Abstract
The thalamus and habenula, two important nodes of the forebrain circuitry, are derived from a single developmental compartment, called prosomere 2, in the diencephalon. Habenular and thalamic neurons display distinct molecular identity, neurochemistry, and connectivity. Furthermore, their progenitors exhibit distinctive neurogenic patterns with a marked delay in the onset of neurogenesis in the thalamus. However, the progenitors in prosomere 2 express many common developmental regulators and the mechanism underlying the specification and differentiation of these two populations of neurons remains unknown. Gbx2, coding for a homeodomain transcription factor, is initially expressed in thalamic neuronal precursors that have just exited the cell cycle, and its expression is maintained in many mature thalamic neurons in adults. Deletion of Gbx2 severely disrupts histogenesis of the thalamus and abolishes thalamocortical projections in mice. Here, by using genome-wide transcriptional profiling, we show that Gbx2 promotes thalamic but inhibits habenular molecular characters. Remarkably, although Gbx2 is expressed in postmitotic neuronal precursors, deletion of Gbx2 changes gene expression and cell proliferation in dividing progenitors in the developing thalamus. These defects are partially rescued by the mosaic presence of wild-type cells, demonstrating a cell non-autonomous role of Gbx2 in regulating the development of thalamic progenitors. Our results suggest that Gbx2 is essential for the acquisition of the thalamic neuronal identity by repressing habenular identity through a feedback signaling from postmitotic neurons to progenitors.
Keywords: Transcription, cell fate, differentiation, neurogenesis, mouse
INTRODUCTION
The thalamus and habenula are two important nodes of the forebrain circuitry. The habenula, which is a paired midline structure that straddles the posterior and dorsal surface of the thalamus, is present in virtually all vertebrates. The habenula receives input from the forebrain limbic system and the basal ganglia through the stria medullaris, and projects to monoaminergic nuclei in the midbrain and hindbrain via the fasciculus retroflexus, also called the habenulopeduncular tract (Lecourtier and Kelly, 2007; Hikosaka et al., 2008). Dysfunction of the habenula has been implicated in psychiatric disorders, such as depression, schizophrenia, and drug-induced psychosis (Hikosaka, 2010). By contrast, the thalamus receives sensory and motor information from the periphery and relays them to the cortex through thalamocortical projections (Jones, 2007). The thalamus is thus considered as the gateway to the cortex and essential for establishing self-consciousness and awareness to one's environment (Sherman and Guillery, 2006). Despite their marked differences in cytoarchitecture and connectivity, thalamic and habenular neurons arise from seemingly homogeneous progenitors. The molecular underpinnings of progenitor differentiation into thalamic and habenular neurons remain elusive.
Based on the pattern of gene expression and axonal tracts, the diencephalon is divided into three developmental compartments called prosomeres (Rubenstein et al., 1994; Puelles and Rubenstein, 2003). Among the three prosomeres (P)1-3 in the diencephalon, P1 gives rise to the pretectum, P2 the thalamus and epithalamus, and P3 the prethalamus. The epithalamus subsequently produces the habenula, pineal gland and choroid plexus. The thalamic progenitor domain can be further divided into rostral and caudal areas: the former gives rise to local GABAergic neuron while the latter to glutamatergic neurons projecting to the cortex (Vue et al., 2007; Jeong et al., 2011). Neurons derived from the caudal thalamus constitute the nuclei complex that is traditionally viewed as the thalamus (Jones, 2007). Significant progress has been made in our understanding of the development of diencephalic prosomeres (Chatterjee and Li, 2012). Extracellular signals, such as Shh (Kiecker and Lumsden, 2004; Scholpp et al., 2006; Jeong et al., 2011) (Vue et al., 2009), FGF(Kataoka and Shimogori, 2008; Martinez-Ferre and Martinez, 2009), and WNT (Zhou et al., 2004; Bluske et al., 2012), which are produced from localized signaling centers, act in concert to pattern the diencephalon. Transcription factors, such as Pax6, Otx2, Fezf2, and Iroquois proteins, also participate in controlling the cell fate of progenitors in different diencephalic prosomeres (Hirata et al., 2006; Puelles et al., 2006; Scholpp et al., 2007; Robertshaw et al., 2013; Chatterjee et al., 2014). We have recently reported the characterization of gene expression in the epithalamus, particularly the habenula in mice and zebrafish (Chatterjee et al., 2014). Interestingly, although postmitotic neuronal precursors for the prospective thalamus and habenula can be readily distinguished by distinct expression patterns of marker genes, P2 progenitors appear remarkably homogenous (Chatterjee et al., 2014). How the P2 domain is partitioned into the future thalamus and epithalamus remains to be determined.
Although it is traditionally believed that cell fate specification occurs in proliferating progenitors, emerging evidence indicate that additional mechanisms determine and/or consolidate cell identity after neural precursors exit the cell cycle (Belliveau and Cepko, 1999; Seuntjens et al., 2009; Hobert, 2011). Experiments in C. elegance have identified a class of transcription factors that play an important role in assigning and/or maintaining the identity of postmitotic neurons and are called “terminal selector genes” (Hobert, 2008; Flames and Hobert, 2009). Furthermore, studies in the neocortex and retina have shown that early-born neurons signal dividing progenitors to ensure the successive generation of cell types (Belliveau and Cepko, 1999; Seuntjens et al., 2009). Gbx2, which encodes a homeodomain transcription factor, is expressed in thalamic neuronal precursors that have just exited the cell cycle; its expression is maintained in mature neurons of thalamic nuclei in adult mice and monkeys (Jones and Rubenstein, 2004; Chen et al., 2009). We have previously demonstrated that the Gbx2 lineage contributes exclusively to thalamic nuclei that project to the cortex (Chen et al., 2009; Li et al., 2012). Deletion of Gbx2 severely disrupts histogenesis of the thalamus and abolishes thalamocortical projections (Miyashita-Lin et al., 1999; Hevner et al., 2002; Chen et al., 2009; Chatterjee et al., 2012; Li et al., 2012). Interestingly, in the absence of Gbx2, thalamic neurons abnormally extend their axons and follow the fasciculus retroflexus tract, reaching the ventral midbrain and hindbrain (Chatterjee et al., 2012). Furthermore, neurons originating from the thalamus aberrantly contribute to the habenula (Chen et al., 2009). These observations raise an intriguing possibility that Gbx2 determines and/or maintains the cell identity of thalamic neurons.
In this study, we explored the potential role of Gbx2 in cell fate specification and/or consolidation in the developing thalamus by identifying alterations in the genome-wide transcriptional profile in the thalamus caused by Gbx2 deletion. We show that Gbx2 is essential for maintaining the thalamic fate and repressing the habenular development. Using genetic mosaics, we demonstrate that Gbx2 exerts its function in part by modulating the developmental program in proliferating progenitors of the thalamus through a feedback mechanism from postmitotic neural precursors.
MATERIALS AND METHODS
Mouse and tissue preparation
All animal procedures described herein were approved by the Animal Care Committee at the University of Connecticut School of Medicine. Mice were housed in a facility with a 12-hour light/dark cycle and have free access to food and water. All mouse strains were maintained in a CD1 genetic background (Charles River Lab, Wilmington, MA). Noon of the day on which a vaginal plug was detected was designated as E0.5 in staging of embryos. The generation of knock-in Gbx2creER allele, which expresses both creER and enhanced green fluorescence protein (EGFP) from the Gbx2 locus, has been described previously (Chen et al., 2009). Embryos carrying the Gbx2creER allele were identified by GFP fluorescence in the spinal cord, cerebellum, and thalamus. The Gbx2 null (Gbx2−) and conditional floxed (Gbx2F) alleles were determined by PCR analysis of tail DNA as described previously (Wassarman et al., 1997; Chen et al., 2009).
Taking advantage of the mosaic nature of creER-mediated recombination induced by tamoxifen, we deleted Gbx2 in a mosaic manner in the thalamus of Gbx2creER/F embryos as described previously (Chen et al., 2009; Chatterjee et al., 2012). As Gbx2 acts cell autonomously to regulate thalamocortical axon guidance, all mosaic deletion embryos were verified by abnormal thalamic axon trajectories examined by GFP fluorescence that was expressed from the Gbx2cerER locus as described previously (Chatterjee et al., 2012). All reported phenotypes were reproduced in at least three mosaic mutant embryos.
Histochemistry, immunofluorescence, and in situ hybridization
Standard protocols were used for immunohistochemistry (ICH) and in situ hybridization (ISH) as described previously (Chatterjee et al., 2012). For bromodeoxyuridine (BrdU) staining, BrdU was dissolved at 0.5 mg/ml in PBS, and injected intraperitoneally into pregnant female mice at 10 μg/g body weights. Embryos were dissected after 7 or 24 hours. Detailed protocols are available on the Li Laboratory website (http://lilab.uchc.edu/protocols/index.html). Primary antibodies used in the study were as follows: rat anti-Ki67 (DAKO); mouse anti-neurofilaments (DSHB hybridoma bank); rabbit anti-pH3 (Merck Millipore); mouse anti-Pou4f1 (Santa Cruz); goat anti-Ox2 (R&D Systems); rabbit anti-GFP (Invitrogen); mouse anti-BrdU (BD Biosciences); rabbit anti-Dbx1 (Vue et al., 2007); rabbit anti-Islr2 (Mandai et al., 2009). Alexa fluorescent secondary antibodies (Invitrogen) were used.
Microarray analysis
The brains of Gbx2creER/+ or Gbx2creER/− embryos at E12.5 were dissected in chilled PBS. Brain slices at 250 μm were produced on a vibratome. Guided by EGFP fluorescence, the thalamus was dissected and quickly frozen for storage before RNA extraction. Total RNA was extracted with TRIzol® (Invitrogen) and purified with RNeasy column (QIAGEN) following manufacturer's instructions. Quality and quantity of the extracted RNA was determined by Bioanalyzer (Agilent Technologies). Hybridizations to the Mouse-6 Expression BeadChip and measurement of hybridization intensities were performed according to the manufacture's procedures (Illumina, San Diego, CA). Summary of raw data was exported from the BeadStudio/GenomeStudio software and processed in the R software environment. Background correction, variance stabilizing transformation and normalization were performed using the background correction, VST and RSN methods implemented in the lumi package (Du et al., 2008). To combine the array data from two generations of BeadChip used in our experiments, we resolved the inconsistency of Illumina identifiers through nuID (Du et al., 2008). A total of 18,662, 19,440, and 16,069 genes in the mouse genome were examined by the first, second, and the combined array assays, respectively. Batch effects were corrected using ComBat (Johnson et al., 2007). Differentially expressed genes (DE) were identified using the limma package (Ritchie et al., 2015) with a cutoff of adjusted p value (false discovery rate-FDR) equal or less than 0.05 and fold change equal or greater than 1.7 (log2 fold change >0.8). All data have been deposited at GEO: GSE71690.
Gene ontology term enrichment analysis was performed using DAVID Bioinformatics Resource (NIH) (Huang da et al., 2009). Gene Set Analysis was performed using the GSA package (Subramanian et al., 2005). Gene sets were downloaded from the monthly updated Baderlab genesets (http://baderlab.org/GeneSets) and an epithalamus-enriched gene set was curated based on published expression data (Quina et al., 2009).
Chick electroporation studies
A full-length mouse Ebf3 cDNA was cloned into a chicken express vector, pMiwIII, under the control of the chicken β-actin promoter. Chick electroporation assays were performed as described previously (Olsen et al., 2006). Briefly, 1.0 μg/μl of pMiwIII-Ebf3 together with 0.5 μg/μl pMiwIII-EGFP, which was used to monitor transfected cells, were injected into the diencephalon of chicken embryos at HH10-12 stage (Hamburger and Hamilton, 1951) and electroporated into the right side of the neural tube with five rectangular electric pulses of 18 V and 50-millisecond intervals. Embryos were dissected at 36 and 55 hours.
RESULTS
Progenitors throughout the P2 compartment express similar developmental regulators
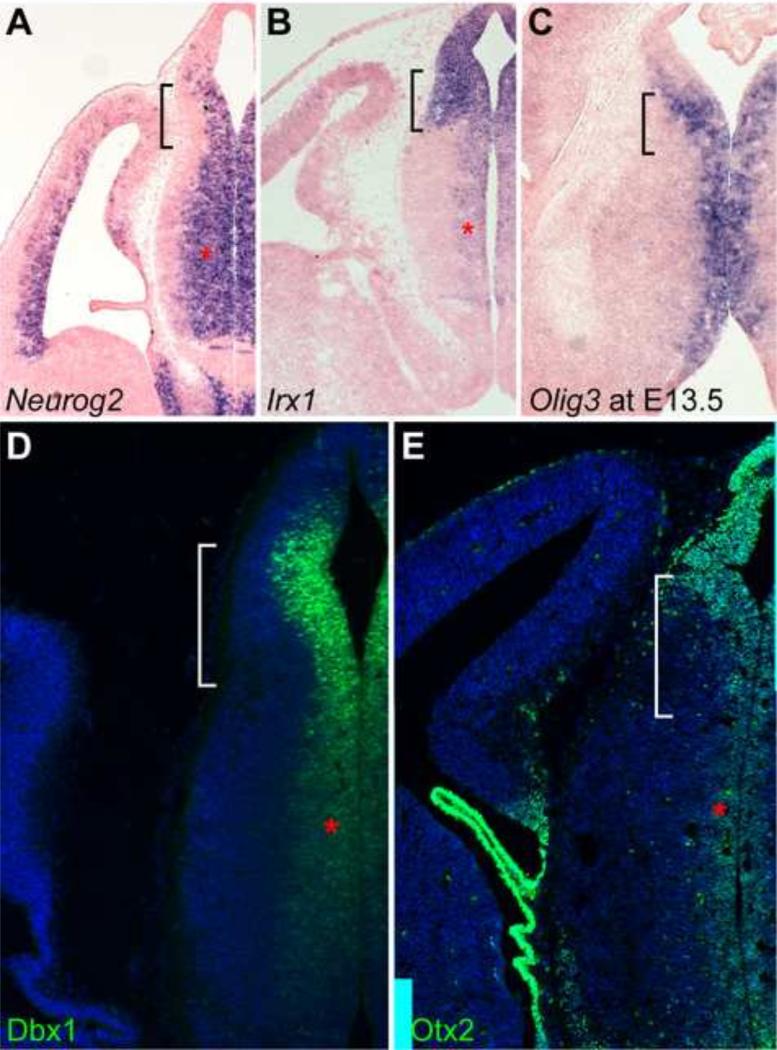
To investigate the developmental programs that establish the thalamic and epithalamic progenitor domains, we examined the expression of several transcription factors that are known as developmental determinants. As described previously (Nakagawa and O'Leary, 2001; Vue et al., 2007), proneural gene Neurog2 (previously named Ngn2) was broadly expressed in the thalamus except for the lateral-most region that is occupied by postmitotic cells at E12.5 (Fig. 1A). Neurog2 expression domain was also extended to the ventricular zone of the epithalamus (Fig. 1A). Irx1, which encodes a transcription factor of the Iroquois family, was expressed in the dorsal part of the diencephalon (Cohen et al., 2000). Transcripts of Irx1 and its paralogous genes Irx2 and Irx3 were found throughout the epithalamus, pretectum and prethalamus, as well as the ventricular zone of the thalamus at E12.5 (Fig. 1B and data not shown). It has been shown that basic helix-loops-helix transcription factor Olig3 defines the entire thalamic VZ at E12.5 (Vue et al., 2007). However, Olig3 transcripts were also detected in the VZ of the epithalamus at E13.5 (Fig. 1C). As described previously (Quina et al., 2009; Chatterjee et al., 2014), robust expression of Dbx1 protein was detected in the epithalamus (Fig. 1D). However, a dorsal-to-ventral gradient of Dbx1 expression was also found in the thalamus at E12.5 (Fig. 1D). Finally, homeodomain transcription factor Otx2, which is essential for the generation of glutamatergic neurons by repressing GABAergic identity in the thalamus (Puelles et al., 2006), was also detected throughout the ventricular zone of the diencephalon (Fig. 1E). Therefore, the progenitor cells of the prospective epithalamus and thalamus share many key developmental regulators.
Figure 1. Progenitor cells in the P2 domain express common developmental regulator genes.
(A-E) In situ hybridization (A-C) and immunostaining (D,E) on coronal sections of mouse brains at E12.5, except for C at E13.5. Probes and antibodies are indicated in the lower left corner of the image. The brackets and asterisks denote the epithalamus and the expression in the ventricular zone of the thalamus, respectively.
Gbx2 is essential for repressing Irx genes in postmitotic cells in the thalamus
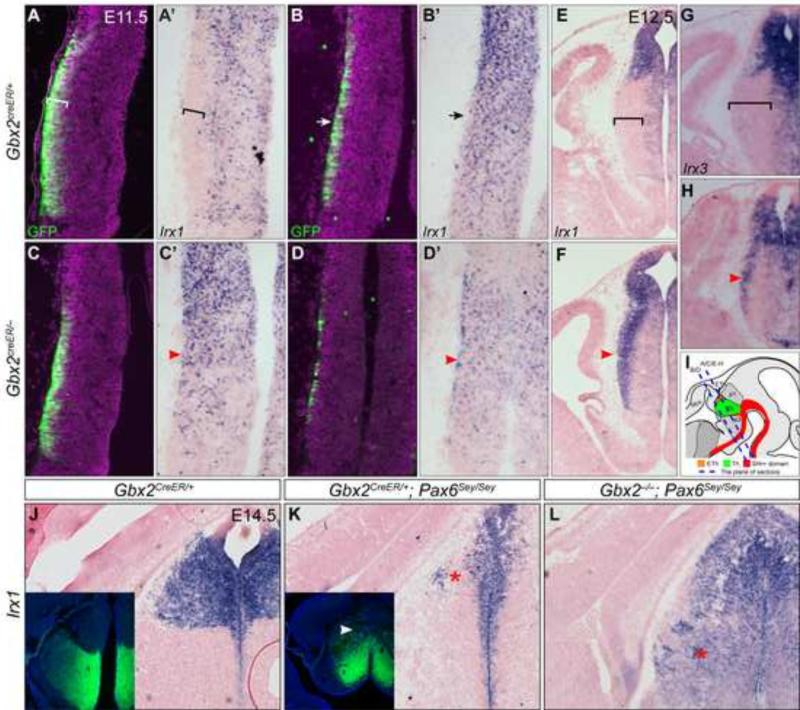
We and others have demonstrated that Gbx2 ablation results in profound defects in the histogenesis of the thalamus and axonal outgrowth and guidance to the neocortex (Miyashita-Lin et al., 1999; Hevner et al., 2002; Chen et al., 2009; Chatterjee et al., 2012; Li et al., 2012). To investigate the mechanism by which Gbx2 controls thalamic development, we analyzed altered gene expression in the thalamus at E11.5, 24 hour after the onset of Gbx2 induction. To monitor the expression of Gbx2, we utilized a Gbx2creER knock-in allele, which contains a creER-ires-GFP cassette in the 5’ untranslated region of the Gbx2 gene (Chen et al., 2009). In mouse embryos carrying this allele, creER and GFP are simultaneously expressed in the same spatiotemporal pattern as the endogenous Gbx2 transcription (Chen et al., 2009). In Gbx2creER/+ embryos at E11.5, GFP was detected only in cells immediately underneath the pial surface in the neural tube wall of the thalamus (Fig. 2A). We have previously shown that these GFP+ cells are postmitotic as they express Tubb3 and fail to incorporate BrdU (Chen et al., 2009). In situ hybridization (ISH) on immediately adjacent sections showed that Irx1 transcripts were absent from the presumed Gbx2-expressing cells but present in the rest of the thalamus and throughout the epithalamus (Fig. 2A’). Because neurogenesis progresses in a caudal-to-rostral gradient in the developing thalamus (Angevine, 1970; McAllister and Das, 1977), significantly fewer GFP+ cells were detected in the rostral part, compared to the caudal, of the thalamus (Fig. 2B and B’). Interestingly, the negative domain of Irx1 was hardly detected in the rostral part of the thalamus at E11.5 (Fig. 2B and B’), suggesting that down-regulation of Irx1 is associated with Gbx2 induction. In agreement with this notion, Irx1 transcripts were absent from the postmitotic thalamic cells that express Gbx2 by E12.5 (Fig. 2E and inset). Importantly, in the absence of Gbx2, Irx1 transcripts were maintained in postmitotic cells in the thalamus at E11.5 and E12.5 (Fig. 2C-F). Three other members of Iroquois genes, Irx2, Irx3, and Irx5, showed a similar expression pattern to that of Irx1 in wild-type embryos, and they were all ectopically expressed in postmitotic thalamic neuronal precursors in Gbx2-KO embryos (Fig. 2G,H, 5B and data not shown). Therefore, Gbx2 is essential for repressing Irx genes in thalamic neural precursors after they exit the cell cycle.
Figure 2. Gbx2 represses Irx genes in postmitotic thalamic neurons.
(A-F) Immunofluorescence and in situ hybridization on coronal sections of control and Gbx2-KO embryos at E11.5 (A-D') and E12.5 (E-H). Antibodies and probes are indicated at the lower left corner; genotypes to the left. (I) Illustration of mouse embryonic brain and the sectioning planes corresponding to A-H. Brackets indicate the GFP/Gbx2 positive and Irx negative domain; arrows show fewer GFP+ cells and remnants of Irx1 transcripts in the anterior part of the thalamus; arrowheads denote the persistent Irx transcripts. (J-L) In situ hybridization for Irx1 on coronal sections of E14.5 brain. Insets show immunostaining for GFP on adjacent sections to J and K respectively. The arrowhead indicates GFP expression in the presumptive habenula; asterisks show the loss (K) or gain (L) of Irx1 expression, respectively.
We have recently shown that Pax6 regulates the partitioning of the epithalamus and thalamus by restricting Shh activity in the ZLI; without Pax6, the thalamus is expanded at the expense of the epithalamus (Chatterjee et al., 2014). Using GFP to follow Gbx2 transcription in Pax6-deficient embryos carrying a Gbx2creER allele, we found that GFP was ectopically expressed in the presumptive epithalamus concomitant with depletion of Irx1 transcripts (Fig. 2K). By contrast, Irx1 was expressed throughout the presumptive epithalamus and thalamus in Pax6 and Gbx2 double mutant embryos (Fig. 2L), demonstrating that the ectopic expression of Gbx2 is responsible for the down-regulation of Irx1. Collectively, our data show that Gbx2 plays a crucial role in repressing Irx1 transcription in the P2 domain.
Gbx2 deletion results in the loss of thalamic markers and ectopic expression of epithalamus-enriched genes
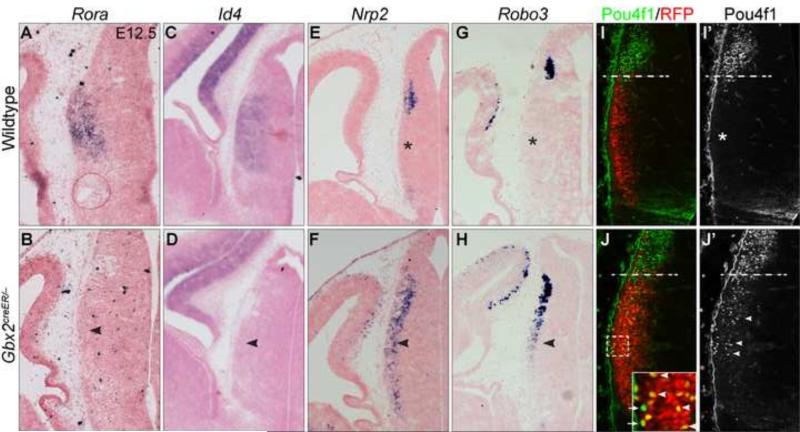
Iroquois transcription factors play important roles in tissue patterning and cell fate specification (Kobayashi et al., 2002; Kiecker and Lumsden, 2004; Robertshaw et al., 2013). To test if the failure to repress multiple Iroquois genes might alter the molecular identity of thalamic precursor cells in Gbx2-KO mice, we examined how Gbx2 deletion affected expression of known molecular markers specific for thalamic or habenular neurons. Rora and Id4 are specifically expressed in thalamic neuronal precursors at E12.5 and later stages (Nakagawa and O'Leary, 2003; Zhou et al., 2004)(Fig. 3A,C). In Gbx2-KO embryos at E12.5, Rora and Id4 transcripts were depleted in the thalamus (Fig. 3B,D). Nrp2, Pou4f1, and Robo3 are specifically expressed in postmitotic neural precursors in the epithalamus at E12.5 (Chatterjee et al., 2014)(Fig. 3E,G,I'). Notably, these habenular markers were ectopically expressed in the Gbx2-deficient thalamus (Fig. 3F,H,J').
Figure 3. Deletion of Gbx2 results in loss of thalamic-specific genes and ectopic expression of habenular markers in the thalamus.
(A-J) In situ hybridization (A-H) and immunofluorescence (I-J’) on coronal sections of the thalamus at E12.5. Probes and antibodies are indicated to the top; genotypes to the left. Asterisks show that absence of Nrp2, Robo3 and Pou4f1 in the wild-type thalamus; arrowheads denote the abnormal expression in the Gbx2-KO thalamus; dashed lines demarcate the border between the thalamus and epithalamus. Note that the majority of Pou4f1+ cells are marked by RFP (arrowheads), and only a few Pou4f1+ cells (arrows) are negative for RFP in the Gbx2-KO thalamus. The boxed area is enlarged in the inset in J.
We have previously shown that cells originating from the thalamus contribute to epithalamus-derived structures in the absence of Gbx2, demonstrating that Gbx2 is essential for the lineage restriction between the thalamus and epithalamus (Chen et al., 2009). To rule out that the ectopic Pou4f1+ cells in the Gbx2-deficient thalamus were caused by abnormal cell movements rather than misregulation of Pou4f1 expression, we genetically marked Gbx2-expressing cells by red fluorescence (RFP) in Gbx2creER/−; R26RRFP/+ embryos by administrating tamoxifen at E10.5. Pou4f1 immunoreactivity was found in the epithalamus but absent from RFP+ cells in the thalamus of E12.5 Gbx2creER/+; R26RRFP/+ embryos that received tamoxifen at E10.5 (Fig. 3I,I'). In Gbx2creER/−; R26RRFP/+ embryos, many Pou4f1+ cells were found in the thalamus and the majority of them were colocalized with RFP, demonstrating that they are descendants of the Gbx2 lineage originating from the thalamus (Fig. 3J,J'). Although some Pou4f1+ cells were negative for RFP, these Pou4f1+/RFP− cells were evenly distributed in both dorsal and ventral parts of the thalamus (Fig. 3J), suggesting that these Pou4f1+/RFP− cells may result from incomplete cre-mediated recombination rather than migration from the epithalamus in Gbx2-deficient embryos. Collectively, our data show that thalamic neurons fail to express thalamic-specific genes but ectopically express epithalamus-specific markers in the absence of Gbx2.
Gbx2 deletion results in global changes of thalamic transcriptome
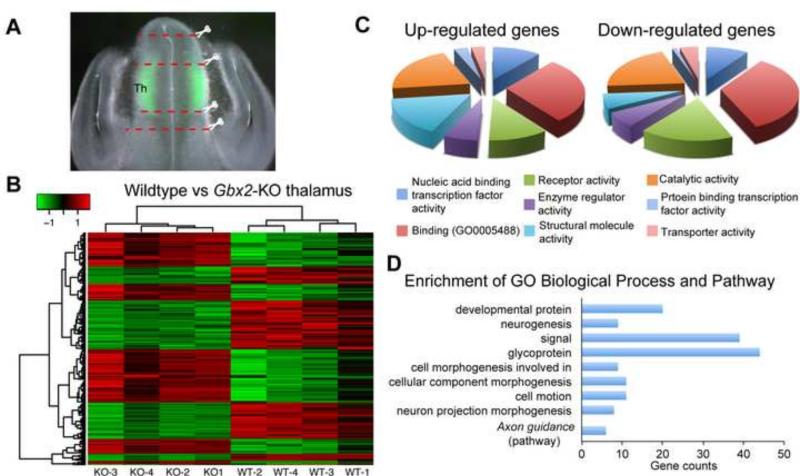
To gain insight into how Gbx2 deletion affects development of the thalamus, we performed genome-wide transcriptional profiling of wild-type and Gbx2-deficient thalamus at E12.5 by microarray. We chose E12.5 because Gbx2 mutant phenotypes were clearly detected at this stage and we could obtain sufficient amount tissue and RNA from a single embryo. We dissected the thalamus from brain slices with the guide of GFP fluorescence that demarcated the thalamus in Gbx2creER/+ (wild type) and Gbx2creER/− (Fig. 4A). We also collected diencephalic tissues dorsal and ventral to the GFP+ domain, containing the epithalamus and prethalamus (as well as the rostral thalamus and ZLI), respectively (Fig. 4A). Using a cutoff of global FDR less than 5% and fold change greater than 1.7, we identified 126 differentially expressed (DE) genes, with 76 decreased and 50 increased, in E12.5 thalamus due to Gbx2 deletion (Tables I and II)(Fig. 4B). Gene ontology and pathway analyses showed that the DE genes are enriched for genes that are involved in brain development, such as neurogenesis, axon projections, and axon guidance (Fig. 4C,D).
Figure 4. Genome-wide transcriptional profiling of wild-type and Gbx2-KO thalamus at E12.5.
(A) Micro-dissection of the thalamus with the guide of GFP in E12.5 embryos carrying the Gbx2creER allele. (B) Heatmap representation of the relative expression level and clustering of differentially expressed genes. (C,D) Diagram representation of enrichment for cellular processes (C) and biological and pathway (D) of Gbx2 downstream genes.
Table I.
Down-regulated genes due to the loss of Gbx2
| key | Symbol | Description | logFC | adj.P.Val | Expression | Verified |
|---|---|---|---|---|---|---|
| Combined | Cyp1b1 | cytochrome P450, family 1, subfamily b, polypeptide 1 | −3.0 | 0.004 | NA | |
| Array2 | Cops8 | COP9 (constitutive photomorphogenic) homolog, subunit 8 (Arabidopsis thaliana) | −2.7 | 0.020 | NA | |
| Combined | Id4 | inhibitor of DNA binding 4 | −2.5 | 0.003 | Yes | Yes |
| Array1 | Chst1 | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | −2.4 | 0.001 | Yes | Yes |
| Combined | Ntng1 | netrin G1 | −2.1 | 0.005 | Yes | Yes* |
| Array1 | Rbfox1 | RNA binding protein, fox-1 homolog (C. elegans) 1 | −2.1 | 0.015 | Yes | |
| Combined | Kirrel3 | kin of IRRE like 3 (Drosophila) | −2.0 | 0.011 | Yes | |
| Combined | Glra2 | glycine receptor, alpha 2 subunit | −2.0 | 0.014 | Yes | Yes |
| Combined | Hs6st2 | heparan sulfate 6-O-sulfotransferase 2 | −1.9 | 0.004 | Yes | Yes |
| Array1 | Sertad4 | SERTA domain containing 4 | −1.9 | 0.002 | NA | IC |
| Array1 | C1ql2 | complement component 1, q subcomponent-like 2 | −1.9 | 0.027 | PTh | |
| Combined | Slc18a2 | solute carrier family 18 (vesicular monoamine), member 2 | −1.8 | 0.008 | Yes | Yes |
| Array2 | Gbx2 | gastrulation brain homeobox 2 | −1.8 | 0.044 | Yes | Yes** |
| Array1 | Hbb-b1 | hemoglobin, beta adult major chain | −1.8 | 0.086 | NA | |
| Combined | Calb2 | calbindin 2 | −1.8 | 0.005 | Yes | |
| Combined | Rora | RAR-related orphan receptor alpha | −1.8 | 0.013 | Yes | Yes |
| Array2 | Sema3a | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | −1.7 | 0.074 | Yes | |
| Combined | Nxph1 | neurexophilin 1 | −1.7 | 0.047 | Yes | |
| Array1 | Wif1 | Wnt inhibitory factor 1 | −1.6 | 0.028 | Yes | Yes* |
| Array1 | Ttc27 | tetratricopeptide repeat domain 27 | −1.6 | 0.026 | NA | |
| Array1 | Sulf2 | sulfatase 2 | −1.6 | 0.003 | ? | |
| Combined | Rit2 | Ras-like without CAAX 2 | −1.6 | 0.011 | Yes | |
| Array1 | Dlk1 | delta-like 1 homolog (Drosophila) | −1.6 | 0.046 | Yes | IC |
| Combined | Samd5 | sterile alpha motif domain containing 5 | −1.5 | 0.005 | Yes | |
| Array1 | Robo2 | roundabout homolog 2 (Drosophila) | −1.5 | 0.034 | Th | Yes** |
| Array2 | Tle1 | transducin-like enhancer of split 1, homolog of Drosophila E(spl) | −1.4 | 0.038 | PTh | |
| Array1 | Dab1 | disabled 1 | −1.3 | 0.007 | Yes | Yes* |
| Combined | Thsd7b | thrombospondin, type I, domain containing 7B | −1.3 | 0.015 | NA | |
| Combined | Sgk1 | serum/glucocorticoid regulated kinase 1 | −1.3 | 0.009 | Yes | |
| Combined | Syt13 | synaptotagmin XIII | −1.3 | 0.005 | Yes | |
| Array1 | Clmp | CXADR-like membrane protein | −1.3 | 0.025 | NA | |
| Array1 | Foxp2 | forkhead box P2 | −1.3 | 0.026 | Yes | Yes* |
| Combined | Gm10304 | predicted gene 10304 | −1.2 | 0.021 | ||
| Combined | Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | −1.2 | 0.030 | Yes | Yes |
| Combined | Ccng2 | cyclin G2 | −1.2 | 0.027 | NA | |
| Combined | Tenm4 | teneurin transmembrane protein 4 | −1.2 | 0.025 | Yes | |
| Array2 | Vegfc | vascular endothelial growth factor C | −1.2 | 0.053 | Yes | |
| Array1 | Lhfp | lipoma HMGIC fusion partner | −1.1 | 0.039 | Yes | |
| Combined | Chn2 | chimerin 2 | −1.1 | 0.024 | ||
| Array2 | Cfap69 | cilia and flagella associated protein 69 | −1.1 | 0.065 | No signals | |
| Combined | Fgfr1op2 | FGFR1 oncogene partner 2 | −1.1 | 0.023 | ||
| Combined | Pcdh9 | protocadherin 9 | −1.1 | 0.041 | Yes | |
| Array1 | Ednrb | endothelin receptor type B | −1.1 | 0.026 | No signals | |
| Array1 | Vcam1 | vascular cell adhesion molecule 1 | −1.1 | 0.061 | No signals | |
| Array2 | Enpp2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | −1.1 | 0.078 | ||
| Combined | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | −1.0 | 0.024 | Yes | |
| Combined | Atrnl1 | attractin like 1 | −1.0 | 0.027 | NA | |
| Array1 | Rnf152 | ring finger protein 152 | −1.0 | 0.041 | Yes | |
| Combined | Arpp21 | cyclic AMP-regulated phosphoprotein, 21 | −1.0 | 0.034 | Yes | |
| Combined | Sel1l3 | sel-1 suppressor of lin-12-like 3 (C. elegans) | −1.0 | 0.011 | ||
| Combined | Rgs10 | regulator of G-protein signalling 10 | −1.0 | 0.044 | NA | |
| Array1 | Brat1 | BRCA1-associated ATM activator 1 | −0.9 | 0.028 | NA | |
| Array1 | Zfhx4 | zinc finger homeodomain 4 | −0.9 | 0.089 | No signals | |
| Array1 | Lxn | latexin | −0.9 | 0.032 | Yes (VZ) | |
| Array1 | Dner | delta/notch-like EGF-related receptor | −0.9 | 0.078 | Not specific | |
| Array1 | Celf2 | CUGBP, Elav-like family member 2 | −0.9 | 0.080 | Yes | |
| Array1 | Fam155a | family with sequence similarity 155, member A | −0.9 | 0.028 | No signals | |
| Array1 | Pmp22 | peripheral myelin protein 22 | −0.9 | 0.041 | NA | |
| Combined | Igsf21 | immunoglobulin superfamily, member 21 | −0.9 | 0.018 | Yes | |
| Array1 | Elmo1 | engulfment and cell motility 1 | −0.9 | 0.026 | Yes | |
| Combined | Glce | glucuronyl C5-epimerase | −0.9 | 0.033 | Yes | |
| Array1 | Dkk3 | dickkopf homolog 3 (Xenopus laevis) | −0.9 | 0.027 | Not specific | |
| Array1 | Tmem130 | transmembrane protein 130 | −0.9 | 0.096 | Not specific | |
| Array1 | Rapgef5 | Rap guanine nucleotide exchange factor (GEF) 5 | −0.9 | 0.028 | Yes | |
| Array1 | Tmem108 | transmembrane protein 108 | −0.9 | 0.030 | No signals | |
| Array1 | Limch1 | LIM and calponin homology domains 1 | −0.9 | 0.063 | No signals | |
| Combined | Epha3 | Eph receptor A3 | −0.9 | 0.046 | Yes | Yes |
| Combined | Gpr83 | G protein-coupled receptor 83 | −0.8 | 0.048 | weak signals | |
| Array1 | Arrdc3 | arrestin domain containing 3 | −0.8 | 0.036 | NA | |
| Combined | Prmt8 | protein arginine N-methyltransferase 8 | −0.8 | 0.025 | NA | |
| Array1 | E130114P18Ri RIKEN cDNA E130114P18 gene | −0.8 | 0.079 | NA | ||
| Array1 | Bcl11a | B cell CLL/lymphoma 11A (zinc finger protein) | −0.8 | 0.057 | Not specific | |
| Combined | Gas7 | growth arrest specific 7 | −0.8 | 0.033 | Yes | Yes |
| Array1 | Esrrb | estrogen related receptor, beta | −0.8 | 0.089 | PT | |
| Array1 | Chrna4 | cholinergic receptor, nicotinic, alpha polypeptide 4 | −0.8 | 0.041 | ET+PT | |
| Array1 | Efna5 | ephrin A5 | −0.8 | 0.057 | Yes 38/76 (50.0%) | Yes** 16/76 (21.1%) |
Table II.
Up-regulated genes due to the loss of Gbx2
| key | Symbol | Description | logFC | adj.P.Val | Expression | Verified |
|---|---|---|---|---|---|---|
| Combined | Hist1h2ai | histone cluster 1, H2ai | 0.8 | 0.049 | NA | |
| Combined | Irx1 | Iroquois related homeobox 1 (Drosophila) | 0.8 | 0.008 | Yes | Yes |
| Array1 | Cenpe | centromere protein E | 0.8 | 0.030 | NA | |
| Array1 | H2-D1 | histocompatibility 2, D region locus 1 | 0.9 | 0.081 | NA | |
| Array1 | Hist2h2aa2 | histone cluster 2, H2aa2 | 0.9 | 0.032 | NA | |
| Combined | Kif4 | kinesin family member 4 | 0.9 | 0.047 | No expression | |
| Array1 | Nrp2 | neuropilin 2 | 0.9 | 0.069 | Yes | Yes |
| Array1 | Rad51ap1 | RAD51 associated protein 1 | 0.9 | 0.089 | No signals | |
| Array1 | Enc1 | ectodermal-neural cortex 1 | 0.9 | 0.053 | Yes | |
| Array1 | Hist1h2ah | histone cluster 1, H2ah | 0.9 | 0.041 | NA | |
| Combined | Dcx | doublecortin | 0.9 | 0.033 | ||
| Combined | Fam111a | family with sequence similarity 111, member A | 0.9 | 0.019 | No expression | |
| Combined | Chd9 | chromodomain helicase DNA binding protein 9 | 0.9 | 0.009 | NA | |
| Array1 | BC003331 | cDNA sequence BC003331 | 1.0 | 0.023 | NA | |
| Combined | Rrm1 | ribonucleotide reductase M1 | 1.0 | 0.047 | Yes? | |
| Combined | Kif26a | kinesin family member 26A | 1.0 | 0.031 | Yes | Yes |
| Array1 | Hist1h2ak | histone cluster 1, H2ak | 1.0 | 0.023 | NA | |
| Combined | Fam65b | family with sequence similarity 65, member B | 1.0 | 0.069 | NA | |
| Combined | Otx1 | orthodenticle homolog 1 | 1.0 | 0.004 | PT | |
| Combined | Angpt1 | angiopoietin 1 | 1.1 | 0.033 | NA | |
| Combined | Smyd2 | SET and MYND domain containing 2 | 1.1 | 0.005 | Ubiquitous | |
| Array1 | Ppp2r2b | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform | 1.1 | 0.025 | Yes | |
| Combined | Tle4 | transducin-like enhancer of split 4, homolog of Drosophila E(spl) | 1.1 | 0.074 | Yes | Yes |
| Combined | Thsd4 | thrombospondin, type I, domain containing 4 | 1.1 | 0.019 | No expression | |
| Combined | Rftn1 | raftlin lipid raft linker 1 | 1.1 | 0.018 | Yes | |
| Combined | Top2a | topoisomerase (DNA) II alpha | 1.1 | 0.065 | Not expressed | |
| Array1 | Ccnd1 | cyclin D1 | 1.2 | 0.075 | Not expressed | |
| Array1 | Col2a1 | collagen, type II, alpha 1 | 1.2 | 0.028 | Yes | |
| Combined | Arhgap12 | Rho GTPase activating protein 12 | 1.2 | 0.044 | Ubiquitous | |
| Combined | Epb4.1 | erythrocyte protein band 4.1 | 1.3 | 0.073 | NA | |
| Array1 | Igfbpl1 | insulin-like growth factor binding protein-like 1 | 1.4 | 0.015 | both ET and Th | |
| Combined | Nefm | neurofilament, medium polypeptide | 1.4 | 0.037 | Yes | Yes |
| Combined | Shisa2 | shisa homolog 2 (Xenopus laevis) | 1.4 | 0.003 | NA | |
| Combined | Fam196b | family with sequence similarity 196, member B | 1.4 | 0.044 | NA | |
| Combined | Mmd2 | monocyte to macrophage differentiation-associated 2 | 1.4 | 0.037 | Yes | |
| Array1 | Map2 | microtubule-associated protein 2 | 1.5 | 0.006 | Ubiquitous | |
| Combined | Eml5 | echinoderm microtubule associated protein like 5 | 1.6 | 0.053 | NA | |
| Combined | Actb | actin, beta | 1.6 | 0.068 | NA | |
| Combined | Ebf3 | early B cell factor 3 | 1.6 | 0.037 | Yes | Yes |
| Combined | Cpne8 | copine VIII | 1.6 | 0.086 | NA | |
| Combined | Olfm1 | olfactomedin 1 | 1.8 | 0.038 | Yes | |
| Combined | Ptx3 | pentraxin related gene | 1.8 | 0.077 | NA | |
| Combined | Ephb1 | Eph receptor B1 | 1.9 | 0.065 | Yes | |
| Combined | Irx2 | Iroquois related homeobox 2 (Drosophila) | 2.0 | 0.069 | Yes | Yes |
| Combined | Cntn2 | contactin 2 | 2.1 | 0.038 | Yes | Yes |
| Combined | Islr2 | immunoglobulin superfamily containing leucine-rich repeat 2 | 2.2 | 0.037 | Yes | Yes |
| Array1 | Zic4 | zinc finger protein of the cerebellum 4 | 2.3 | 0.017 | NC | |
| Combined | Lmo3 | LIM domain only 3 | 2.8 | 0.020 | Yes 18/48 (37.5%) | Yes 10/48 (20.8%) |
To validate the microarray data, we first inspected the expression pattern of the DE genes in gene expression databases such as Mouse Genomic Informatics, Allen Brain Atlas and GenePaint (Visel et al., 2004; Finger et al., 2011; Thompson et al., 2014). Among those DE genes of available expression information, 50% of them (38/76) were found to be either specifically or strongly expressed in the thalamus between E13.5 or E15.5 (Table I). By comparing the thalamus to the neighboring diencephalic tissues (see Methods), we also identified 35 genes that were enriched in the thalamus (FDR≤0.05 and log2FC≥1), and the majority of them were indeed expressed in the thalamus based on data in the expression databases (Table I). Remarkably, 16 of these thalamus-enriched genes (45.7%, Fisher's exact test p<0.001) were down regulated in Gbx2-deficient thalamus, supporting the notion that Gbx2 is essential for acquisition or maintenance of the molecular character of the thalamus.
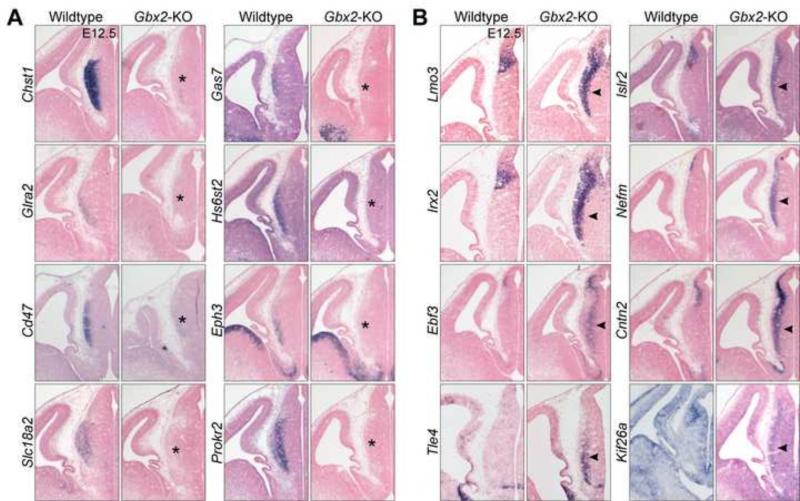
We next performed ISH to confirm that Gbx2 deletion reduced expression of the presumptive thalamus-specific genes. Out of 18 DE genes that were examined, 16 were notably downregulated in the thalamus of Gbx2-KO embryos at E12.5 or E13.5 (Fig. 5A and data not shown; the other two probes gave inconclusive results). Remarkably, Chst1, Hs6st2, Glra2, and Cd47 were specifically expressed in the thalamus at E12.5, but their transcripts were mostly depleted in Gbx2-deficient thalamus (Fig. 5A). Taken together, our expression studies show that Gbx2 deletion results in a loss of the molecular signature of the developing thalamus.
Figure 5. Verification of DE genes identified by microarrays.
(A,B) In situ hybridization on coronal sections of the thalamus at E12.5. Probes and genotypes are indicated to the left and top, respectively. Note the loss of thalamic markers (asterisks in A) and gain of epithalamic ones (arrowheads in B) due to the loss of Gbx2.
Gbx2-deficient thalamus adopts an expression profile of the developing habenula
To investigate the hypothesis that thalamic neurons lacking Gbx2 assumed a molecular identity similar to that of the habenula, we first inspected the expression pattern of the up-regulated DE genes in the expression databases. Among the 50 up-regulated DE genes, 18 (36.0%) were highly expressed in the epithalamus (Table II). To avoid an arbitrary cut-off in selecting DE genes, we performed Gene Set Analysis (Subramanian et al., 2005) and found that epithalamus-enriched genes compiled from microarray experiments (see details in Methods) were significantly increased in the Gbx2-deficient thalamus (enrichment score=0.67, FDR<0.001). Conversely, levels of thalamus-enriched genes were significantly reduced due to Gbx2 deletion (enrichment score = −2.57, FDR<0.001). ISH confirmed ectopic expression of epithalamus-enriched genes, such as Lmo3, Irx2, Ebf3, Islr2, Nefm, Cntn2 and Kif26a, in the thalamus due to Gbx2 ablation (Fig. 5B).
Besides Pou4f1, which is specific to the epithalamus (Quina et al., 2009), many other genes that are inhibited by Gbx2, such as Lmo3, Irx1, Irx2, Ebf3, Islr2, and Cntn2, are expressed in both the epithalamus and pretectum. To investigate the alternative explanation of de-repression of pretectal fate due to Gbx2 deletion, we identified 12 pretectal markers, including anterior pretectal genes (Bhlhe22, C1ql2, Cux2, Esrrb, Neurod6, Pax3, Pik3r1, and Tfap2d) and posterior pretectal genes (Gata3, Six3, and Tal1) using the Allen Developmental Mouse Brain reference atlas (Thompson et al., 2014)(Fig. S1A). Importantly, we have previously shown that the expression of Bhlhe22 and Pax3 are unaffected by Gbx2 deletion (Chen et al., 2009), and none of these pretectal markers were up-regulated due to Gbx2 deletion (Fig. S1B), suggesting that Gbx2 deletion does not convert the thalamus to a pretectal fate. Together, our data shows that thalamic neurons adopt molecular features resembling that of the epithalamus in the absence of Gbx2.
Gbx2 deletion alters neurogenic progression in the thalamus
Despite the ambiguity of the molecular signatures between the thalamic and epithalamic progenitors, they exhibit distinctive patterns of neurogenic progression with a markedly delayed neurogenesis in the thalamus (Jones, 2007). For example, the habenular nuclei become distinguishable at E14.5, while the neighboring thalamic domain that gives rise to the parataenial, rhomboid, reunions, mediodorsal and anterior nuclei are not discernible until E18.5 in rats (McAllister and Das, 1977). In sharp contrast to the neighboring diencephalic structure, the developing thalamus contains a remarkably large number of basal progenitor cells (Smart, 1972; Wang et al., 2011). Therefore, the prospective thalamus and epithalamus have distinctive developmental programs that control the proliferation and progression of neural progenitors. Indeed, we detected the mRNA and protein of Nefm and Dcx, whose expression is associated with generation of postmitotic neurons, in the epithalamus at E12.5, at least 24 hours before they were found in the thalamus (Fig. 5B and data not shown). Remarkably, Nefm and Dcx, which were absent from the wild-type thalamus until E13.5, were expressed in the mantle zone of the thalamus in Gbx2-KO embryos at E12.5 (Fig. 5B and Table II), suggesting that inactivation of Gbx2 accelerates neurogenesis in the thalamus. Moreover, Id4, which codes for a basic helix-loop-helix negative regulator of neurogenesis(Yun et al., 2004; Bedford et al., 2005), was specifically expressed in the wild-type but lost in Gbx2-KO thalamus (Fig. 3C,D). Conversely, Ebf3 was strongly expressed in the epithalamus but its expression was markedly increased in the Gbx2-KO thalamus (Fig. 5A). It has been shown that forced expression of Ebf3 inhibits cell proliferation and increase apoptosis or tumor cells in vitro (Garel et al., 1999; Bai et al., 2007). Using a chick electroporation assay, we showed that expression of Ebf3 indeed promoted neural progenitor cells to exit the cell cycle in the developing thalamus (Fig. S2). These observations suggest that Gbx2 regulates neurogenic progression in the thalamus, possibly through the control of Id4 and Ebf3. In the absence of Gbx2, the neurogenesis is accelerated in the thalamus matching that in the epithalamus.
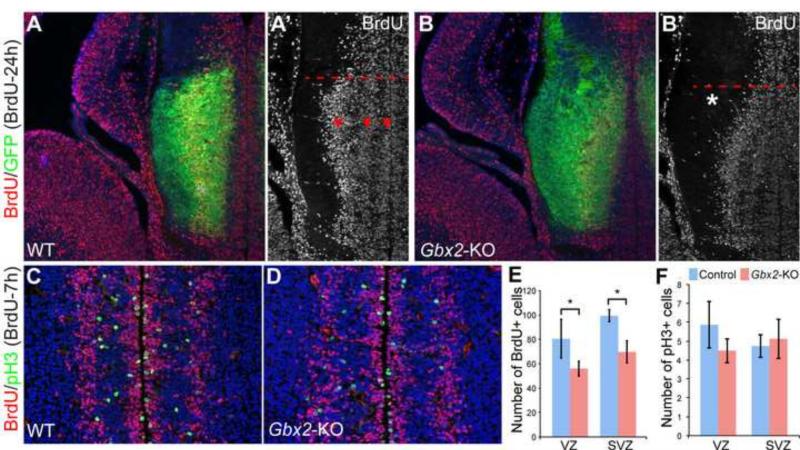
To investigate if Gbx2 deletion affected cell proliferation, we performed 24-hour pulse-chase labeling of BrdU at E12.5. We detected three stripes of BrdU+ cell aggregates parallel to the ventricular surface in the thalamus in wild-type embryos at E13.5 (Fig. 6A,A’). In Gbx2-KO embryos, although the three stripes of BrdU+ cells were still discernible in the ventral/anterior part of the thalamus, they collapsed into one, similar to that found in the epithalamus, in the dorsal/posterior part of the thalamus (Fig. 6B,B’). Furthermore, the innermost stripe of BrdU+ cells in the ventricular zone was markedly thinner in the Gbx2-KO thalamus at E13.5 (Fig. 6B’). By chase labeling for 7 hours with BrdU, we showed that numbers of BrdU+ cells were significantly reduced in both the VZ and SVZ of the thalamus, particularly prominent near the junction between the thalamus and epithalamus (Fig. 6C-G). The number of cells marked by phosphorylated histone 3 (pH3), which marks cells undergoing mitotic division, was not statistically different between the wild-type and Gbx2-KO thalamus (Fig. 6C-D,H). Collectively, our results show that loss of Gbx2 alters the proliferation and neurogenic progression in the thalamus.
Figure 6. Loss of Gbx2 causes abnormal cell proliferation in the thalamus.
(A-F) Immunofluorescence on coronal sections of control and Gbx2-KO thalamus at E13.5 following BrdU chase or pulse labeling as indicated. Dashed lines demarcate the boundary between the thalamus and the epithalamus; arrowheads indicate the three stripes of BrdU+ cells parallel to the ventricular surface; the asterisk shows the abnormal curvature of the outer band of BrdU+ cells in the dorsal-most region of the thalamus due to the loss of Gbx2. (G-I) Histogram representation of quantification of proliferating and newly generated postmitotic cells. Asterisks indicate statistical significance (p<0.05, Student's t-test).
Cell non-autonomous function of Gbx2 in regulating neurogenic progression in the thalamus
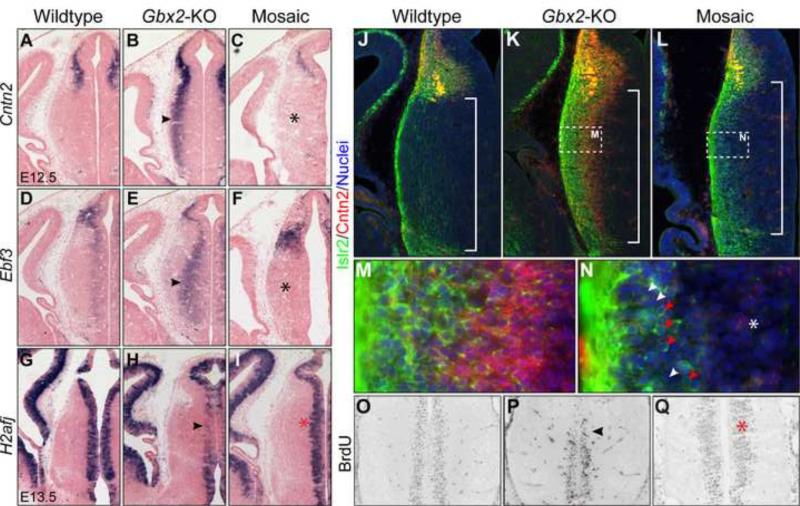
Given that Gbx2 is induced in postmitotic neural precursors, the abnormal cell proliferation in the Gbx2-KO thalamus suggests that Gbx2 regulates progression of thalamic progenitors by feedback signaling from postmitotic cells. Indeed, our gene expression analyses revealed that Gbx2 deletion altered gene expression not only in the mantle zone, but also the intermediate and even the VZ of the thalamus (Fig. 5 and 7). For example, loss of Gbx2 increased the expression of Cntn2 and Ebf3 in the intermediate domains between the mantle and ventricular zone of the thalamus in Gbx2-KO embryos at E12.5 (Fig. 7A,B,D,E). H2afj, which codes for an atypical histone protein (Nishida et al., 2005), is expressed throughout the VZ of the neural tube (Fig. 7G). H2afj transcripts were notably reduced in the thalamus particularly in the dorsal-most region of the Gbx2-KO thalamus at E13.5 (Fig. 7H). We have previously demonstrated a cell non-autonomous function of Gbx2 in the establishment of the compartment boundary between the thalamus and epithalamus using chimeras and mosaics (Chen et al., 2009). Using the same approach as described previously (Chen et al., 2009; Chatterjee et al., 2012), we administrated tamoxifen to E10.5 Gbx2creER/F embryos so that creER-mediated recombination rendered the genotype of Gbx2-expressing cells from Gbx2creER/F to Gbx2creER/− in a mosaic manner. Remarkably, the normal expression of Cntn2, Ebf3 and H2afj was restored in E13.5 Gbx2creER/F embryos that received tamoxifen at E10.5 (Fig. 7C,F,I). We were unable to directly verify the loss of Gbx2 protein because of a lack of workable anti-Gbx2 antibodies. We thus explored a different approach by analyzing downstream targets of Gbx2. In agreement with the ISH data (Fig. 5B), immunostaining showed elevated expression of Cntn2 and Islr2 in the thalamus of Gbx2-KO embryos at E12.5 (Fig. 7J,K,M). Importantly, Cntn2 immunoreactivity was mostly absent in the thalamus of E12.5 Gbx2creER/F embryos that were given tamoxifen at E10.5 (Fig. 7L,N). By contrast, Islr2+ cells persisted and exhibited the salt-and-pepper mosaic pattern in the thalamus (Fig. 7L,N). Using a Gbx2 RNA probe that recognizes sequence that is flanked by the two loxP sites of the Gbx2F allele, we showed mosaic deletion of Gbx2 transcripts in the mantle zone following tamoxifen administration, including the area where ectopic Cntn2 and Islr2 expression was detected in Gbx2-null embryos at E12.5 (Fig. 7 and S4). These findings demonstrate that Islr2 and Cntn2 are differentially regulated by the cell autonomous and non-autonomous function of Gbx2, respectively. Therefore, our data indirectly verify the presence of wild-type and Gbx2-deficient cells, which are respectively indicated by Islr2− and Islr2+ cells, in the thalamus of Gbx2creER/F embryos that were given tamoxifen at E10.5. Finally, BrdU pulse labeling revealed that cell proliferation was also rescued in Gbx2 mosaic-deletion embryos at E13.5 (Fig. 7O-Q). Collectively, these data demonstrate that Gbx2 plays a cell non-autonomous role in regulating proliferation and progression of progenitors in the developing thalamus.
Figure 7. Gbx2 act cell non-autonomously to regulate gene expression and cell proliferation.
(A-I) In situ hybridization on coronal sections of the thalamus of indicated genotypes between E12.5 and E13.5. (J-N) Immunostaining of Islr2 and Cntn2 on coronal sections of the E12.5 thalamus. Brackets demarcate the thalamus; red arrowheads indicate Islr2+, presumably Gbx2-deficient cells; white arrowheads indicate Islr2−, presumably wild-type cells. (O-W) Immunostaining of BrdU on coronal sections of E13.5 thalamus following 1-hour pulse labeling. The arrowhead indicates the abnormal gene expression and cell proliferation in Gbx2-KO thalamus; the asterisk shows the rescue in the thalamus that is composed of wild-type and Gbx2-deficient cells.
DISCUSSION
In the current study, we have performed microarray analysis to compare the transcriptome of the thalamus between wild-type and Gbx2-deficient embryos at E12.5. To the best of our knowledge (based on information in the GEO database), this is the first attempt to capture the expression profile of the embryonic thalamus in mice. We show that Gbx2 is essential for acquisition of the molecular identity of the thalamus. Interestingly, the loss of the thalamic identity due to Gbx2 ablation is associated with a partial switch to the habenular identity. Moreover, Gbx2 controls a feedback mechanism from postmitotic cells in the mantle zone to regulate the development of thalamic progenitor cells. Our new findings explain the profound defect in the developing thalamus due to the loss of Gbx2, and shed light on the molecular mechanism underlying the subdivision of the epithalamus and thalamus from a single developmental compartment in the diencephalon.
Despite the differences in cytoarchitecture and connectivity between thalamic and habenular neurons, the molecular mechanism underlying their specification and differentiation remains largely unknown. Our efforts through microarray analysis and expression database inspection have failed to identify molecular markers that distinguish the progenitor domains between the epithalamus and thalamus. By contrast, neural precursors arising from the prospective epithalamus and thalamus express markedly different transcriptional profiles at the onset of neurogenesis (Chatterjee et al., 2014)(Fig. 2,3,5). Gbx2 is one of the earliest genes that are induced as neural cells exit the cell cycle in the thalamus (Bulfone et al., 1993; Nakagawa and O'Leary, 2003; Chen et al., 2009). Here, we show that inactivation of Gbx2 prevents thalamic neurons from acquiring the thalamic identity and results in a partial conversion from thalamic to habenular molecular features. This cell-identity switch may account for the profound defects found in the thalamus lacking Gbx2, particularly in the misrouting of thalamic axons to the ventral midbrain and hindbrain (Chatterjee et al., 2012), and in the abnormal contribution of thalamus-derived neurons to the habenula due to the loss of Gbx2 (Chen et al., 2009). Therefore, Gbx2 plays a crucial role in the subdivision of the epithalamus and thalamus within the P2 compartment by suppressing the habenular identity.
Irx genes are important for pattern formation and cell fate decision (Scholpp and Lumsden, 2010; Chatterjee and Li, 2012). Prior studies have demonstrated the important roles of Irx1 and Irx3 in development of the diencephalon (Scholpp et al., 2007; Rodriguez-Seguel et al., 2009; Robertshaw et al., 2013). By comparing Irx1 expression at different positions of the developing thalamus along the anterior and posterior axis, we show that Irx1 is initially expressed throughout the P2 domain and down-regulated in neuronal precursors as they exit the cell cycle and turn on Gbx2 transcription. Importantly, by studying mouse embryos lacking Gbx2 or both Gbx2 and Pax6, we demonstrate that Gbx2 inhibits Irx genes in the thalamus (Fig. 2). In both mice and humans, the six Irx genes are grouped into two genomic clusters, IrxA (Irx1/2/4) and IrxB (Irx3/5/6) (Peters et al., 2000). Interestingly, Irx1/2/3/5 are similarly expressed in the epithalamus and the ventricular zone of the thalamus; in the absence of Gbx2, these four genes were ectopically expressed in postmitotic cells of the thalamus (Fig. 2 and data not shown), suggesting that Irx1/2/3/5, but not Irx4/6, may be controlled by common regulatory elements present in both clusters. One of the recurring themes of Irx function is to define distinct competence for inductive signals in regionalization of the neural epithelium (Kobayashi et al., 2002; Robertshaw et al., 2013). Therefore, the repression of Irx genes from the postmitotic thalamic cells may be essential for a switch from the habenular to thalamic developmental program, probably by changing the competence of thalamic precursor cells to inductive signals. This notion will be tested in future studies.
In agreement with a delay in the onset of neurogenesis in the thalamus compared to the epithalamus, robust expression of Nefm and Dcx, indicative of postmitotic neurons, is present in the epithalamus but not the thalamus at E12.5 (Fig. 3,5). Furthermore, Id4 and Ebf3, which have opposing roles in neurogenesis, are differentially expressed in the epithalamus and thalamus (Fig. 3,5). Deletion of Gbx2 leads to precocious expression of Nefm, Dcx, and Ebf3 and blocks Id4 expression in the thalamus. The loss of Gbx2 also affects cell proliferation in the thalamic VZ, particularly in the area near the epithalamus and thalamus border (Fig. 6). Therefore, the thalamus adopts not only a molecular signature but also neurogenic pattern that resemble those of the epithalamus in the absence of Gbx2. In zebrafish embryos, the neurogenetic gradient in the thalamus is controlled by her6, through interacting with neurog1 and coe2 (an Ebf gene) (Scholpp et al., 2009). We found no significant changes in the mRNA level of Hes genes, including Hes1–the her6 orthologue, in the thalamus due to Gbx2 deletion. ICH showed that Hes1 expression in the thalamus was indistinguishable between wild-type and Gbx2-KO embryos at E12.5 (Fig. S3). Therefore, Gbx2 regulates neurogenesis in the thalamus through a Hes1-independent pathway. Using genetic mosaics, we demonstrate that Gbx2 has a cell non-autonomous function probably by controlling a feedback mechanism to regulate cell proliferation and neurogenic progression in the thalamus. The precise mechanism underlying the cell non-autonomous function of Gbx2 remains to be determined. Examination of the list of DE genes has not revealed an obvious candidate. Interestingly, we have recently shown that Shh signaling has a long-range effect in the partitioning of P2 into the epithalamus and thalamus, and reduction of Shh signaling results in expansion of the epithalamus at the expense of the thalamus (Chatterjee et al., 2014). Therefore, Gbx2 may participate in the regulation of the range of Shh function during differentiation between the thalamus and epithalamus. To explore this hypothesis further, more extensive transcriptome study using RNA-Seq technology may be required to discover Gbx2 downstream targets that are involved in modulation of Shh signaling.
Supplementary Material
HIGHLIGHTS.
Genome-wide transcriptome studies identify putative Gbx2 downstream targets.
Gbx2 inhibits expression of multiple Iroquois genes in the diencephalon.
Gbx2 promotes thalamic identity and represses habenular identity.
Gbx2 controls a feedback signaling from postmitotic cells to dividing progenitors.
Table III.
Thalamus-enriched genes
| Symbol | Description | logFC | adj.P.Val | Loss in Gbx2-KO |
|---|---|---|---|---|
| Gbx2 | gastrulation brain homeobox 2 | −6.16 | 8.85E-05 | Yes |
| Sema5a | sema domain, seven thrombospondin repeats (type 1 and type 1-like), trans | −3.94 | 6.22E-05 | |
| Chst1 | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | −3.37 | 1.14E-03 | Yes |
| Syt13 | synaptotagmin XIII | −3.03 | 1.94E-03 | Yes |
| Cyp1b1 | cytochrome P450, family 1, subfamily b, polypeptide 1 | −2.99 | 2.58E-04 | Yes |
| Gng8 | guanine nucleotide binding protein (G protein), gamma 8 | −2.77 | 4.30E-03 | |
| Crabp2 | cellular retinoic acid binding protein II | −2.48 | 1.29E-03 | |
| Nrn1 | neuritin 1 | −2.24 | 2.23E-02 | |
| Id4 | inhibitor of DNA binding 4 | −2.00 | 2.58E-04 | Yes |
| Hs6st2 | heparan sulfate 6-O-sulfotransferase 2 | −2.00 | 1.26E-02 | Yes |
| Ntng1 | netrin G1 | −1.98 | 1.36E-02 | Yes |
| Sertad4 | SERTA domain containing 4 | −1.84 | 8.95E-03 | Yes |
| Robo2 | roundabout homolog 2 (Drosophila) | −1.64 | 2.59E-02 | Yes |
| Sorcs2 | sortilin-related VPS10 domain containing receptor 2 | −1.55 | 2.58E-04 | |
| Gm10304 | predicted gene 10304 | −1.54 | 4.92E-02 | Yes |
| Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | −1.53 | 2.38E-03 | Yes |
| Rora | RAR-related orphan receptor alpha | −1.53 | 1.65E-02 | Yes |
| Nr5a2 | nuclear receptor subfamily 5, group A, member 2 | −1.51 | 4.30E-03 | |
| Prmt8 | protein arginine N-methyltransferase 8 | −1.43 | 2.07E-02 | Yes |
| Osbpl3 | oxysterol binding protein-like 3 | −1.39 | 1.61E-03 | |
| Hs3st1 | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | −1.31 | 4.30E-03 | |
| Pou2f2 | POU domain, class 2, transcription factor 2 | −1.31 | 2.22E-02 | |
| Ldb2 | LIM domain binding 2 | −1.31 | 4.29E-02 | |
| Thsd7b | thrombospondin, type I, domain containing 7B | −1.28 | 1.39E-02 | Yes |
| Sel1l3 | sel-1 suppressor of lin-12-like 3 (C. elegans) | −1.21 | 2.85E-02 | Yes |
| Arhgap18 | Rho GTPase activating protein 18 | −1.12 | 1.27E-02 | |
| Tmtc2 | transmembrane and tetratricopeptide repeat containing 2 | −1.11 | 8.32E-03 | |
| Tmem132a | transmembrane protein 132A | −1.09 | 3.48E-02 | |
| Snx10 | sorting nexin 10 | −1.08 | 2.22E-02 | |
| Lima1 | LIM domain and actin binding 1 | −1.07 | 3.00E-02 | |
| 1700025G04Rik | RIKEN cDNA 1700025G04 gene | −1.05 | 3.79E-03 | |
| Sgk1 | serum/glucocorticoid regulated kinase 1 | −1.01 | 2.02E-02 | Yes |
| AI504432 | expressed sequence AI504432 | −1.01 | 2.69E-02 | |
| Fign | fidgetin | −1.01 | 1.08E-02 | |
| Fam78b | family with sequence similarity 78, member B | −1.01 | 4.65E-03 | |
| Lhx9 | LIM homeobox protein 9 | −3.94 | 2.24E-07 | |
| Olig3 | oligodendrocyte transcription factor 3 | −3.32 | 1.04E-03 | |
| Necab2 | N-terminal EF-hand calcium binding protein 2 | −1.82 | 1.10E-02 | |
| Kirrel3 | kin of IRRE like 3 (Drosophila) | −1.52 | 1.31E-02 | Yes 17/39 (43.6%) |
ACKNOWLEDGEMENTS
We thank Drs. Alan W. Leung and Iftekher Naser for their critical reading and comments on the manuscript. We thank Dr. Yashushi Nakagawa, David Ginty for providing the antibodies against Dbx1 and Islr2, Drs. Eric Turner and Chi-Chung Hui for providing Ebf3 expressing vector and cDNAs for Irx1,Irx2 and Irx3, respectively. The monoclonal anti-neurofilament antibody (2H3) was developed by T.M. Jessell and J. Dodd, and obtained through the Developmental Studies Hybridoma Bank under the auspices of the NICHD and maintained by The University of Iowa (Iowa City, IA). This work was supported by a grant from the National Institute of Health (R01MH094914).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Angevine JB., Jr. Time of neuron origin in the diencephalon of the mouse. An autoradiographic study. J Comp Neurol. 1970;139:129–187. doi: 10.1002/cne.901390202. [DOI] [PubMed] [Google Scholar]
- Bai G, Sheng N, Xie Z, Bian W, Yokota Y, Benezra R, Kageyama R, Guillemot F, Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Bedford L, Walker R, Kondo T, van Cruchten I, King ER, Sablitzky F. Id4 is required for the correct timing of neural differentiation. Dev Biol. 2005;280:386–395. doi: 10.1016/j.ydbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126:555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- Bluske KK, Vue TY, Kawakami Y, Taketo MM, Yoshikawa K, Johnson JE, Nakagawa Y. beta-Catenin signaling specifies progenitor cell identity in parallel with Shh signaling in the developing mammalian thalamus. Development. 2012;139:2692–2702. doi: 10.1242/dev.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Li JY. Patterning and compartment formation in the diencephalon. Front Neurosci. 2012;6:66. doi: 10.3389/fnins.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Guo Q, Weber S, Scholpp S, Li JY. Pax6 regulates the formation of the habenular nuclei by controlling the temporospatial expression of Shh in the diencephalon in vertebrates. BMC Biol. 2014;12:13. doi: 10.1186/1741-7007-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Li K, Chen L, Maisano X, Guo Q, Gan L, Li JY. Gbx2 regulates thalamocortical axon guidance by modifying the LIM and Robo codes. Development. 2012;139:4633–4643. doi: 10.1242/dev.086991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guo Q, Li JY. Transcription factor Gbx2 acts cell-nonautonomously to regulate the formation of lineage-restriction boundaries of the thalamus. Development. 2009;136:1317–1326. doi: 10.1242/dev.030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DR, Cheng CW, Cheng SH, Hui CC. Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech Dev. 2000;91:317–321. doi: 10.1016/s0925-4773(99)00263-4. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2011;39:D835–841. doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Marin F, Grosschedl R, Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hevner RF, Miyashita-Lin E, Rubenstein JL. Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: evidence that cortical and thalamic axons interact and guide each other. J Comp Neurol. 2002;447:8–17. doi: 10.1002/cne.10219. [DOI] [PubMed] [Google Scholar]
- Hikosaka O. The habenula: from stress evasion to value-based decision-making. Nature reviews Neuroscience. 2010;11:503–513. doi: 10.1038/nrn2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka O, Sesack SR, Lecourtier L, Shepard PD. Habenula: crossroad between the basal ganglia and the limbic system. J Neurosci. 2008;28:11825–11829. doi: 10.1523/JNEUROSCI.3463-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Nakazawa M, Muraoka O, Nakayama R, Suda Y, Hibi M. Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development. 2006;133:3993–4004. doi: 10.1242/dev.02585. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A. 2008;105:20067–20071. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Maintaining a memory by transcriptional autoregulation. Curr Biol. 2011;21:R146–147. doi: 10.1016/j.cub.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Dolson DK, Waclaw RR, Matise MP, Sussel L, Campbell K, Kaestner KH, Epstein DJ. Spatial and temporal requirements for sonic hedgehog in the regulation of thalamic interneuron identity. Development. 2011;138:531–541. doi: 10.1242/dev.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones EG. The thalamus. 2nd Edition. Cambridge University Press; Cambridge ; New York: 2007. [Google Scholar]
- Jones EG, Rubenstein JL. Expression of regulatory genes during differentiation of thalamic nuclei in mouse and monkey. J Comp Neurol. 2004;477:55–80. doi: 10.1002/cne.20234. [DOI] [PubMed] [Google Scholar]
- Kataoka A, Shimogori T. Fgf8 controls regional identity in the developing thalamus. Development. 2008;135:2873–2881. doi: 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7:1242–1249. doi: 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- Lecourtier L, Kelly PH. A conductor hidden in the orchestra? Role of the habenular complex in monoamine transmission and cognition. Neuroscience and biobehavioral reviews. 2007;31:658–672. doi: 10.1016/j.neubiorev.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Li K, Zhang J, Li JY. Gbx2 plays an essential but transient role in the formation of thalamic nuclei. PLoS One. 2012;7:e47111. doi: 10.1371/journal.pone.0047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K, Guo T, St Hillaire C, Meabon JS, Kanning KC, Bothwell M, Ginty DD. LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development. Neuron. 2009;63:614–627. doi: 10.1016/j.neuron.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ferre A, Martinez S. The development of the thalamic motor learning area is regulated by Fgf8 expression. J Neurosci. 2009;29:13389–13400. doi: 10.1523/JNEUROSCI.2625-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister JP, II, Das GD. Neurogenesis in the epithalamus, dorsal thalamus and ventral thalamus of the rat: an autoradiographic and cytological study. J Comp Neurol. 1977;172:647–686. doi: 10.1002/cne.901720407. [DOI] [PubMed] [Google Scholar]
- Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, Rubenstein JL. Early neocortical regionalization in the absence of thalamic innervation. Science. 1999;285:906–909. doi: 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci. 2001;21:2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, O'Leary DD. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- Nishida H, Suzuki T, Tomaru Y, Hayashizaki Y. A novel replication-independent histone H2a gene in mouse. BMC Genet. 2005;6:10. doi: 10.1186/1471-2156-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T, Dildrop R, Ausmeier K, Ruther U. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 2000;10:1453–1462. doi: 10.1101/gr.144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles E, Acampora D, Gogoi R, Tuorto F, Papalia A, Guillemot F, Ang SL, Simeone A. Otx2 controls identity and fate of glutamatergic progenitors of the thalamus by repressing GABAergic differentiation. J Neurosci. 2006;26:5955–5964. doi: 10.1523/JNEUROSCI.1097-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, Turner EE. Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. J Neurosci. 2009;29:14309–14322. doi: 10.1523/JNEUROSCI.2430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertshaw E, Matsumoto K, Lumsden A, Kiecker C. Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc Natl Acad Sci U S A. 2013;110:E3919–3926. doi: 10.1073/pnas.1304311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Seguel E, Alarcon P, Gomez-Skarmeta JL. The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev Biol. 2009;329:258–268. doi: 10.1016/j.ydbio.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Martinez S, Shimamura K, Puelles L. The embryonic vertebrate forebrain: the prosomeric model. Science. 1994;266:578–580. doi: 10.1126/science.7939711. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33:373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Wolf O, Brand M, Lumsden A. Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development. 2006;133:855–864. doi: 10.1242/dev.02248. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 2007;134:3167–3176. doi: 10.1242/dev.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Delogu A, Gilthorpe J, Peukert D, Schindler S, Lumsden A. Her6 regulates the neurogenetic gradient and neuronal identity in the thalamus. Proc Natl Acad Sci U S A. 2009;106:19895–19900. doi: 10.1073/pnas.0910894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 2009;12:1373–1380. doi: 10.1038/nn.2409. [DOI] [PubMed] [Google Scholar]
- Sherman SM, Guillery RW. Exploring the thalamus and its role in cortical function. 2nd Edition. MIT Press; Cambridge, Mass.: 2006. [Google Scholar]
- Smart IH. Proliferative characteristics of the ependymal layer during the early development of the mouse diencephalon, as revealed by recording the number, location, and plane of cleavage of mitotic figures. J Anat. 1972;113:109–129. [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–323. doi: 10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vue TY, Bluske K, Alishahi A, Yang LL, Koyano-Nakagawa N, Novitch B, Nakagawa Y. Sonic hedgehog signaling controls thalamic progenitor identity and nuclei specification in mice. J Neurosci. 2009;29:4484–4497. doi: 10.1523/JNEUROSCI.0656-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, Nakagawa Y. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol. 2007;505:73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- Wang L, Bluske KK, Dickel LK, Nakagawa Y. Basal progenitor cells in the embryonic mouse thalamus - their molecular characterization and the role of neurogenins and Pax6. Neural Dev. 2011;6:35. doi: 10.1186/1749-8104-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Lewandoski M, Campbell K, Joyner AL, Rubenstein JL, Martinez S, Martin GR. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development. 1997;124:2923–2934. doi: 10.1242/dev.124.15.2923. [DOI] [PubMed] [Google Scholar]
- Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- Zhou CJ, Pinson KI, Pleasure SJ. Severe defects in dorsal thalamic development in low-density lipoprotein receptor-related protein-6 mutants. J Neurosci. 2004;24:7632–7639. doi: 10.1523/JNEUROSCI.2123-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.