Background: Triphosphate tunnel metalloenzymes carry out diverse enzymatic reactions.
Results: Two metal co-factors are identified involved in substrate binding and in catalysis.
Conclusion: A unified catalytic mechanism is proposed and biochemically investigated.
Significance: The functional diversity of TTM enzymes is rationalized by a common mechanism that allows very different substrates to be bound and processed.
Keywords: adenylate cyclase (adenylyl cyclase), enzyme mechanism, manganese, nuclear magnetic resonance (NMR), protein structure, x-ray crystallography, inorganic polyphosphatase, thiamine triphosphatase
Abstract
Triphosphate tunnel metalloenzymes (TTMs) are present in all kingdoms of life and catalyze diverse enzymatic reactions such as mRNA capping, the cyclization of adenosine triphosphate, the hydrolysis of thiamine triphosphate, and the synthesis and breakdown of inorganic polyphosphates. TTMs have an unusual tunnel domain fold that harbors substrate- and metal co-factor binding sites. It is presently poorly understood how TTMs specifically sense different triphosphate-containing substrates and how catalysis occurs in the tunnel center. Here we describe substrate-bound structures of inorganic polyphosphatases from Arabidopsis and Escherichia coli, which reveal an unorthodox yet conserved mode of triphosphate and metal co-factor binding. We identify two metal binding sites in these enzymes, with one co-factor involved in substrate coordination and the other in catalysis. Structural comparisons with a substrate- and product-bound mammalian thiamine triphosphatase and with previously reported structures of mRNA capping enzymes, adenylate cyclases, and polyphosphate polymerases suggest that directionality of substrate binding defines TTM catalytic activity. Our work provides insight into the evolution and functional diversification of an ancient enzyme family.
Introduction
Inorganic polyphosphate (polyP) is a linear polymer of orthophosphate units joined by phosphoanhydride bonds. It occurs ubiquitously and abundantly in all life forms (1). In bacteria, polyP kinases generate polyP from ATP, but it is presently unknown how the cellular polyP stores of higher organisms are being synthesized (2, 3). We previously reported a fungal polyP polymerase that is distinct from bacterial polyP kinases (4, 5). The yeast enzyme resides in the vacuolar transporter chaperone (VTC)2 membrane protein complex, which generates polyP from ATP in the cytosol and translocates the growing chain into the vacuole (4, 6). The catalytic core of VTC maps to a cytoplasmic 8-stranded β-tunnel domain in Vtc4p, which harbors binding sites for the nucleotide substrate, for a manganese metal co-factor and for an orthophosphate priming the polymerase reaction (4).
The Vtc4p β-tunnel domain is not unique to eukaryotic polyP polymerases but is a structural hallmark of triphosphate tunnel metalloenzymes (TTMs), whose characteristic features are the presence of a topologically closed hydrophilic β-barrel, the preference for triphosphate-containing substrates, and for a divalent metal co-factor (PFAM (7) families CYTH (8) and VTC (9, 10)). Founding members of the TTM family were fungal (11, 12), protozoal (9), and viral RNA triphosphatases (13–16). Subsequently, other enzymes with very similar tunnel topologies were discovered, including the bacterial class IV adenylate cyclase cyaB (17, 18), mammalian thiamine triphosphatases (ThTPases) (19–21), and long- (22) and short-chain (23–26) inorganic polyphosphatases. Finally, the tunnel domain fold is also found in the mediator head complex subunits Med18 and Med20, but these proteins appear to have lost catalytic activity (27).
A staggering number of different catalytic activities and substrate preferences has been reported within the TTM family (10). However, the lack of substrate- and product-bound structures has made it difficult to define the sequence-fingerprints responsible for a specific enzyme activity in individual family members.
Experimental Procedures
Protein Expression and Purification
AtTTM3 (Uniprot ID Q9SIY3) was cloned into vector pMH-HT providing an N-terminal His6 tag and a tobacco etch virus protease cleavage site. Protein expression in Escherichia coli BL21 (DE3) RIL to A600 nm = 0.6 was induced with 0.25 mm isopropyl β-d-galactoside in terrific broth at 16 °C for 16 h. Cells were collected by centrifugation at 4500 × g for 30 min, washed in PBS buffer, centrifuged again at 4500 × g for 15 min, and snap-frozen in liquid nitrogen. For protein purification cells were resuspended in lysis buffer (50 mm Tris-Cl (pH 8.0), 500 mm NaCl, 5 mm 2-mercaptoethanol), homogenized (Emulsiflex C-3, Avestin), and centrifuged at 7000 × g for 60 min. The supernatant was loaded onto a Ni2+ affinity column (HisTrap HP 5 ml, GE Healthcare), washed with 50 mm Tris (pH 8), 1 m NaCl, 5 mm 2-mercaptoethanol, and eluted in lysis buffer supplemented with 200 mm imidazole (pH 8.0). The His6 tag was cleaved with tobacco etch virus for 16 h at 4 °C during dialysis against lysis buffer. AtTTM3 was further purified by a second Ni2+ affinity step and by gel filtration on a Superdex 75 HR26/60 column (GE Healthcare) equilibrated in 25 mm Tris (pH 7.2), 250 mm NaCl, 5 mm 2-mercaptoethanol. Monomeric peak fractions were dialyzed against 20 mm Hepes (pH 7.4), 50 mm NaCl, 0.5 mm tris(2-carboxyethyl)phosphine and concentrated to 35 mg/ml for crystallization. Protein concentrations were estimated by protein absorption at 280 nm using the calculated molecular extinction coefficient. Site-specific mutations were introduced by PCR, and mutant proteins were purified like wild type.
The coding sequence of ygiF was amplified from genomic DNA (E. coli Mach 1 cells, Life Technologies) by PCR, and a synthetic gene coding for full-length mouse ThTPase (Uniprot ID Q8JZL3) and codon-optimized for expression in E. coli was obtained from Geneart (Life Technologies). Coding sequences were cloned into plasmid pMH-HT, and expression and purification were performed as described for AtTTM3.
Crystallization and Data Collection
Tetragonal AtTTM3 crystals (form A) developed at room temperature from hanging drops composed of 1.5 μl of protein solution and 1.5 μl of crystallization buffer (2.6 m NaCl, 0.1 m citric acid/NaOH (pH 5.0)) suspended over 1.0 ml of the latter as reservoir solution. Crystals were transferred in reservoir solution supplemented with 20% (v/v) ethylene glycol and 0.5 m KI for 20 s and snap-frozen in liquid nitrogen. Single-wavelength anomalous diffraction (SAD) data were collected on a Rigaku MicroMax rotating anode equipped with a copper filament, osmic mirrors, and an R-AXIS IV++ detector. Subsequently, an isomorphous native dataset was collected on a crystal originating from the same crystallization drop (see Table 1). A monoclinic crystal form (form B) developed in 20% (w/v) PEG 3350, 0.2 m NaCl, 0.1 m Bis-Tris (pH 7.0) and diffracted up to 1.3 Å resolution. Data processing and scaling were done with XDS (November 2014 version) (28). Hexagonal crystals for full-length ygiF grew at room temperature in hanging drops (1.5 μl and 1.5 μl) containing 0.2 m NaCl, 20 (w/v) PEG 3350. Crystals were transferred into crystallization buffer supplemented with 20% (v/v) ethylene glycol and snap-frozen in liquid nitrogen. For phasing, 0.1 m NaI was added to the cryo solution, and crystals were soaked for 5 min. SAD data were collected at a wavelength of 1.9 Å (see Table 2). Tetragonal mouse ThTPase crystals (form A) developed at room temperature from hanging drops composed of 1.5 μl of protein solution and 1.5 μl of crystallization buffer (27% (w/v) PEG 3350, 0.1 m Tris (pH 9.0), 0.2 m MgCl2) suspended over 1.0 ml of the latter as reservoir solution. Crystals were transferred into a reservoir solution supplemented with 20% (v/v) ethylene glycol and snap-frozen in liquid nitrogen. Monoclinic crystals (form B) developed in 1.6 m sodium/potassium phosphate buffer pH 6.8 (1:1 ratio) and were cryo-protected by stepwise transfer into a solution containing 1.6 m sodium/potassium phosphate buffer pH 6.8 and 20% (v/v) ethylene glycol.
TABLE 1.
Crystallographic data collection and refinement statistics for AtTTM3
Highest resolution shell is shown in parenthesis.
| AtTTM3 NaI soak (Co Kα) | AtTTM3/PPPi/Mg2+ (form A) | AtTTM3/PPPi/Mn2+ (form A) | AtTTM3/PPPi/Mn2+ (form A; Mn-K edge) | AtTTM3/PPPi/Mn2+ (form B) | AtTTM3/PPPi/Mn2+ (form B, Mn-K edge) | AtTTM3/Pi/Mn2+ | |
|---|---|---|---|---|---|---|---|
| PDB ID | 5a5y | 5a66 | 5a67 | 5a68 | |||
| Data collection | |||||||
| Beam line | In house | SLS PXII | SLS PXII | SLS PXII | SLS PXIII | SLS PXIII | SLS PXII |
| Wavelength (Å) | 1.54 | 1.0 | 1.0 | 1.8 | 1.0 | 1.82 | 1.0 |
| Space group | I422 | I422 | I422 | I422 | P21 | P21 | P21 |
| Cell dimensions | |||||||
| a, b, c (Å) | 136.03, 136.03, 145.64 | 136.33, 136.33, 144.20 | 136.27, 136.27, 145.48 | 136.32, 136.32, 145.99 | 44.37, 33.87, 72.22 | 44.36, 33.87, 72.32 | 44.22, 33.81, 71.81 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 94.96, 90 | 90, 94.98, 90 | 90, 94.4, 90 |
| Resolution (Å) | 19.76-2.60 | 45.33-1.92 | 48.18-2.05 | 48.20-2.76 | 19.61-1.30 | 44.19-2.52 | 43.20-1.67 |
| (2.76-2.60) | (2.04-1.92) | (2.17-2.05) | (2.93-2.76) | (1.37-1.30) | (2.67-2.52) | (1.77-1.67) | |
| Rsym | 0.145 (0.763) | 0.076 (1.212) | 0.088 (0.74) | 0.266 (1.117) | 0.046 (0.772) | 0.038 (0.061) | 0.085 (0.615) |
| I/σI | 12.11 (1.96) | 20.06 (2.18) | 19.13 (2.20) | 12.02 (1.82) | 20.04 (1.94) | 25.29 (15.68) | 9.5 (1.73) |
| Completeness (%) | 98.8 (93.9) | 98.5 (90.8) | 98.0 (88.1) | 98.1 (88.6) | 98.3 (93.3) | 98.4 (95.0) | 94.4 (76.8) |
| Redundancy | 5.02 (3.75) | 10.59 (8.35) | 11.73 (6.47) | 8.34 (6.52) | 6.24 (5.27) | 3.30 (3.08) | 4.1 (2.95) |
| Refinement | |||||||
| Resolution (Å) | 45.33-1.92 | 48.18–2.05 | 19.61-1.30 | 43.20-1.67 | |||
| No. reflections | 46,150 | 38,451 | 50,172 | 21,931 | |||
| Rwork/Rfree | 0.153/0.198 | 0.182/0.220 | 0.138/0.174 | 0.182/0.222 | |||
| No. atoms | |||||||
| Protein | 3455 | 3455 | 1775 | 1670 | |||
| Pi/PPi/PPPi | 26 | 26 | 13 | 10 | |||
| Mg2+/Mn2+ | 2 | 2 | 2 | 3 | |||
| Water | 239 | 227 | 248 | 135 | |||
| B-factors | |||||||
| Protein | 38.60 | 30.07 | 15.07 | 15.65 | |||
| Pi/PPi/PPPi | 34.78 | 29.41 | 18.34 | 34.16 | |||
| Mg2+/Mn2+ | 36.00 | 29.97 | 20.50 | 21.43 | |||
| Water | 44.82 | 40.89 | 35.33 | 39.39 | |||
| r.m.s.d. | |||||||
| Bond lengths (Å) | 0.007 | 0.006 | 0.013 | 0.010 | |||
| Bond angles (°) | 1.268 | 1.098 | 1.377 | 1.486 | |||
| Ramachandran plot (%) | |||||||
| Favored | 99.3 | 99.3 | 100 | 100 | |||
| Outliers | 0 | 0 | 0 | 0 | |||
TABLE 2.
Crystallographic data collection and refinement statistics for ygiF
Highest resolution shell is shown in parenthesis.
| ygiF (NaI soak) | ygiF/PPPi/Mg2+ | ygiF/PPPi/Mn2+ | ygiF/PPPi/Mn2+ (Mn-K edge) | |
|---|---|---|---|---|
| PDB ID | 5a60 | 5a61 | ||
| Data collection | ||||
| Beam line | SLS PXII | SLS PXII | SLS PXII | SLS PXII |
| Wavelength (Å) | 1.9 | 1.0 | 1.0 | 1.7 |
| Space group | P65 | P65 | P65 | P65 |
| Cell dimensions | ||||
| a = b, c (Å) | 91.11, 125.56 | 89.92, 125.58 | 89.68, 125.33 | 89.95, 125.31 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 49.127-2.70 | 48.88-1.82 | 48.77-1.50 | 48.74-1.86 |
| (2.87-2.70) | (1.93-1.82) | (1.59-1.50) | (1.97-1.86) | |
| Rsym | 0.077 (0.186) | 0.068 (0.944) | 0.036 (1.20) | 0.037 (0.313) |
| I/σI | 31.08 (11.28) | 14.91 (2.18) | 23.90 (1.93) | 26.80 (3.91) |
| Completeness (%) | 95.8 (90.2) | 98.8 (96.9) | 99.7 (98.3) | 97.9 (90.2) |
| Redundancy | 19.63 (18.47) | 9.70 (9.06) | 15.48 (14.18) | 8.30 (3.55) |
| Refinement | ||||
| Resolution (Å) | 48.88-1.82 | 48.77–1.50 | ||
| No. reflections | 47,520 | 86,573 | ||
| Rwork/ Rfree | 0.150/0.189 | 0.151/0.171 | ||
| No. atoms | ||||
| Protein | 3398 | 3395 | ||
| Pi/PPi/PPPi | 13 | 13 | ||
| Mg2+/Mn2+ | 2 | 2 | ||
| Water | 289 | 295 | ||
| B-factors | ||||
| Protein | 55.83 | 52.41 | ||
| Pi/PPi/PPPi | 46.83 | 42.84 | ||
| Mg2+/Mn2+ | 46.65 | 42.53 | ||
| Water | 58.34 | 58.82 | ||
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.007 | 0.006 | ||
| Bond angles (°) | 1.149 | 1.186 | ||
| Ramachandran plot (%) | ||||
| Favored | 99.1 | 99.3 | ||
| Outliers | 0 | 0 | ||
Co-crystallization and Soaking Experiments
AtTTM3-PPPi-Mg2+/Mn2+ form A crystals were transferred into the soaking solution (2.6 m NaCl, 0.1 Bis-Tris (pH 5), 10 mm sodium tripolyphosphate (Sigma), 10 mm MgCl2 (or MnCl2), 20% (v/v) ethylene glycol) by serial transfer to replace the citrate otherwise bound to the tunnel center.
For AtTTM3-PPPi-Mg2+/Mn2+, form B crystals were soaked for 20–30 min in 20% (w/v) PEG 3350, 0.2 m NaCl, 0.1 m Bis-Tris pH 7.0, 20% (v/v) ethylene glycol, 10 mm sodium tripolyphosphate, 5 mm MnCl2 using the same procedure as outlined above (substrate-bound structure).
For AtTTM3-Pi-Mn2+, AtTTM3 was co-crystallized with 5 mm sodium tripolyphosphate and 10 mm MnCl2 (product-bound structure).
For ygiF-PPPi-Mg2+/Mn2+, the substrate-bound complex was obtained by soaking (20–30 min) ygiF apo crystals in their crystallization buffer supplemented with 20% (v/v) glycerol, 10 mm sodium tripolyphosphate and 5 mm MgCl2 or MnCl2. ThTPase-ThTP-Mg2+. Form B crystals were soaked in crystallization buffer containing 20% (v/v) glycerol, 10 mm thiamine triphosphate and 10 mm MgCl2.
For ThTPase-ThDP-Mg2+, the product-bound structure was obtained by soaking form A crystals in a solution containing 10 mm sodium tripolyphosphate and 10 mm MnCl2 for 30 min. Two data sets were collected, one with λ = 1.0 Å and one close to the Mn-K edge (λ = 1.9 Å). No anomalous signal was found in the latter data set, possibly due to the high MgCl2 (0.2 m) concentration present in the crystallization buffer.
Structure Solution and Refinement
To solve the structure of AtTTM3, SAD and native data were scaled together with the program XPREP (Bruker AXS, Madison, WI) for SIRAS (single isomorphous replacement with anomalous scattering) phasing. The program SHELXD (29) was used to locate 37 iodine sites. Consistent sites were input in the program SHARP (30) for site refinement and phasing at 3.0 Å resolution. Density modification and phase extension to 2.6 Å was carried out with PHENIX.RESOLVE (31). The structure was built in alternating cycles of model correction in COOT (32) and restrained refinement in refmac5 (33) against a high resolution native data set (Table 1). The structure of crystal form B was determined by molecular replacement with the program PHASER (34).
The structure of ygiF was solved by scaling redundant SAD data using XPREP. SHELXD located 15 consistent iodine and sulfur sites, which were input into SHARP for SAD site refinement and phasing at 2.7 Å resolution. Density modification and phase extension to 2.15 Å was carried out with PHENIX.RESOLVE (Table 2). The structure of mouse ThTPase was solved using the molecular replacement methods as implemented in the program PHASER and using the structure of the human ThTPase (PDB ID 3BHD) as a search model. Analysis with MolProbity (35) indicated excellent stereochemistry for all refined models. Phasing and refinement statistics are summarized in Tables 1–3.
TABLE 3.
Crystallographic data collection and refinement statistics for mouse ThTPase
Highest resolution shell is shown in parenthesis.
| mThTPase/ThTP/Mg2+ | mThTPase/ThDP/Pi/Mg2+ | |||
|---|---|---|---|---|
| PDB ID | 5a64 | 5a65 | ||
| Data collection | ||||
| Beam line | ESRF ID29 | SLS 5PXIII | ||
| Wavelength (Å) | 0.98 | 1.0 | ||
| Space group | C2 | P43212 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 103.18, 93.58, 70.78 | 105.48, 105.48, 111.81 | ||
| α, β, γ (°) | 90, 93.04, 90 | 90, 90, 90 | ||
| Resolution (Å) | 70.68-2.10 (2.23-2.10) | 49.40-1.98 (2.10-1.98) | ||
| Rsym | 0.059 (0.581) | 0.11 (2.03) | ||
| I/σI | 19.34 (2.36) | 19.66 (1.90) | ||
| Completeness (%) | 99.5 (98.3) | 99.7 (98.6) | ||
| Redundancy | 4.58 (4.49) | 25.60 (23.85) | ||
| Refinement | ||||
| Resolution (Å) | 70.68-2.10 | 49.40-1.98 | ||
| No. reflections | 37,148 | 42,091 | ||
| Rwork/ Rfree | 0.212/0.232 | 0.208/0.258 | ||
| No. atoms | ||||
| Protein | 3102 | 3117 | ||
| ThTP/Pi | 60 | 62 | ||
| Mg2+/Mn2+ | 2 | 4 | ||
| Water | 148 | 142 | ||
| B-factors | ||||
| Protein | 13.54 | 38.56 | ||
| Pi/PPi/PPPi | 50.08 | 60.08 | ||
| Mg2+/Mn2+ | 44.18 | 43.38 | ||
| Water | 44.88 | 53.05 | ||
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.012 | 0.010 | ||
| Bond angles (°) | 1.46 | 1.515 | ||
| Ramachandran plot (%) | ||||
| Favored | 98.0 | 99.0 | ||
| Outliers | 0 | 0 | ||
Synthesis of Thiamine Triphosphate
Thiamine triphosphate was synthesized as described (36) and purified by preparative high performance liquid chromatography.
NMR Time-course Experiment
A series of one-dimensional 31P NMR experiments was acquired at 310 K with a 600-MHz Bruker Avance-III spectrometer using a QXI probe-head allowing direct detection of 31P and equipped with a z-spoil gradient coil. 31P spectra were recorded using a relaxation delay of 1 s and an acquisition time of 42.6 ms (spectral width = 12,019.23 Hz). 128 scans were collected, resulting in a measurement time of 140 s per spectrum. 512 spectra were collected over a total acquisition time of ∼20 h. The enzymatic reaction was performed using 50 nm AtTTM3 and 5 mm sodium tripolyphosphate in 20 mm Bis-Tris propane (pH 8.5), 250 mm NaCl, 5 mm MgCl2 mixture. Deuterated water was added to a final concentration of 20% to the reaction mix before starting the experiment. Spectral parameters were calibrated and optimized on a 5 mm sodium tripolyphosphate sample to minimize time loss between the beginning of the reaction and the beginning of its observation by NMR. Spectra were processed using Topspin (version 2.1.6) (Bruker).
Phosphohydrolase Activities of AtTTM3 and ygiF Mutant Proteins
For the determination of the phosphohydrolase activity 2.5 nm AtTTM3 were incubated with 0.5 μm concentrations of the different phosphate-containing substrates in reaction buffer (150 mm NaCl, 20 mm Bis-Tris propane (pH 8.5), 5 mm MgCl2) at 37 °C. The reaction was stopped after 10 min, and the amount of free inorganic phosphate released was measured using the malachite green assay with minor modifications (37). 100 μl of reaction solution were mixed with 28 μl of dye mix (3 mm malachite green, 15% (v/v) sulfuric acid, 1.5% molybdate (w/v), 0.2% (v/v) Tween 20). After 5 min of incubation with the dye, the absorption at 595 nm was measured using a synergy H4 plate reader (Biotek). For each substrate a blank reaction was prepared lacking the enzyme. Controls either contained EDTA to a final concentration of 5 mm, or the enzyme was heat-inactivated at 95 °C for 5 min. To compare wild-type and mutant versions of AtTTM3 and ygiF, enzyme concentrations were increased to 180 nm to detect residual activity for some of the mutant proteins. Experiments were performed in triplicate; average values are plotted ±S.D.
Thermal shift assays were performed as previously described (38). Sypro Orange (Sigma) was added to wild-type and mutant AtTTM3 proteins in 25 mm Tris (pH 8.0), 250 mm NaCl, 5 mm 2-mercaptoethanol mixed with Sypro Orange to a final protein concentration of 10 μm. The protein-dye mixtures were loaded into a 96-well RT-PCR plate (Thermo Scientific), and measurements were performed using a C1000 thermal cycler (Bio-Rad). The fluorescence of SYPRO Orange was continously monitored at 570 nm, as a temperature gradient was applied (0.05 °C/s from 10 °C to 95 °C). Data were analyzed using the CFX Manager software (Bio-Rad). The maximum of the first derivative for each melting curve indicates the melting point of the protein. Experiments were performed in triplicate; average values are plotted ±S.D.
Polyphosphate Detection by UREA PAGE
Reactions contained 50 μm concentrations of the respective enzyme (AtTTM3, ygiF, mouse ThTPase, Vtc4p) in 150 mm NaCl, 20 mm Bis-Tris propane (pH 8.3), 1 mm MnCl2, and 10 mm ATP as substrate. Reactions were incubated at room temperature overnight. The reaction was stopped by adding Proteinase K (Sigma), and the resulting samples were mixed 1:1 with sample buffer (50% (w/v) urea, 2× Tris borate-EDTA, 20 mm EDTA (pH 8.0), 20% (v/v) glycerol, 0.25% (w/v) bromphenol blue). Samples were loaded onto a Tris borate-EDTA (TBE)-urea polyacrylamide gel (1× TBE, 7 m urea, 15% (v/v) polyacrylamide (19:1 acrylamide/bisacrylamide), 0.06% (w/v) tetramethylethylenediamine, 0.6% (w/v) ammonium persulfate) and stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI).
Results and Discussion
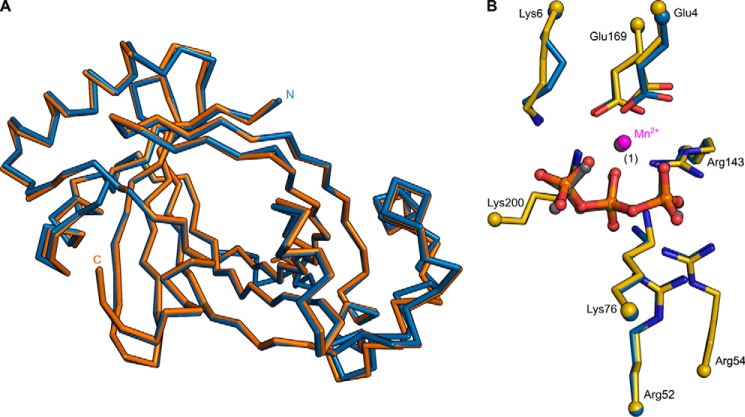
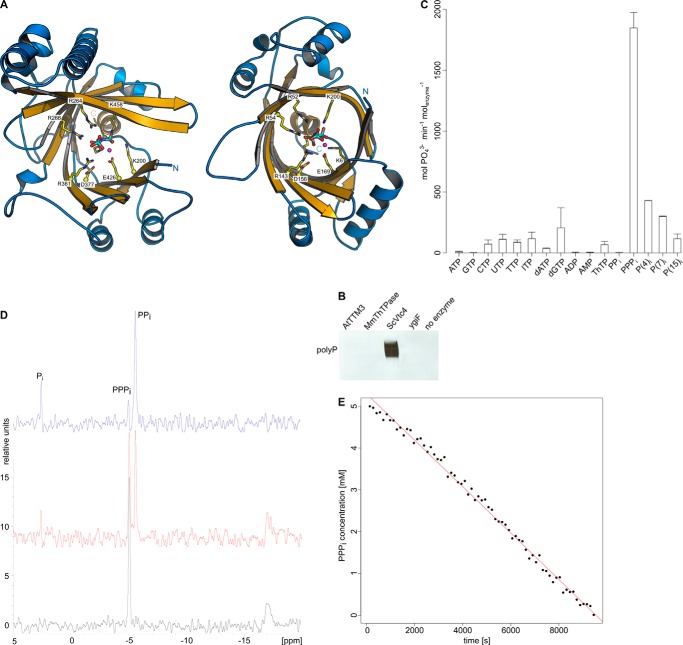
Our initial aim was to identify a polyP polymerase in plants using a combined structural and biochemical screen. We located three putative TTM proteins in Arabidopsis thaliana. AtTTM3 shares 12% sequence identity with Vtc4p (r.m.s.d. is 2.6 Å, comparing 149 corresponding Cα atoms and 1.1 Å comparing 77 Cα atoms in the tunnel center) and contains a conserved β-tunnel domain (26). We determined co-crystal structures of AtTTM3 with tripolyphosphate and Mg2+ or Mn2+ metal co-factors in two independent crystal lattices (Fig. 1). Crystal form B corresponds to the previously reported structure of AtTTM3 (PDB ID 3v85) in complex with a citric acid molecule (r.m.s.d is 0.3 Å comparing 203 corresponding Cα atoms) (26). Our structures reveal a conserved mode of substrate and metal co-factor binding in AtTTM3 and Vtc4p and an invariant arrangement of residues in the tunnel center (Fig. 2A) (4). We thus tested whether AtTTM3 can polymerize polyP from ATP or other nucleotide substrates but could not detect such an activity (Fig. 2B). Instead, AtTTM3 has specific, Mg2+ ion-dependent, short-chain polyphosphatase activity (Fig. 2C) as previously reported (26). We find that AtTTM3 hydrolyzes tripolyphosphate (PPPi) into pyro (PPi)- and orthophosphate (Pi) with a turnover rate of ∼10/s (Fig. 2, D and E). The Pi release from PPPi amounts to 27 ± 6/s when assayed by malachite green (see “Experimental Procedures”). AtTTM3 is not able to hydrolyze PPi but appears to catalyze the asymmetric cleavage of inorganic polyPn (n = 3–15), releasing Pi (Fig. 2, C and D).
FIGURE 1.
A, structural superposition of AtTTM3 crystals forms A (blue) and B (orange). The structures align with an r.m.s.d. of 0.5 Å comparing 202 corresponding Cα atoms. B, close-up of the tunnel domain centers in forms A and B (with selected side chains shown in bond representation), both bound to PPPi (in bond representation) and Mn2+ (magenta/gray spheres).
FIGURE 2.
AtTTM3 is a short-chain inorganic polyphosphatase. A, ribbon diagrams of the yeast inorganic polyphosphate polymerase Vtc4p (left panel) and Arabidopsis TTM3 (right panel). Tunnel domain β-strands are shown in yellow, and the α-helices are in blue. The TTM domain is topologically closed on one side by a C-terminal plug helix. Conserved residues in the tunnel center are shown (in bond representation, in yellow) alongside with the triphosphate substrates and Mn2+ ions (magenta spheres). B, DAPI stained UREA page gel reveals that only Vtc4p can generate inorganic polyphosphate from ATP. C, substrate specificity of AtTTM3. Pi release is measured for a range of different di- and triphosphate containing potential substrates. D, a one-dimensional 31P NMR time-course experiment reveals that AtTTM3 specifically generates PPi and Pi from PPPi. t = 0 min (black line), 94 min (red line), and 141 min (blue line). E, decay of the PPPi substrate concentration during the NMR time-course experiment indicates a turnover number of ∼10/s.
Plant and Bacterial Tripolyphosphatases Contain Two Metal Coordination Sites
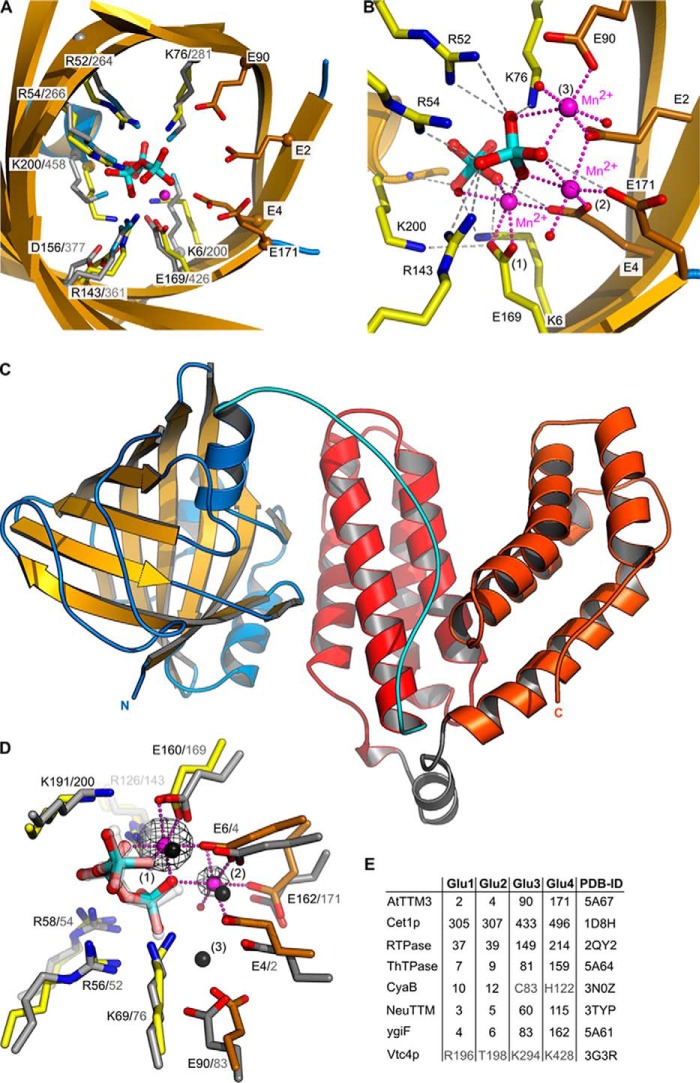
Comparison of AtTTM3 and Vtc4p revealed that four basic residues, which lead the growing polyP chain away from the active site in Vtc4p, are specifically replaced by four glutamate residues (Glu-2AtTTM3, Glu-4AtTTM3, Glu-90AtTTM3, and Glu171AtTTM3) in AtTTM3 (Fig. 3, A and E) (4). These residues, which form an acidic patch in the vicinity of the AtTTM3 tripolyphosphate substrate, are conserved among diverse TTM proteins with different catalytic activities (Figs. 3E and 4). The corresponding residues in the RNA triphosphatase Cet1p (Glu305Cet1p, Glu307Cet1p, and Glu-496Cet1p) coordinate a Mn2+ ion (11), and mutation of these residues to alanine or aspartate impairs catalysis in fungal (12, 39), viral (13–16, 40,), and protozoan (9, 41) RNA triphosphatases, in a broad-range polyphosphatase from Clostridium thermocellum (22) and in a TTM adenylate cyclase (18).
FIGURE 3.
An acidic patch in TTM proteins allows for the coordination of two metal ions. A, structural superposition of AtTTM3 (in yellow) and Vtc4p (in gray) (r.m.s.d. is 1.1 Å comparing 77 corresponding Cα atoms in the tunnel center) identifies an acidic patch in AtTTM3 (in orange, in bond representation), which is absent Vtc4p. The AtTTM3 tripolyphosphate substrate and a Mn2+ ion (magenta sphere) are shown alongside. B, structure of a AtTTM3 post-catalysis complex reveals two orthophosphates and three Mn2+ ions bound to the tunnel center. C, ribbon diagram of full-length ygiF from E. coli. The tunnel domain (residues 1–200) is shown in yellow (β-strands) and blue (α-helices), the linker region (residues 201–220) is highlighted in cyan, and the four-helix bundles of the C-terminal CHAD domain are depicted in red (residues 221–324) and orange (residues 383–433), respectively. D, structural superposition (r.m.s.d. is 0.65 Å comparing 75 corresponding Cα atoms) of the ygiF tunnel core (in yellow, the acidic patch is in orange) bound to PPPi and Mn2+ ions (magenta spheres) with the AtTTM3 product complex (in gray). A phased omit difference density map contoured at 25 σ is shown alongside (black mesh). Note that two of the three AtTTM3 metal coordination sites are also found in ygiF. E, table comparison of acidic patch residues in different TTM proteins. RTPase, RNA triphosphatase.
FIGURE 4.
TTM proteins with different enzymatic activities harbor two metal ion centers. A, structural superposition of a AtTTM3-PPPi-Mn2+ complex (in yellow, in bond representation) with the Vtc4p-ANP-Mn2+ complex (PDB ID 3G3R, in gray) (r.m.s.d. is 2.7 Å comparing 151 corresponding Cα atoms) reveals a similar mode of substrate and metal co-factor binding to site 1. Glu-4 from the acidic patch in AtTTM3 is, however, not found in Vtc4p. B, comparison of AtTTM3-PPPi-Mn2+ (in yellow) with the product bound state (r.m.s.d. is 0.3 Å comparing 203 corresponding Cα atoms) reveals that the two orthophosphates in the product complex align with the terminal phosphates of the PPPi substrate. One of the three Mn2+ ions (bound to site 1) is found consistently in both structures. C, structural superposition (r.m.s.d. is 2.2 Å comparing 138 corresponding Cα atoms) of ygiF bound to PPPi and to 2 Mn2+ ions located in sites 1 and 2 with the RNA triphosphatase Cet1p (PDB ID 1D8H, in gray) reveals that the Cet1p manganese ion previously reported maps to metal binding site 2. D, comparisons of the ygiF-PPPi-Mn2+ complex with the product-bound state of the adenylate cyclase cyaB (PDB ID 3N10, in gray) again reveals two conserved metal binding sites. The Mn2+ ion bound to site 2 in the case of cyaB is coordinated by His-122.
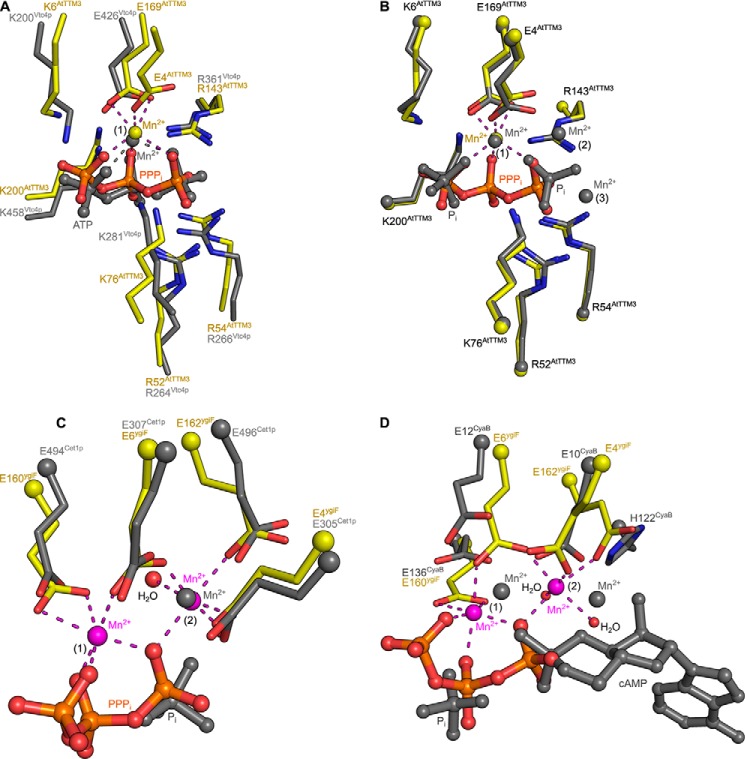
Based on kinetic and mutational studies, one (23) and two-metal (40, 41) mechanisms have been proposed for TTMs, but only a few substrate complexes in the presence of metal co-factors have been reported thus far (4, 18). As AtTTM3 efficiently hydrolyzes PPPi in the presence of Mg2+ ions only at neutral or basic pH, we used two crystal forms grown at pH 5.0 (form A) and pH 7.5 (form B) to crystallize substrate-metal complexes under non-hydrolyzable and hydrolyzable conditions, respectively. In both crystal forms, we determined structures of AtTTM3 in the presence of PPPi and either MgCl2 or MnCl2. In all our structures the catalytic Mg2+ ions can be substituted by Mn2+ ions, for which we calculated phased anomalous difference electron density maps from data collected near the Mn-K edge to confirm their modeled positions (Tables 1 and 2). At pH 5.0 (form A) we find a triphosphate moiety bound in the AtTTM3 active site. Here, PPPi coordinates a Mg2+/Mn2+ ion that acts as a bridge between the substrate and the invariant Glu-169AtTTM3 (Fig. 3A). The same metal co-factor coordination has been previously found in a Vtc4p AppNHp-Mn2+ complex (4) (Figs. 3A and 4A). A short soak at pH 7.0 (form B) shows the same arrangement of PPPi and Mn2+ in the active site (Fig. 1). However, when we co-crystallized AtTTM3 with its substrate at neutral pH, we found PPPi hydrolyzed and the tunnel center occupied by two orthophosphates and three Mn2+ ions (Fig. 3B). One of these Mn2+ ions is again found coordinated by the two phosphates (which correspond to the α and γ positions in PPPi; Fig. 4B) and by Glu-169AtTTM3, whereas the remaining metal coordination centers are formed by Glu-2AtTTM3, Glu-4AtTTM3, Glu-90AtTTM3, and Glu171AtTTM3 from the acidic patch conserved among many TTMs (Fig. 3, B and E).
To assess the functional relevance of the observed metal co-factors in AtTTM3, we studied substrate binding and metal ion coordination in the evolutionary distant TTM ygiF from E. coli. YgiF shares ∼18% sequence identity with AtTTM3 and has the same catalytic activity and substrate specificity (25, 26). The structure of full-length ygiF was solved using iodide/sulfur SAD phasing and reveals the conserved TTM fold (r.m.s.d. with the AtTTM3 tunnel domain is 2.0 Å compared with 172 corresponding Cα atoms, and 0.65 Å comparing 75 corresponding Cα in the tunnel center) (Fig. 3C). ygiF contains a second, α-helical domain, which is connected to the TTM by a well ordered linker (residues 201–221, cyan in Fig. 3C) and which has been previously annotated as CHAD domain (8). The domain folds into two four-helix bundles of similar architecture and connectivity forming a V-shaped assembly (shown in red and orange in Fig. 3C). A structural homology search with the program DALI (42) returned no significant hits, and the molecular function of this domain, which is often found in bacterial metallophosphoesterases (8), remains to be elucidated.
We next solved structures of ygiF in complex with PPPi and in the presence of either MgCl2 or MnCl2 (Table 2). The ygiF triphosphate substrate is bound in the same conformation as observed in AtTTM3, and we could identify two metal co-factors. One Mg2+/Mn2+ ion is again found coordinated by tripolyphosphate and by Glu-160ygiF, which corresponds to Glu-169 in AtTTM3 (position 1 in Fig. 3D). The second Mg2+/Mn2+ ion is coordinated by three glutamate residues from the acidic patch in ygiF (Glu-4ygiF, Glu-6ygiF, Glu-162ygiF) and by a water molecule that is positioned close to a terminal phosphate of the tripolyphosphate substrate (position 2, Fig. 3, D and E). When we calculated phased anomalous difference maps from diffraction data collected near the Mn-K edge, we found that the ygiF metal binding site 1 shows a higher occupancy compared with the second site (peak heights are 125 and 45 σ, respectively), whereas we cannot detect difference density at the third metal position found in AtTTM3 (Fig. 3D). Taken together, our experiments define two consistent Mg2+/Mn2+ coordination centers in plant and bacterial TTM tripolyphosphatases.
We compared our structures to other TTM enzymes for which metal ion-bound complexes have been reported. In the RNA triphosphatase Cet1p, a Mn2+ ion is bound by the acidic patch, and its position corresponds to site 2 in ygiF and AtTTM3 (Figs. 4C and 3E) (11). In a bacterial TTM protein with adenylate cyclase activity, both positions 1 and 2 are occupied by Mn2+ ions (Figs. 4D and 3E) (18). The acidic patch in mouse ThTPase (corresponding to site 2 in AtTTM3 and ygiF) again allows for Mg2+ ion binding, as concluded from NMR titration experiments (20). These findings together indicate that many TTM proteins contain two metal ion binding sites, as previously proposed (40, 41). It is likely that site 1 is generated by a triphosphate substrate-divalent metal ion complex binding to the tunnel center (21, 23). Here, the octahedral coordination of the Mg2+/Mn2+ ion is completed by an glutamate residue (Glu-169AtTTM3, Glu-160ygiF), which is invariant in all TTM proteins characterized thus far (Figs. 3D and 4). A second binding site is formed by three glutamate residues originating from an acidic patch conserved among many but not all TTM enzymes (Figs. 3, A, D, and E, and 4) (4, 12, 24). This metal ion is positioned close to a terminal phosphate in our AtTTM3 and ygiF substrate complex structures (Fig. 3D) and may thus be involved in catalysis rather than in substrate binding.
Structures of Mammalian Thiamine Triphosphatase Reveal the Location of the γ-Phosphate
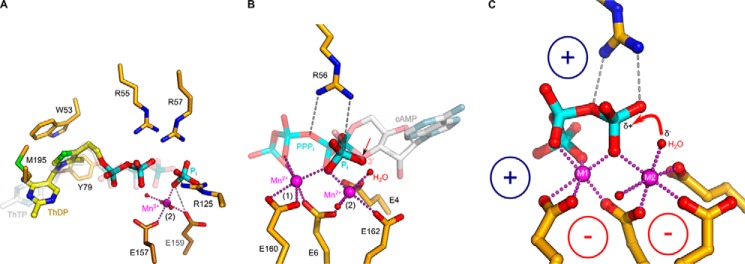
We investigated the contributions of the two metal ion centers to TTM substrate binding and catalysis. A conceptual problem with the analysis of our plant and bacterial tripolyphosphatases is that they catalyze the asymmetric cleavage of a symmetric substrate (Fig. 2) (25, 26). It is thus difficult to assess in crystal structures, which terminal phosphate represents the γ-phosphate that is being hydrolyzed (Fig. 3, A and B). We thus structurally characterized a mammalian TTM ThTPase, which was previously shown to specifically hydrolyze thiamine triphosphate (ThTP) into ThDP and Pi (19–21, 43). We synthesized ThTP from ThDP and produced co-crystal structures of mouse ThTPase with its substrate at pH 6, where ThTPase catalytic activity is minimal (20). Consequently, we found an intact ThTP molecule bound in the tunnel center of ThTPase (Fig. 5A). The thiamine portion of the substrate binds to a pocket generated by the tunnel walls and the C-terminal plug helix, with the thiazole ring making a stacking interaction with Trp-53 and with Met-195 from the plug helix (Fig. 5A). The ThTP triphosphate moiety binds in the same conformation as outlined for the PPPi-bound structures of AtTTM3 and ygiF above. Our substrate-bound mouse ThTPase structure supports an earlier docking model of human ThTPase (21). We next solved a crystal structure of mouse ThTPase in the presence of ThTP and Mg2+ in a second crystal form grown at pH 9.0, where substrate hydrolysis can occur (20). Indeed, we found a product complex trapped in the active site of the enzyme, with a ThDP molecule and an orthophosphate located in the tunnel center (Fig. 5A). ThDP is coordinated by Arg-55 and Arg-57 in the substrate binding site but no longer allows for the coordination of a Mg2+/Mn2+ ion in metal binding site 1, possibly because the missing γ-phosphate would be required for Mg2+/Mn2+ coordination (Fig. 5A). The γ-phosphate in our structure apparently has been hydrolyzed, and the resulting Pi has slightly moved away from the tunnel center (Fig. 5A). It is now found coordinated by Arg-125 and in direct contact with a Mn2+ ion located in metal binding site 2, reinforcing the notion that this metal ion may play a crucial role in catalysis (Fig. 5A).
FIGURE 5.
TTM proteins use a two-metal catalytic mechanism. A, close-up view of the mouse ThTPase tunnel center with either the ThTP substrate bound (transparent gray) or the ThDP/Pi products post-catalysis (in yellow, in bond representation). The thiamine part of ThTP is buried in a pocket close to the C-terminal plug helix of the TTM domain formed by Tyr-79 and Met-195; the thiazole ring makes a stacking interaction with Trp-53 (in yellow, in bond representation). The product Pi is coordinated by a Mn2+ ion bound to site 2 and by Arg-125. B, close-up of the ygiF active site (in yellow, in bond representation) bound to PPPi and two Mn2+ ions (magenta spheres) in sites 1 and 2. The Mn2+ ion in site 2 coordinates a water molecule (red sphere), which is well positioned to act as nucleophile. Structural superposition with a product-bound class IV adenylate cyclase (PDB ID 3N10) reveals the O3′ of cAMP in the same position as the water molecule in ygiF. This position is also occupied by an oxygen atom of the product Pi located in the AtTTM3 post-catalysis complex. C, the suggested mechanism for acidic-patch containing TTM proteins. The metal ion in site 1 coordinates the triphosphate moiety of the substrate to the tunnel center by interacting with a conserved Glu residue. Three additional glutamates form metal binding site 2, which coordinates and polarizes a water molecule to attack the γ-phosphate of the substrate. Conserved basic residues in the tunnel center are involved in substrate binding and potentially stabilize the pyrophosphate leaving group.
TTM Proteins Use a Two-metal Catalytic Mechanism
The structural features surrounding metal binding site 2 in our structures allow proposing a unified catalytic mechanism for triphosphate tunnel metalloenzymes. In our PPPi-bound ygiF structure we find a water molecule coordinated by the second Mn2+ ion, which is in an ideal position to serve as the activated nucleophile (Fig. 5, B and C). Indeed, structural superposition with a bacterial adenylate cyclase (18) reveals that its cAMP O3′ group, which acts as the nucleophile in the cyclization reaction, is located in the same position as the water molecule in our ygiF structure (Fig. 5B). Consistently, this position also is occupied by an oxygen atom of a product orthophosphate, which we located in our AtTTM3 post-catalysis structure (see above, Figs. 3B and 5B). Based on these findings, we propose that metal binding site 1 is required for proper substrate coordination in TTM proteins (21, 23), and metal ion 2 activates a water molecule to allow for a nucleophilic attack on the triphosphate substrate (Fig. 5C). This would rationalize why the glutamate residues from the acidic patch, which are involved in the coordination of the second metal ion, are well conserved among TTM proteins (Figs. 3E and 4). The basic residues in the tunnel center appear to be mainly involved in substrate coordination (Figs. 2A and 3A); however, the invariant Arg-56ygiF (Arg-52AtTTM3) possibly activates the substrate for the nucleophilic attack by forming a hydrogen bond with the oxygen atom connecting the β- and γ-phosphate of the substrate (Fig. 5, B and C). The suggested TTM reaction mechanism is reminiscent of the one found in mammalian type V adenylate cyclases (44) and in nucleic acid polymerases (45, 46) as previously speculated (8).
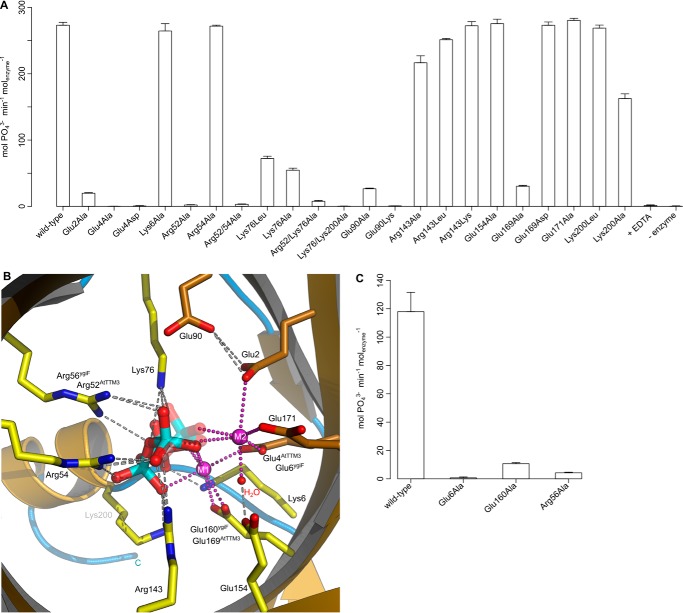
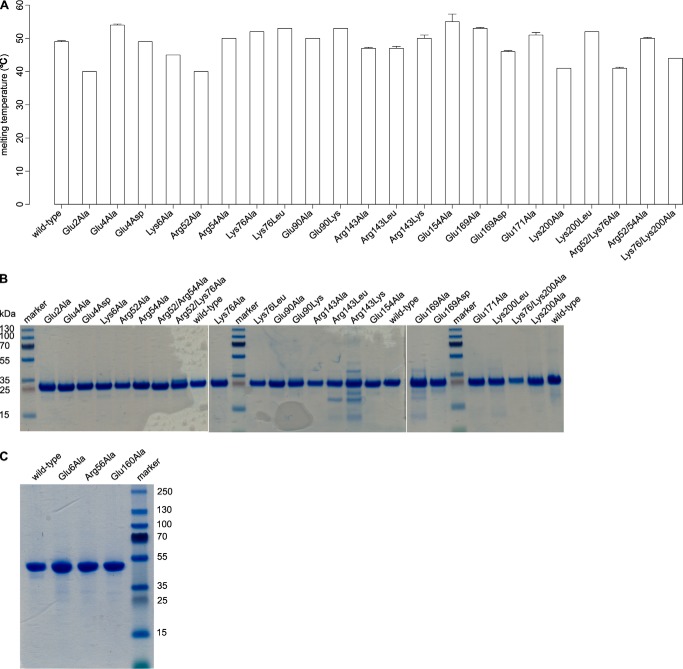
We next performed a mutational analysis of substrate- and metal co-factor-interacting residues in AtTTM3 and ygiF (Fig. 6). Mutation of Glu-2AtTTM3, Glu-4AtTTM3, and Glu-169AtTTM3 to Asp or Ala strongly reduces the PPPi hydrolysis of the plant enzyme (Fig. 6, A and B; see Fig. 7 for mutant protein stability). Consistently, changing the corresponding Glu-6ygiF and Glu-160ygiF to Ala impairs the enzymatic activity of ygiF, suggesting that the proper arrangement of metal binding sites 1 and 2 in the tunnel center is essential for catalysis (Fig. 6, A–C). Mutation of Arg-52AtTTM3 or Arg-56ygiF, but not of the neighboring Arg-54AtTTM3, to Ala again strongly inhibits catalysis, highlighting its potential role as proton donor (Fig. 6, A–C, see above). In addition, we find moderately reduced enzymatic activities upon the mutation of Lys-76AtTTM3 to Leu or Ala (Fig. 6, A and B). This residue and the corresponding Lys-69ygiF appear to be involved in substrate binding and orient Glu-2AtTTM3/Glu-4ygiF to coordinate metal ion 2 (Fig. 6, A and B). Glu-2AtTTM3 is also in contact with Glu-90AtTTM3, mutation of which to Ala again reduces the enzymatic activity of the Arabidopsis enzyme (Fig. 6, A and B). Taken together, our and previous mutational analyses (22, 24, 26) consistently suggest that the proper establishment of two metal centers appears to be critical for catalysis in TTM tripolyphosphatases and other TTM proteins (40, 41). The basic residues, with the exception of the catalytic Arg-52AtTTM3 appear to be mainly involved in substrate coordination in the tunnel center (Fig. 6, A–C). Further experimentation will be required to rationalize the specific effects of certain point mutations on substrate/metal co-factor binding or catalysis itself.
FIGURE 6.
Reduced enzyme activities for several AtTTM3 and ygiF mutant proteins are consistent with the proposed catalytic mechanism. A, PPPase activity of structure-based point AtTTM3 point mutants found in direct contact with the PPPi substrate or the two metal co-factors as shown in B. C, PPPase activity of the corresponding residues in ygiF.
FIGURE 7.
Structural integrity and purity of AtTTM3 and ygiF mutant proteins. A, melting temperatures for wild-type and mutant AtTTM3 recombinant proteins. SDS-PAGE analysis of purified wild-type and mutant AtTTM3 (B) and ygiF (C) proteins is shown. The calculated molecular weights for AtTTM3 and ygiF are 24.3 and 48.6 kDa, respectively.
Comparison with Vtc4p suggests that the first but not the second metal binding site are present in the yeast polyphosphate polymerase (Figs. 2, A and B, and 4A) (4). Importantly, mutation of Lys-200AtTTM3, which corresponds to the catalytic Lys-458Vtc4p, to either Leu or Ala has little effect on tripolyphosphatase activity of the plant enzyme (Fig. 6, A and B) (4). This suggests that there are significant mechanistic differences between TTM polyphoshatases and polyphosphate polymerases despite their strong structural homology (Figs. 2, A and B, and 3A).
Directionality of Substrate Binding Defines TTM Catalytic Activity
To better understand how acidic-patch containing TTMs are able to carry out very different enzyme reactions, we superimposed our substrate-bound structures of AtTTM3, ygiF, and mouse ThTPase with an ATP-analog complex of a bacterial TTM adenylate cyclase (18) (Fig. 8). We found that although the triphosphate parts of all ligands closely align in the tunnel center, their “tail” moieties can enter the tunnel domain from opposite sites in different enzymes (Fig. 8). The unique modes of substrate binding in ThTPases and adenylate cyclases allow these enzymes to carry out rather different reactions and to produce different leaving groups (ortho- and pyrophosphate, respectively) while maintaining a unified cleavage site in close proximity of metal binding site 2. Both the N- and C-terminal sides of the tunnel domain have evolved to recognize specific substrates, as exemplified by our ThTPase structure and by the class IV adenylate cyclase complexes (Figs. 5, A and B, and 8) (18). The observed substrate binding mode in mouse ThTPase reinforces the notion that to bind their substrates, TTMs require opening of their closed tunnel domains into a cup-shaped hand, as previously shown by NMR spectroscopy (20).
FIGURE 8.
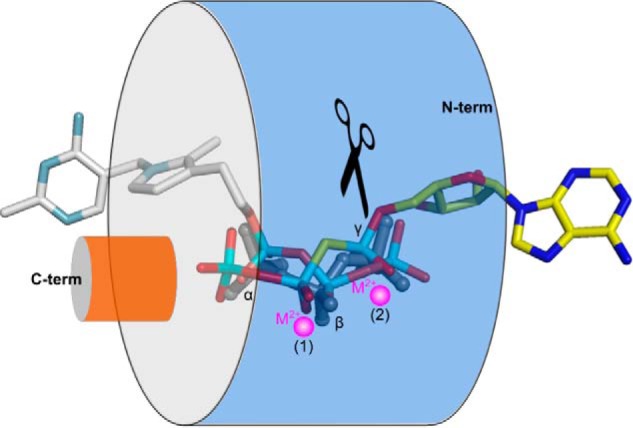
Directionality of substrate binding defines TTM catalytic activity. Schematic representation of PPPi (ygiF), ThTP (mThTPase, in gray), and ATP-analog (PDB ID 3N0Y, in yellow) binding to the tunnel domain. Different substrates can bind to the tunnel from opposite sites. The respective triphosphate moieties are well aligned, and the cleavage site is maintained (black scissors), leading to different reaction products.
Members of the ancient triphosphate tunnel metalloenzyme family can be found in all kingdoms of life. Our comparative analysis defines that most of these enzymes share a common catalytic mechanism in their tunnel centers, yet they have evolved different substrate recognition modes on their tunnel entries. The observed substrate plasticity apparently allows TTM proteins to act on a wide array of enzyme substrates and to perform very different reactions, many of which likely remain to be discovered.
Author Contributions
J. M. and M. H. designed the study and wrote the paper. V. T. performed NMR titrations, J. M. performed all other experiments. All authors analyzed data and approved the final version of the manuscript.
Acknowledgments
We thank T. Baiga for help with ThTP synthesis, members of the Hothorn laboratory for discussion, and W. Saenger for suggestions and critical reading of the manuscript. Diffraction experiments were performed on the PXII and PXIII beam lines at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland and at beam line ID29 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France.
This work was supported by the European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013)/ERC Grant Agreement 310856, the Max Planck Society, and the European Molecular Biology Organisation (EMBO) Young Investigator Programme (to MH). The authors declare that they have no conflicts of interest with the contents of this article.
- VTC
- vacuolar transporter chaperone
- TTM
- triphosphate tunnel metalloenzymes
- AtTTM3
- A. thaliana triphosphate tunnel metalloenzyme
- PPPi
- tripolyphosphate
- ThTP
- thiamine triphosphate
- ThDP
- thiamine diphosphate
- ThTPase
- thiamine triphosphatase
- mThTPase
- mouse ThTPase
- SAD
- single-wavelength anomalous diffraction
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Bis-Tris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- r.m.s.d.
- root mean square deviation
- Mn-K
- manganese K x-ray absorption edge.
References
- 1. Rao N. N., Gómez-García M. R., Kornberg A. (2009) Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605–647 [DOI] [PubMed] [Google Scholar]
- 2. Kornberg S. R. (1957) Adenosine triphosphate synthesis from polyphosphate by an enzyme from Escherichia coli. Biochim. Biophys. Acta. 26, 294–300 [DOI] [PubMed] [Google Scholar]
- 3. Ahn K., Kornberg A. (1990) Polyphosphate kinase from Escherichia coli: purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265, 11734–11739 [PubMed] [Google Scholar]
- 4. Hothorn M., Neumann H., Lenherr E. D., Wehner M., Rybin V., Hassa P. O., Uttenweiler A., Reinhardt M., Schmidt A., Seiler J., Ladurner A. G., Herrmann C., Scheffzek K., Mayer A. (2009) Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516 [DOI] [PubMed] [Google Scholar]
- 5. Zhu Y., Huang W., Lee S. S., Xu W. (2005) Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep. 6, 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerasimaite R., Sharma S., Desfougères Y., Schmidt A., Mayer A. (2014) Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 127, 5093–5104 [DOI] [PubMed] [Google Scholar]
- 7. Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H.-R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) The Pfam protein families database. Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iyer L. M., Aravind L. (2002) The catalytic domains of thiamine triphosphatase and CyaB-like adenylyl cyclase define a novel superfamily of domains that bind organic phosphates. BMC Genomics 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gong C., Smith P., Shuman S. (2006) Structure-function analysis of Plasmodium RNA triphosphatase and description of a triphosphate tunnel metalloenzyme superfamily that includes Cet1-like RNA triphosphatases and CYTH proteins. RNA 12, 1468–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bettendorff L., Wins P. (2013) Thiamine triphosphatase and the CYTH superfamily of proteins. FEBS J. 280, 6443–6455 [DOI] [PubMed] [Google Scholar]
- 11. Lima C. D., Wang L. K., Shuman S. (1999) Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99, 533–543 [DOI] [PubMed] [Google Scholar]
- 12. Bisaillon M., Shuman S. (2001) Structure-function analysis of the active site tunnel of yeast RNA triphosphatase. J. Biol. Chem. 276, 17261–17266 [DOI] [PubMed] [Google Scholar]
- 13. Gong C., Shuman S. (2003) Mapping the active site of vaccinia virus RNA triphosphatase. Virology 309, 125–134 [DOI] [PubMed] [Google Scholar]
- 14. Gong C., Shuman S. (2002) Chlorella virus RNA triphosphatase: mutational analysis and mechanism of inhibition by tripolyphosphate. J. Biol. Chem. 277, 15317–15324 [DOI] [PubMed] [Google Scholar]
- 15. Benarroch D., Smith P., Shuman S. (2008) Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure 16, 501–512 [DOI] [PubMed] [Google Scholar]
- 16. Kyrieleis O. J., Chang J., de la Peña M., Shuman S., Cusack S. (2014) Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure 22, 452–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gallagher D. T., Smith N. N., Kim S.-K., Heroux A., Robinson H., Reddy P. T. (2006) Structure of the class IV adenylyl cyclase reveals a novel fold. J. Mol. Biol. 362, 114–122 [DOI] [PubMed] [Google Scholar]
- 18. Gallagher D. T., Kim S.-K., Robinson H., Reddy P. T. (2011) Active-site structure of class IV adenylyl cyclase and transphyletic mechanism. J. Mol. Biol. 405, 787–803 [DOI] [PubMed] [Google Scholar]
- 19. Lakaye B., Makarchikov A. F., Antunes A. F., Zorzi W., Coumans B., De Pauw E., Wins P., Grisar T., Bettendorff L. (2002) Molecular characterization of a specific thiamine triphosphatase widely expressed in mammalian tissues. J. Biol. Chem. 277, 13771–13777 [DOI] [PubMed] [Google Scholar]
- 20. Song J., Bettendorff L., Tonelli M., Markley J. L. (2008) Structural basis for the catalytic mechanism of mammalian 25-kDa thiamine triphosphatase. J. Biol. Chem. 283, 10939–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Delvaux D., Kerff F., Murty M. R., Lakaye B., Czerniecki J., Kohn G., Wins P., Herman R., Gabelica V., Heuze F., Tordoir X., Marée R., Matagne A., Charlier P., De Pauw E., Bettendorff L. (2013) Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase. Biochim. Biophys. Acta. 1830, 4513–4523 [DOI] [PubMed] [Google Scholar]
- 22. Jain R., Shuman S. (2008) Polyphosphatase activity of CthTTM, a bacterial triphosphate tunnel metalloenzyme. J. Biol. Chem. 283, 31047–31057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delvaux D., Murty M. R., Gabelica V., Lakaye B., Lunin V. V., Skarina T., Onopriyenko O., Kohn G., Wins P., De Pauw E., Bettendorff L. (2011) A specific inorganic triphosphatase from Nitrosomonas europaea: Structure and catalytic mechanism. J. Biol. Chem. 286, 34023–34035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keppetipola N., Jain R., Shuman S. (2007) Novel triphosphate phosphohydrolase activity of Clostridium thermocellum TTM, a member of the triphosphate tunnel metalloenzyme superfamily. J. Biol. Chem. 282, 11941–11949 [DOI] [PubMed] [Google Scholar]
- 25. Kohn G., Delvaux D., Lakaye B., Servais A.-C., Scholer G., Fillet M., Elias B., Derochette J.-M., Crommen J., Wins P., Bettendorff L. (2012) High inorganic triphosphatase activities in bacteria and mammalian cells: Identification of the enzymes involved. PLoS ONE 7, e43879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moeder W., Garcia-Petit C., Ung H., Fucile G., Samuel M. A., Christendat D., Yoshioka K. (2013) Crystal structure and biochemical analyses reveal that the Arabidopsis triphosphate tunnel metalloenzyme AtTTM3 is a tripolyphosphatase involved in root development. Plant J. 76, 615–626 [DOI] [PubMed] [Google Scholar]
- 27. Larivière L., Geiger S., Hoeppner S., Röther S., Strässer K., Cramer P. (2006) Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 13, 895–901 [DOI] [PubMed] [Google Scholar]
- 28. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 29. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A. 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 30. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D. Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 31. Terwilliger T. C. (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bettendorff L., Nghiêm H. O., Wins P., Lakaye B. (2003) A general method for the chemical synthesis of γ-32P-labeled or unlabeled nucleoside 5′-triphosphates and thiamine triphosphate. Anal. Biochem. 322, 190–197 [DOI] [PubMed] [Google Scholar]
- 37. Baykov A. A., Evtushenko O. A., Avaeva S. M. (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171, 266–270 [DOI] [PubMed] [Google Scholar]
- 38. Cummings M. D., Farnum M. A., Nelen M. I. (2006) Universal screening methods and applications of ThermoFluor. J. Biomol. Screen. 11, 854–863 [DOI] [PubMed] [Google Scholar]
- 39. Pei Y., Schwer B., Hausmann S., Shuman S. (2001) Characterization of Schizosaccharomyces pombe RNA triphosphatase. Nucleic Acids Res. 29, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martins A., Shuman S. (2003) Mapping the triphosphatase active site of baculovirus mRNA capping enzyme LEF4 and evidence for a two-metal mechanism. Nucleic Acids Res. 31, 1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gong C., Martins A., Shuman S. (2003) Structure-function analysis of Trypanosoma brucei RNA triphosphatase and evidence for a two-metal mechanism. J. Biol. Chem. 278, 50843–50852 [DOI] [PubMed] [Google Scholar]
- 42. Holm L., Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 43. Lakaye B., Makarchikov A. F., Wins P., Margineanu I., Roland S., Lins L., Aichour R., Lebeau L., El Moualij B., Zorzi W., Coumans B., Grisar T., Bettendorff L. (2004) Human recombinant thiamine triphosphatase: purification, secondary structure, and catalytic properties. Int. J. Biochem. Cell Biol. 36, 1348–1364 [DOI] [PubMed] [Google Scholar]
- 44. Tesmer J. J., Sunahara R. K., Johnson R. A., Gosselin G., Gilman A. G., Sprang S. R. (1999) Two-metal-ion catalysis in adenylyl cyclase. Science 285, 756–760 [DOI] [PubMed] [Google Scholar]
- 45. Steitz T. A. (1998) Structural biology: a mechanism for all polymerases. Nature 391, 231–232 [DOI] [PubMed] [Google Scholar]
- 46. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C. E. (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]