radiation, and exposure to protons and heavy ions as radiation sources may lead to the development of degenerative disease and cancer. In this study, we evaluated the pro-fibrotic and epigenetic effects of exposure to protons (150 MeV/nucleon, 0.1 Gy) and heavy iron ions (56Fe, 600 MeV/nucleon, 0.5 Gy) alone or in combination (protons on Day 1 and 56Fe on Day 2) in C57BL/6 male mice 4 weeks after irradiation). Exposure to 56Fe, proton or in combination, did not result in histopathological changes in the murine lung. At the same time, combined exposure to protons and 56Fe resulted in pronounced molecular alterations in comparison with either source of radiation alone. Specifically, we observed a substantial increase in the expression of cytokine Il13, loss of expression of DNA methyltransferase Dnmt1, and reactivation of LINE-1, SINE B1 retrotransposons, and major and minor satellites. Given the deleterious potential of the observed effects that may lead to development of chronic lung injury, pulmonary fibrosis, and cancer, future studies devoted to the investigation of the long-term effects of combined exposures to proton and heavy ions are clearly needed.
Keywords: pulmonary fibrosis, repetitive elements, space radiation
1. Introduction
Understanding the health effects of exposure to space radiation is of particular importance because of the mounting interest in deep space exploration. Two main sources of radiation exposure to astronauts are the solar particle events (SPE) and galactic cosmic rays (GCR). High-energy protons constitute ~85% of space radiation with the high-energy charged nuclear particles, such as heavy ions, comprising the rest [1]. While previous studies have been devoted primarily to the effects of single source irradiation, there is an imperative need to utilize exposures to two or more sources to better simulate the occupational exposure that will be encountered by astronauts.
The lung is a target organ for ionizing radiation, and exposure to it may result in pulmonary fibrosis and cancer [2,3]. The latter is considered as one of the largest potential risks for astronauts [4]. This consideration was further substantiated by a recent study reporting that exposure to relatively low doses of protons and heavy ions (56Fe or 28Si) led to the development of chronic lung injury and lung tumors in mice [5].
Epigenetics define somatically heritable changes in the expression of genetic information without alterations in the DNA sequence. DNA methylation is the most studied epigenetic event. It plays a critical role during development, in the maintenance of cellular homeostasis, and is one of the primary regulators of the proper expression of genetic information in a sex-, tissue-, and cell type-dependent manner. DNA methylation also serves as a key mechanism in the silencing of repetitive elements - the most highly methylated sequences in mammalian genomes, and are represented as transposable elements and satellite repeats [6,7]. Long Interspersed Nucleotide Element 1 (LINE-1), Short Interspersed Nucleotide Element B1 (SINE B1) and Endogenous Retroviruses (ERV) are the most abundant transposable elements in mammalian genomes and comprise 23%, 5%, and 10% of the mouse genome, respectively [8]. Satellite repeats are the centromere-associated repetitive sequences that in the mouse are represented as major satellites (6 Mb of 234 bp units located primarily at the pericentromeric regions) and minor satellites (~600 Kb of 120 bp units, located at centromeres) [9].
It has become increasingly evident that exposure to protons or heavy ions, aside from their strong genotoxic potential, can also affect the cellular epigenome, DNA methylation, in particular [10–15]. Alterations in global DNA methylation caused by exposure to radiation primarily originate from the repetitive elements rather than from specific genes [13] and may result in the aberrant expression of the former. Loss of repetitive elements-associated DNA methylation and their reactivation leads to genomic instability and is currently considered not only as a hallmark of cancer, but as one of the driving forces of carcinogenesis [16–18]. Furthermore, the role of epigenetic events in radiation-induced pulmonary fibrosis has been recently recognized [19].
In this study, we aimed to investigate the pro-fibrotic and epigenetic effects of exposure to protons and/or 56Fe ions in the dose range relevant to a space mission in the mouse lung 1 month after irradiation. Previous studies showed that this time-point is sufficient to eliminate the residual damage induced by total body irradiation [12]. On the other hand, this time-point is characteristic of a pneumatic phase in pulmonary fibrosis development and also enables evaluation of the persistence of alterations in DNA methylation and expression of repetitive elements in the radiation target tissue.
2. Materials and Methods
2.1. Animals and radiation exposures
Six-month-old male C57BL/6J mice (n = 64) purchased from the Jackson Laboratory (Bar Harbor, ME) were shipped to Brookhaven National Laboratories (BNL) in Upton, NY. After a one-week acclimation period, the mice were either sham irradiated or received whole-body irradiation [protons 0.1 Gy, 150 MeV/n, 56Fe 600 MeV/n; 0.5 Gy, or combined sequential exposure to 0.1 Gy of protons (Day 1) and 0.5 Gy of 56Fe (Day 2); n = 16 mice per group] at the dose rate of 0.5 Gy/min. The dose of protons was chosen as likely during an SPE. The energy of 150 MeV is commonly used in a therapeutic setting and also represents energy near the maximum abundance of protons expected in most SPEs [20]. The dose of 56Fe was selected as the lowest dose previously shown to cause cell loss of neural precursor cells in the hippocampus [21]. Sequential irradiation with protons before iron reflects the likely exposure of cells in the space environment where daily traversals by protons are accompanied by infrequent traversals by heavy ions. At the selected energy of 600 MeV/n, thorough penetration of the animals with a relatively flat Bragg peak entrance region is expected. Dosimetry was performed by the NASA Space Radiation Laboratory physics dosimetry group at BNL to ensure the quality of exposure. During the entire experiment, sham-irradiated mice were not housed together with irradiated mice. For each exposure, animals were individually placed into clear Lucite cubes (3 in x 1½ in x 1½ in) with breathing holes. The focused beam of high-energy 56Fe particles was generated by the Booster accelerator at BNL and transferred to the experimental beam line at the NSRL facility. Dose calibration was performed with three parallel plate ion chambers that were positioned upstream of the target and a NIST traceable Far West thimble chamber. The values of the thimble chamber were then compared with the upstream ion chambers so that the desired dose could be delivered to the samples based on upstream ion chamber measurements. Sham irradiated mice served as controls and were placed into the same enclosures and for the same amount of time, since previous studies report no effect of sham irradiation on molecular end-points [12]. One week after irradiation, the mice were shipped to Oregon Health and Science University (OHSU) for hippocampus-dependent cognitive testing and biochemical markers of hippocampal function [22]. At BNL and OHSU, the mice were housed under a constant 12 h light:dark cycle. Food (PicoLab Rodent Diet 20, no. 5053; PMI Nutrition International, St. Louis, MO) and water were provided ad libitum.
All animals were killed by cervical dislocation 4 weeks after irradiation; lungs were excised and immediately frozen in liquid nitrogen for further molecular analysis or fixed in 4% PFA for immunohistochemical analysis. All procedures were approved by the Institutional Animal Care and Use Committee at OHSU and BNL.
2.2. Sample coding
The researchers were blinded throughout all phases of the experiments; decoding only occurred after the final analyses were performed.
2.3. Immunohistochemistry
Lungs were inflated in situ with neutral buffered formalin using a 25 gauge needle via the trachea, and the trachea was tied off with suture to retain the formalin. The perfused lungs were then placed into a container containing formalin at a 1:10 ratio of tissue:formalin by volume. Tissues were fixed for 24 hours and sliced for immunohistochemical analysis. Lung samples were stained with hematoxylin/eosin (H&E) and Masson’s Trichrome at the University of Arkansas for Medical Sciences Pathology Core Facility.
2.4. Nucleic Acids Extraction
RNA and DNA were extracted simultaneously from flash-frozen tissue using the AllPrep DNA/RNA extraction kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. DNA concentrations and integrity were analyzed by the Nanodrop 2000 (Thermo Scientific, Waltham, MA) and 1% agarose gel.
2.5. Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from frozen lung tissue using the AllPrep DNA/RNA extraction kit (QIAGEN) according to the manufacturer’s protocol. RNA concentrations and integrity were analyzed by the Nanodrop 2000 (ThermoScientific). Only RNA samples with the 260/280 ratios between 1.95 and 2.05 and the 260/230 ratios above 1.5 were considered for further molecular analyses. cDNA was synthesized using random primers and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol (Life Technologies). The levels of gene transcripts were determined by quantitative Real Time PCR (qRT-PCR) using TaqMan Gene Expression Assays (Life Technologies). Assays for determination of mRNA abundance are provided in Supplementary Table 1. Assays for determination of expression of repetitive elements are provided in Supplementary Table 2. Each plate contained one experimental gene and a housekeeping gene. The cycle threshold (Ct) for each sample was determined from the linear region of the amplification plot. The ΔCt values for all genes were determined relative to the control gene Gapdh (Mm 99999915_g1, Life Technologies). The ΔΔCt were calculated using each exposed group means relative to control group means. The fold change data were calculated from the ΔΔCt values. All qRT-PCR reactions were conducted in triplicate and repeated twice.
2.6. Methylation analysis of repetitive elements
The methylation status of repetitive elements was analyzed by methylation sensitive quantitative polymerase chain reaction (MS-qPCR) as previously described. Briefly, 1 μg of genomic DNA was digested with 1 U of SmaI enzyme in 1X CutSmart buffer at 25°C for 2 h. This was followed by a 16 h digestion at 37°C in the presence of 1 U of the enzymes HpaII, HhaI, and AciI in 1X CutSmart buffer. The digestion was finalized by adding 0.5 U of BstUI enzyme in 1X CutSmart buffer for 4 h at 60°C. All enzymes were purchased from New England Biolabs (Ipswich, MA, USA). Digested DNA was then analyzed by qRT-PCR on a ViiA 7 Real-Time PCR System (Applied Biosystems, Forrest City, CA, USA). DNA samples not digested with the restriction enzyme mix served as positive control, while samples 1) lacking the specific primers for DNA amplification and/or DNA template and 2) RAW264.7 murine macrophage-derived DNA pre-treated with 5-azacytidine, a potent demethylating agent, served as negative controls. The threshold cycle (Ct) was defined as the fractional cycle number that passes the fixed threshold. The Ct values were converted into the absolute amount of input DNA using the absolute standard curve method and further normalized towards rDNA readings. Assays for determination of repetitive elements methylation are provided in Supplementary Table 2.
2.7. LINE-1 copy numbers analysis
LINE-1 copy number was assessed as previously described [12]. Briefly, LINE-1 ORF1 was amplified by real-time quantitative PCR from 10 ng of gDNA. Relative abundance of the target in gDNA was normalized to 5S ribosomal DNA using the ΔΔCt method. The FAM/ZEN-conjugated primers with the probe sequence are shown in Supplementary Table 2 (Integrated DNA Technologies, Coralville, IA) and were used at a final concentration of 5 μM. Amplification was performed for 40 cycles using conditions for the 2X Taqman Universal Master Mix as recommended by the manufacturer (Life Technologies). The total reaction volume was 20 μL.
2.8. Statistical analysis
All data are presented as mean ± standard error of mean(s). All assessed parameters were measured within the same batch of animals. Statistically significant differences for each treatment compared to the control (at α=95%) were assessed using one-way ANOVAs followed by Dunnett’s or Tukey’s posthoc tests. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software Inc. LaJolla, CA).
3. Results
3.1. Histopathological evaluation
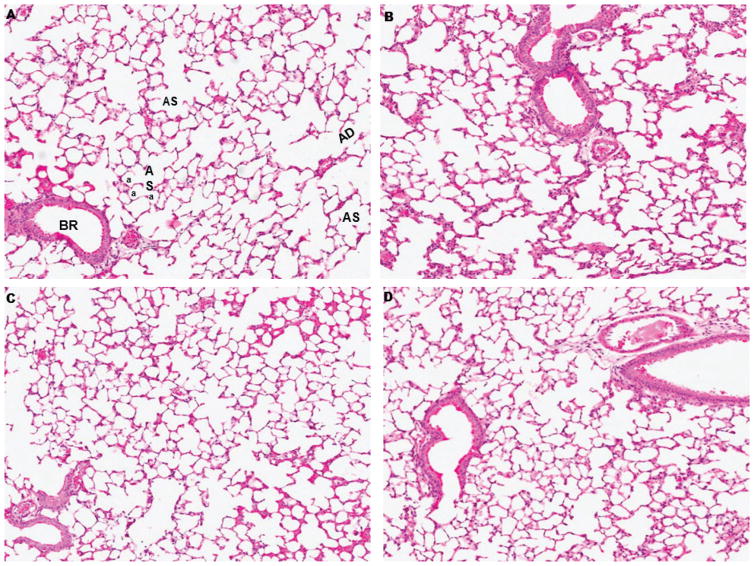
First, histopathological evaluation under the light microscope was performed on the control and exposed to protons, 56Fe, and protons+56Fe lungs stained with H&E. Figure 1 illustrates normal lung parenchyma with various structural elements, including bronchioles, alveolar ducts, alveolar sacks, and thin walled alveoli that were not disturbed by irradiation.
Figure 1.
Representative histological sections of mouse lung 1 month after exposure to space radiation. (A) Sham irradiated mice illustrating normal lung parenchyma: bronchiole (BR), alveolar duct (AD), alveolar sac (AS) and alveoli (a). (B) Protons irradiated, (C) 56Fe irradiated, (D) protons+56Fe irradiated. A-D H&E stained. Original magnification x275.
3.2. Assessment of pulmonary fibrosis by Mason’s trichrome staining
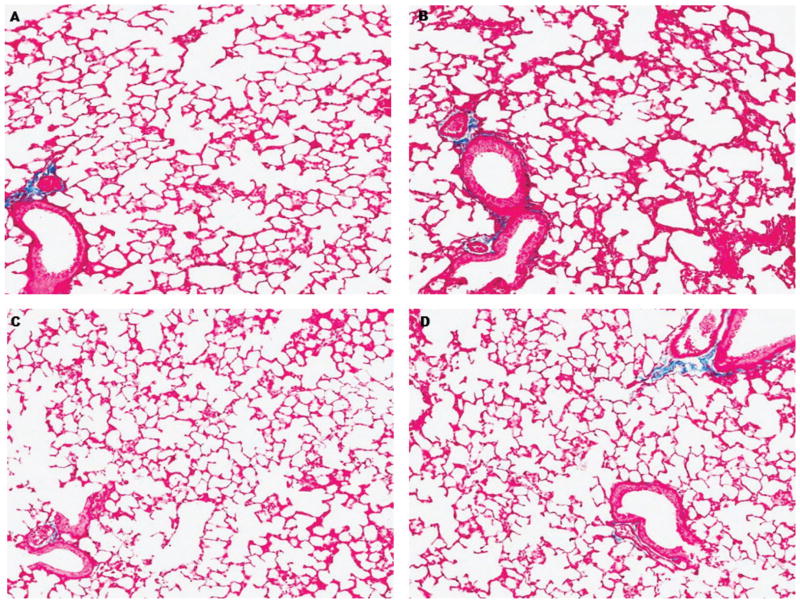
Taking into account pro-fibrotic changes observed in mice after a 13-day space mission [23] and 5 months after exposure to low mean absorbed doses of 56Fe [13], next we addressed the collagen deposition in the lungs of control and exposed mice. As demonstrated in Figure 2, no evidence of Masson’s trichrome stain in the alveolar walls of the lung was observed, suggesting absence of pulmonary fibrosis-associated events characteristic of the pneumonic phase of the disease.
Figure 2.
Representative histological sections of mouse lung 1 month after exposure to space radiation and stained with Masson’s Trichrome for collagen (blue). (A) Sham irradiated mice, (B) protons irradiated, (C) 56Fe irradiated, (D) protons+56Fe irradiated. Original magnification x275.
3.3. Analysis of molecular pro-fibrotic biomarkers
Molecular biomarkers may have higher sensitivity to pro-fibrotic changes than histopathological, especially at early phases. Therefore, next we analyzed the expression of a panel of genes that were previously reported to be deregulated in the pneumatic phase of radiation-induced pulmonary fibrosis.
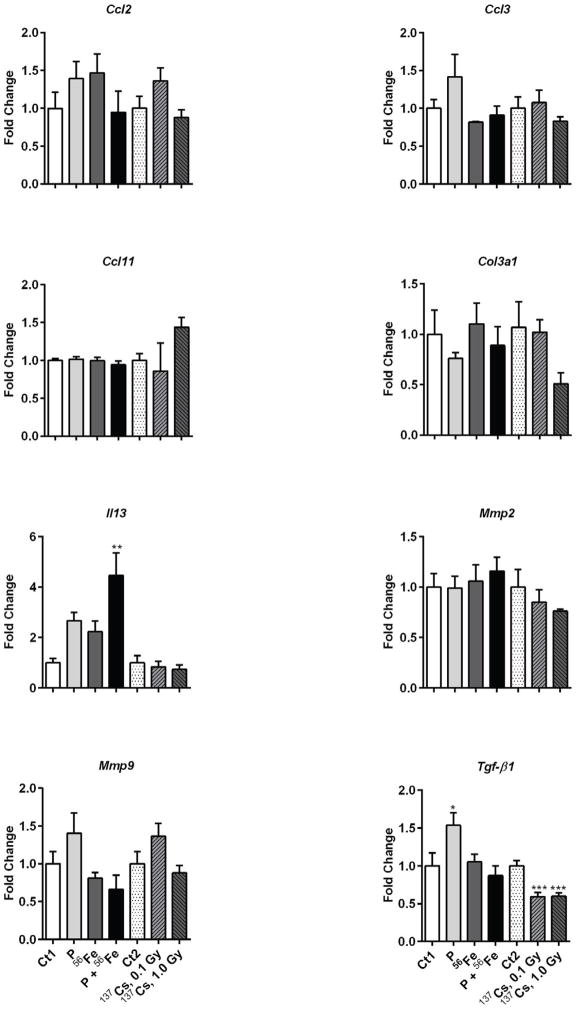
We identified a significant increase in the expression of the Tgfβ1 gene after exposure to protons (1.5-fold, p<0.05) (Figure 3). The most pronounced changes were identified in the expression of Il13, where a 4.1-fold increase was observed after combined exposures to protons and 56Fe (p<0.01) and non-significant 2.2- and 2.3-fold increases after exposure to 56Fe and protons, respectively. An insignificant decrease in the mRNA levels of the matrix metalloproteinase, Mmp9, was identified after exposure to 56Fe or protons+56Fe.
Figure 3.
Effects of space and terrestrial radiation on the expression of pro-fibrotic genes. The differential gene expression was determined by quantitative RT-PCR. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test. Ct – sham irradiated mice, P – mice exposed to protons, 56Fe – mice exposed to heavy iron ions, P+56Fe – mice exposed to protons (Day 1) and heavy iron ions (Day 2).
3.4. Analysis of DNA methyltransferases expression
Ionizing radiation is a potent epigenotoxic stressor that targets DNA methyltransferases, genes involved in the maintenance of DNA methylation. Accumulating evidence also indicates involvement of Dnmt1, the major maintenance DNA methyltransferase, in pulmonary fibrosis [19,24]. Expression of Dnmt1 was significantly decreased in the lungs of mice exposed to combined protons+56Fe irradiation (−1.4-fold, p<0.05) (Figure 4). No changes were detected in the expression of Dnmt3a and Dnmt3b de novo methyltransferases.
Figure 4.
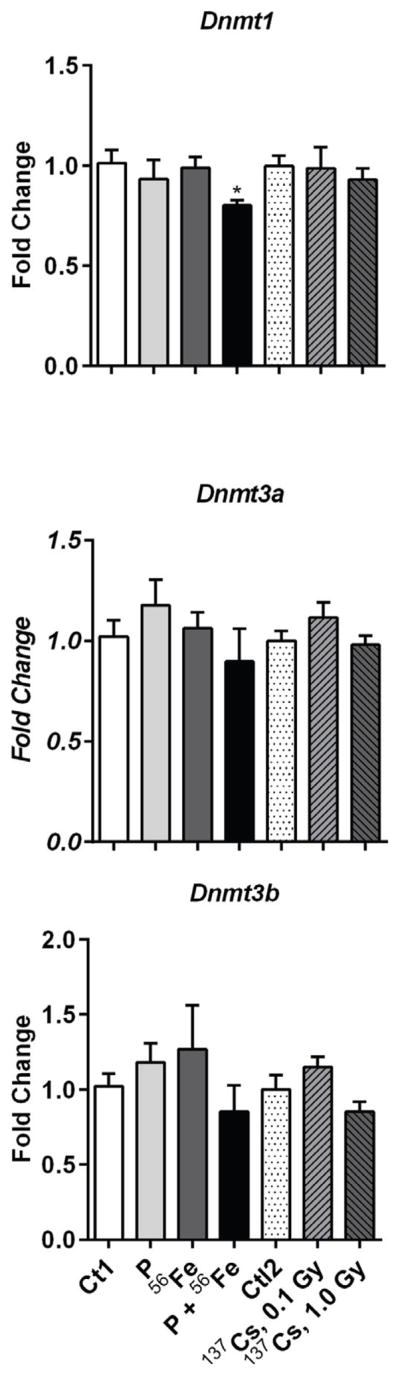
Effects of space and terrestrial radiation on DNA methylation machinery. The differential gene expression was determined by quantitative RT-PCR. Data are presented as mean ± SE. *p≤0.05, ANOVA with Dunnett’s test. Ct – sham irradiated mice, P – mice exposed to protons, 56Fe – mice exposed to heavy iron ions, P+56Fe – mice exposed to protons (Day 1) and heavy iron ions (Day 2).
3.5. Analysis of repetitive elements-associated DNA methylation
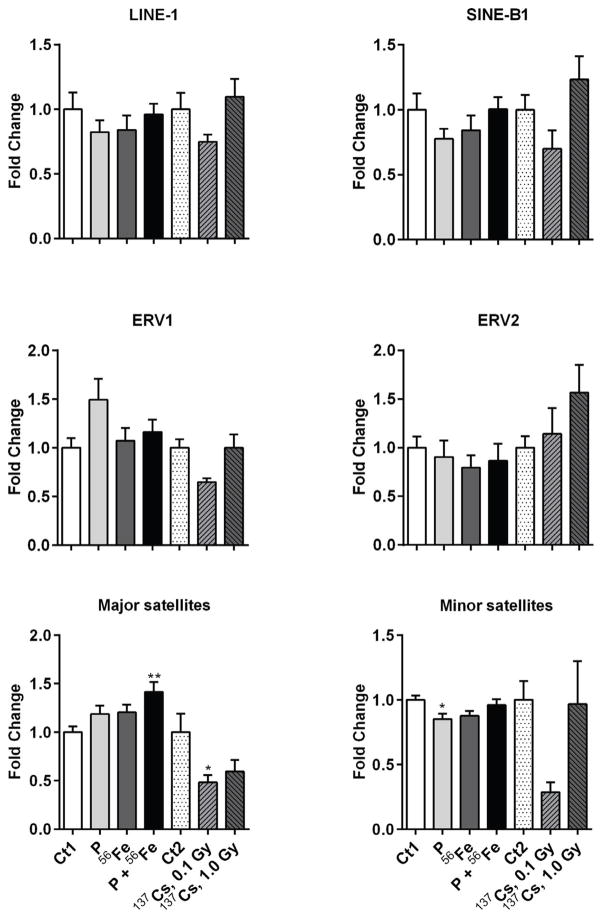
Loss in expression of Dnmt1 may lead to the loss of global DNA methylation. Repetitive elements comprise up to two-thirds of mammalian genomes and are considered as surrogate biomarkers of alterations in DNA methylation. Previous studies confirmed that exposure to space radiation leads to alterations in the methylation of repetitive elements rather than alterations in gene-specific methylation [13]. Therefore, we analyzed the methylation status in a panel of the most abundant repetitive elements in mammalian genomes: retrotransposons LINE-1, SINE B1, ERV1, ERV2, and major and minor satellites. We did not identify significant changes in the methylation status in any of the evaluated repetitive elements in response to space radiation, except for the weak hypermethylation of major satellites after exposure to protons+56Fe (1.35-fold, p<0.01) and weak hypomethylation of minor satellites in response to protons irradiation (−1.2-fold, p<0.05) (Figure 5).
Figure 5.
Effects of space and terrestrial radiation on repetitive elements-associated DNA methylation. The methylation status of DNA repetitive elements was measured by methylation-sensitive qPCR assay. Data are presented as mean ± SE. *p≤ 0.05, **p≤0.01, ANOVA with Dunnett’s test. Ct – sham irradiated mice, P – mice exposed to protons, 56Fe – mice exposed to heavy iron ions, P+56Fe – mice exposed to protons (Day 1) and heavy iron ions (Day 2).
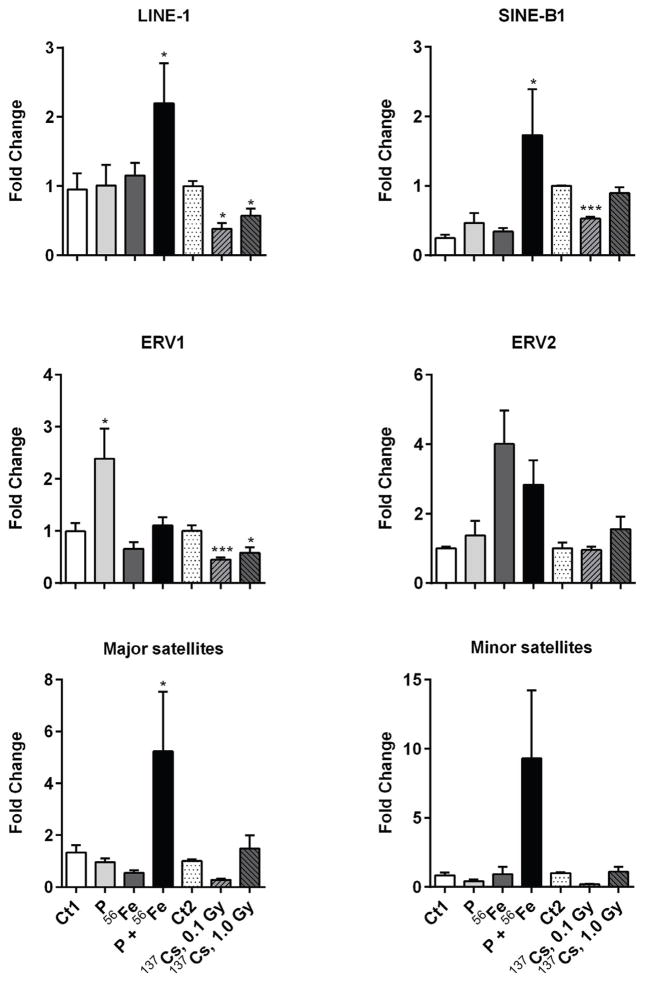
3.6. Analysis of expression of repetitive elements
Exposure to environmental stressors, including radiation, may lead to reactivation of repetitive elements [18]. In this study, we observed a substantial increase in the mRNA levels of LINE-1 (2.2-fold, p<0.05), and SINE B1 (1.7-fold, p<0.05), associated with combined exposure to protons and 56Fe (Figure 6). The most pronounced effects were observed in the satellite DNA, where exposure to protons+56Fe led to a 5-fold increase in the expression of major satellites (p<0.05) and an 8.9-fold increase in the expression of minor satellites (p=0.065). Exposure to protons alone led to a 2.2-fold increase in ERV1 expression (p<0.05), while exposure to 56Fe alone or to protons+56Fe was characterized by reactivation of ERV2, although insignificant (Figure 6).
Figure 6.
Effects of space and terrestrial radiation on the expression of repetitive elements. The differential expression of repetitive elements was determined by quantitative RT-PCR. Data are presented as mean ± SE. *p≤0.05, **p≤0.01, ANOVA with Dunnett’s test. Ct – sham irradiated mice, P – mice exposed to protons, 56Fe – mice exposed to heavy iron ions, P+56Fe – mice exposed to protons (Day 1) and heavy iron ions (Day 2).
3.7. Analysis of LINE-1 repetitive element copy number
Reactivation of repetitive elements capable of transposition, such as LINE-1 retrotransposon, may lead to their aberrant activity and insertional mutagenesis in a “copy-paste” manner [18]. Therefore, as a final step in this study, we addressed the copy number of LINE-1 in the lungs of control and exposed animals. We did not identify any significant changes in copy number in any of the experimental groups (Supplementary Figure 1).
4. Discussion
In this study, we address the pro-fibrotic and epigenetic effects in the murine lung 1 month after space trip-relevant exposure to protons, 56Fe, or protons+56Fe irradiation. This is the first study, to our knowledge, reporting the effects of combined irradiation in the lung tissue using the in vivo model.
In our study, exposure to protons, 56Fe or a combination of protons and 56Fe did not lead to detectable histopathological changes, including an absence of pro-fibrotic alterations, in the lung tissue. The recent study by Christofidou-Solomidou et al. also reported an absence of fibrotic changes in response to 56Fe irradiation, but reported significant structural changes, such as air space enlargement [5]. The discrepancies, observed between these studies can be explained by differences in the age at which mice were irradiated (2.5 months vs 6 months in our study), differences at time-points at which mice were evaluated (23.5 months vs 1 month in our study) and different mouse strains utilized (C3H/HeNCrl vs C57BL/6 in our study).
Transforming growth factor-beta 1 Tgfβ1 is a cytokine and one of the major drivers of radiation-induced fibrosis [2]. In our study, we found increased expression of Tgfβ1 after exposure to protons. This finding is in good agreement with the earlier study that reported increased Tgfβ1 mRNA levels in the lungs of mice exposed to simulated SPE [23]. At the same time, 56Fe or combined protons+56Fe irradiation did not affect Tgfβ1. We have previously shown lack of alteration in Tgfβ1 in the lungs of mice 5 months after exposure to 56Fe (600 MeV/n; dose range 0.1–0.4 Gy) [13]. Interestingly, expression of Tgfβ1 was increased in the lungs of mice after a 13-day mission on the Space Shuttle Endeavour [23], in an environment characterized primarily by the presence of protons, but not heavy ions. Altogether, these findings suggest that Tgfβ1 is a target for protons, but possibly not for heavy ions.
Il-13 is another pro-inflammatory cytokine that plays a central role in the pathogenesis of pulmonary fibrosis [25] and asthma [26,27]. It regulates eosinophilic inflammation, mucus secretion, and airway hyperresponsiveness and is considered as a key regulator of the extracellular matrix. Substantial up-regulation of Il-13 in mice exposed to protons+56Fe suggests gross immunological repercussions in the lung tissue, as overexpression of Il13 alone is sufficient to induce non-allergic asthma [27]. Furthermore, it has been shown that Il13 can induce fibrosis independently of Tgfβ1 [28]. We identified a significant and substantial up-regulation of Il13 in the lung of mice exposed to protons and 56Fe (4.1-fold increase, p<0.001), although non-significant increases were also observed after single exposures. Therefore, long-term studies investigating the pro-fibrotic potential of combined exposure to protons and 56Fe are warranted.
Combined exposure to protons and 56Fe has also led to epigenetic alterations. DNA methyltransferase Dnmt1 is critical for the maintenance of DNA methylation patterns during replication and for silencing of repetitive elements [7]. Although decreased expression of Dnmt1 did not lead to marked changes in the methylation of repetitive elements, the expression of the latter was substantially increased, specifically after exposure to protons+56Fe. LINE-1, SINE B1 and ERV belong to retrotransposons, mobile DNA sequences, capable of moving and invading genomes via a “copy and paste” mechanism. Both low- and high-LET radiation may disrupt the epigenetic control over these sequences exhibited as loss of DNA methyltransferases function and DNA hypomethylation and lead to subsequent reactivation of transposable elements [12,29]. The observed increases in LINE-1, SINE B1, and ERV transcripts in the lung may result in retrotransposition events, associated with them insertional mutagenesis, genome amplification, and genomic instability and may further serve as a causative factor in life-threatening disorders, including cancer. Indeed, Iskow and colleagues report new somatic L1 insertions detected at high frequencies in human lung cancer genomes [30]. In this study, we did not identify increased copy numbers of the most abundant transposable element LINE-1. This may be explained by the relatively short time after exposure and, possibly, by low sensitivity of the assay given the high number (over 500,000 copies) of LINE-1 elements in the genome.
Another potentially dangerous outcome of combined exposure to protons and 56Fe is the accumulation of satellite DNA mRNA transcripts. The major and minor satellites are the centromeric- and pericentromeric-specific sequences that act as centromere-building elements and serve as major components of heterochromatin [9]. Accumulation of mRNA transcripts associated with satellite DNA is reported in both cancer and non-cancerous disease. For instance, overexpression of pericentromeric satellite repeats was detected in pancreatic ductal adenocarcinomas in humans and in a mouse model [31], in human heart failure [32], and in response to exposure to environmental stressors [33,34]. As centromeric and pericentromeric RNAs serve as key players for heterochromatin formation, the observed increase in major and minor satellites transcripts in response to combined exposure to protons and 56Fe may significantly affect normal chromatin assembly and result in chromosomal aberrations.
In conclusion, in our experimental conditions, combined exposure to protons and 56Fe did not lead to the accumulation of collagen and increased expression of Tgfβ1 in the mouse lung, but resulted in substantially increased levels of Il13, decreased expression of Dnmt1 and reactivation of retrotransposons LINE-1 and SINE B1 and dramatic increases in mRNA levels of major and minor satellites. Given the deleterious potential of the observed effects that may lead to development of chronic lung injury, pulmonary fibrosis, and cancer, future studies devoted to the investigation of the long-term effects of combined exposures to proton and heavy ions are clearly needed.
Supplementary Material
Supplementary Figure 1. Effects of space radiation on LINE-1 repetitive elements copy number. LINE-1 Open Reading Frame 1 (ORF1) copy number was assessed by real-time quantitative PCR. Data are presented as mean ± SE.
Supplementary Table 1. Gene-specific assays used for qRT-PCR amplification.
Supplementary Table 2. Assays used in the analysis of methylation and expression of repetitive elements.
Acknowledgments
We are thankful to Dr. Kristy Kutanzi for critical reading and Rebecca Helm for proofreading and editing the manuscript. The study was supported in part by National Institute of Health Center of Biological Research Excellence, grant# 1P20GM109005; National Aeronautics and Space Administration, grant# NNX10AD59G (GAN); Arkansas Space Grant Consortium through National Aeronautics and Space Administration, grant# NNX13AB29A (RP); NNJ12ZSA001N (JR), NIH/UAMS Clinical and Translational Science Award UL1TR000039 and KL2TR000063, the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000, and the National Space Biomedical Research Institute through the National Aeronautics and Space Administration NCC 9-58, grant# RE03701.
Footnotes
Declaration of interests
The authors declare no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Badhwar GD, Cucinotta FA. Depth dependence of absorbed dose, dose equivalent and linear energy transfer spectra of galactic and trapped particles in polyethylene and comparison with calculations of models. Radiat Res. 1998;149:209–218. [PubMed] [Google Scholar]
- 2.Yarnold J, Brotons MC. Pathogenetic mechanisms in radiation fibrosis. Radiother Oncol. 2010;97:149–161. doi: 10.1016/j.radonc.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Maddams J, Parkin DM, Darby SC. The cancer burden in the United Kingdom in 2007 due to radiotherapy. Int J Cancer. 2011;129:2885–2893. doi: 10.1002/ijc.26240. [DOI] [PubMed] [Google Scholar]
- 4.Shay JW, Cucinotta FA, Sulzman FM, Coleman CN, Minna JD. From mice and men to earth and space: joint NASA-NCI workshop on lung cancer risk resulting from space and terrestrial radiation. Cancer Res. 2011;71:6926–6929. doi: 10.1158/0008-5472.CAN-11-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christofidou-Solomidou M, Pietrofesa RA, Arguiri E, Schweitzer KS, Berdyshev EV, McCarthy M, et al. Space radiation-associated lung injury in a murine model. Am J Physiol Lung Cell Mol Physiol. 2015;308:L416–28. doi: 10.1152/ajplung.00260.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross JP, Rand KN, Molloy PL. Hypomethylation of repeated DNA sequences in cancer. Epigenomics. 2010;2:245–269. doi: 10.2217/epi.10.2. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 8.Miousse IR, Chalbot MC, Lumen A, Ferguson A, Kavouras I, Koturbash I. Response of transposable elements to environmental stressors. Mutat Res Rev Mutat Res. 2015 doi: 10.1016/j.mrrev.2015.05.003. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ugarkovic D. Functional elements residing within satellite DNAs. EMBO Rep. 2005;6:1035–1039. doi: 10.1038/sj.embor.7400558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goetz W, Morgan MN, Baulch JE. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat Res. 2011;175:575–587. doi: 10.1667/RR2390.1. [DOI] [PubMed] [Google Scholar]
- 11.Lima F, Ding D, Goetz W, Yang AJ, Baulch JE. High LET (56)Fe ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environ Mol Mutagen. 2014;55:266–277. doi: 10.1002/em.21832. [DOI] [PubMed] [Google Scholar]
- 12.Miousse IR, Shao L, Chang J, Feng W, Wang Y, Allen AR, et al. Exposure to low-dose 56Fe-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res. 2014;182:92–101. doi: 10.1667/RR13580.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nzabarushimana E, Miousse IR, Shao L, Chang J, Allen AR, Turner J, et al. Long-term epigenetic effects of exposure to low doses of 56Fe in the mouse lung. J Radiat Res. 2014;55:823–828. doi: 10.1093/jrr/rru010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raber J, Rosi S, Zuloaga D, Jopsom T, Marzulla T, Stewart B, et al. The relation between cognitive injury, network stability, and epigenetic change following exposure to space radiation. NASA Human Research Program Investigators’ Workshop; 2014; Galveston Texas, U.S.A. [Google Scholar]
- 15.Jangiam W, Tungjai M, Rithidech KN. Induction of chronic oxidative stress, chronic inflammation and aberrant patterns of DNA methylation in the liver of titanium-exposed CBA/CaJ mice. Int J Radiat Biol. 2015;91:389–398. doi: 10.3109/09553002.2015.1001882. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 17.Tommasi S, Zheng A, Yoon JI, Besaratinia A. Epigenetic targeting of the Nanog pathway and signaling networks during chemical carcinogenesis. Carcinogenesis. 2014;35:1726–1736. doi: 10.1093/carcin/bgu026. [DOI] [PubMed] [Google Scholar]
- 18.Miousse I, Koturbash I. The Fine LINE: Drawing the Cancer Landscape. BioMed Research International. 2015 doi: 10.1155/2015/131547. e-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigel C, Schmezer P, Plass C, Popanda O. Epigenetics in radiation-induced fibrosis. Oncogene. 2015;34:2145–2155. doi: 10.1038/onc.2014.145. [DOI] [PubMed] [Google Scholar]
- 20.Townsend LW, Badhwar GD, Braby LA, Blakely EA, Cucinotta FA, Curtis SB, et al. Information Needed to Make Radiation Protection Recommendations for Space Missions Beyond Low-Earth Orbit. National Council on Radiation Protection & Measurements Report. 2006;153 [Google Scholar]
- 21.Rola R, Fishman K, Baure J, Rosi S, Lamborn KR, Obenaus A, et al. Hippocampal neurogenesis and neuroinflammation after cranial irradiation with 56Fe particles. Radiat Res. 2008;169:626–632. doi: 10.1667/RR1263.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raber J, Allen AR, Sharma S, Allen B, Rosi S, Olsen RHJ, et al. Effects of combined proton and 56Fe irradiation on hippocampal function. NASA Human Research Program Investigators’ Workshop; 2014; Galveston, Texas, U.S.A. [Google Scholar]
- 23.Tian J, Pecaut MJ, Slater JM, Gridley DS. Spaceflight modulates expression of extracellular matrix, adhesion, and profibrotic molecules in mouse lung. J Appl Physiol (1985) 2010;108:162–171. doi: 10.1152/japplphysiol.00730.2009. [DOI] [PubMed] [Google Scholar]
- 24.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson MS, Wynn TA. Pulmonary fibrosis: pathogenesis, etiology and regulation. Mucosal Immunol. 2009;2:103–121. doi: 10.1038/mi.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 27.Chapmana AM, Malkin DJ, Camacho J, Schiestl RH. IL-13 overexpression in mouse lungs triggers systemic genotoxicity in peripheral blood. Mutat Res. 2014;769:100–107. doi: 10.1016/j.mrfmmm.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaviratne M, Hesse M, Leusink M, Cheever AW, Davies SJ, McKerrow JH, et al. IL-13 activates a mechanism of tissue fibrosis that is completely TGF-beta independent. J Immunol. 2004;173:4020–4029. doi: 10.4049/jimmunol.173.6.4020. [DOI] [PubMed] [Google Scholar]
- 29.Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, et al. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis. 2007;28:1831–1838. doi: 10.1093/carcin/bgm053. [DOI] [PubMed] [Google Scholar]
- 30.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haider S, Cordeddu L, Robinson E, Movassagh M, Siggens L, Vujic A, et al. The landscape of DNA repeat elements in human heart failure. Genome Biol. 2012;13 doi: 10.1186/gb-2012-13-10-r90. R90-2012-13-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Tryndyak V, et al. Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene. Environ Health Perspect. 2011;119:635–640. doi: 10.1289/ehp.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miousse IR, Chalbot MC, Aykin-Burns N, Wang X, Basnakian A, Kavouras IG, et al. Epigenetic alterations induced by ambient particulate matter in mouse macrophages. Environ Mol Mutagen. 2014;55:428–435. doi: 10.1002/em.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Effects of space radiation on LINE-1 repetitive elements copy number. LINE-1 Open Reading Frame 1 (ORF1) copy number was assessed by real-time quantitative PCR. Data are presented as mean ± SE.
Supplementary Table 1. Gene-specific assays used for qRT-PCR amplification.
Supplementary Table 2. Assays used in the analysis of methylation and expression of repetitive elements.