Abstract
Endophytes are novel sources of natural bioactive compounds. This study seeks endophytes that produce the anticancer enzyme l-asparaginase, to harness their potential for mass production. Four plants with anticancer properties; Cymbopogon citratus, Murraya koenigii, Oldenlandia diffusa and Pereskia bleo, were selected as host plants. l-Asparaginase-producing endophytes were detected by the formation of pink zones on agar, a result of hydrolyzes of asparagine into aspartic acid and ammonia that converts the phenol red dye indicator from yellow (acidic condition) to pink (alkaline condition). The anticancer enzyme asparaginase was further quantified via Nesslerization. Results revealed that a total of 89 morphotypes were isolated; mostly from P. bleo (40), followed by O. diffusa (25), C. citratus (14) and M. koenigii (10). Only 25 of these morphotypes produced l-asparaginase, mostly from P. bleo and their asparaginase activities were between 0.0069 and 0.025 μM mL−1 min−1. l-Asparaginase producing isolates were identified as probable species of the genus Colletotrichum, Fusarium, Phoma and Penicillium. Studies here revealed that endophytes are good alternative sources for l-asparaginase production and they can be sourced from anticancer plants, particularly P. bleo.
Keywords: Anticancer plants, Colletotrichum, Endophytes, Fusarium, l-Asparaginase
Introduction
Endophytes are microorganisms that reside inside the internal tissues of living plants without causing any symptoms and obvious harm to the host plants [1]. Endophytes are ubiquitously found in all plants and are valued for their ability to synthesize various useful bioactive compounds [2]. These bioactive compounds were originally involved in defense mechanisms against phytopathogens [2]. However, in the recent years, endophytic bioactive compounds were gradually integrated in novel drug discoveries due to their wide variety of biological activities as antibiotic, anticancer, antioxidant, and anti-inflammatory agents [3]. Research on endophyte-derived bioactive compounds was escalated with the discovery of taxol from endophytic Taxomyces andrenae [4]. This anticancer agent was formerly extracted from the Pacific yew tree, and with this discovery, more taxol can be produced without the mass destruction of the yew trees. With the discovery of taxol-producing endophyte T. andrenae from yew trees, many researchers hypothesize that endophytes from anticancer plants have potential to synthesize compounds with anticancer properties. This prompted investigations on the study of anticancer compounds produced by various endophytes, such as campthothecin [5], maytansine [6] and cajanol [7]. In this study, the interest is to source for l-asparaginase-producing endophytes.
l-Asparaginase (E.C.3.5.1.1) is an enzyme that catalyzes the hydrolysis of l-asparagine to l-aspartate and ammonia [8]. In cancer treatment, l-asparaginase removes l-asparagine in the serum, depriving tumor cells from large amounts of asparagine required for growth [9], thus controlling tumor growth effectively [10]. In fact, l-asparaginase is a clinically acceptable anticancer agent for the treatment of Acute Lymphoblastic Leukemia (ALL) [9]. l-Asparaginase was conventionally derived from bacteria, typically from Escherichia coli and Erwinia carotovora due to their cost-effective nature [8]. However, l-asparaginase from prokaryotic sources causes has now been found to have many side effects, primarily hypersensitivity which leads to allergic reactions and anaphylaxis. Thus, in the recent years, eukaryotic fungi have been investigated as sources of l-asparaginase [2] as the interest in endophytes as sources of l-asparaginase is relatively new and only few studies have been reported.
In this study, endophytic fungi were isolated from four common Malaysian medicinal plants; Pereskia bleo (Seven star needle or Qi Xing Zhen), Murraya koenigii (Curry plant), Oldenlandia diffusa (Chinese herb Bai Hua She She Cao) and Cymbopogon citratus (Lemon grass). These plants are known to have ethno-botanical properties. P. bleo is used to treat diabetes, hypertension and also cancer [11]. The curry plant, M. koenigii, is often used as traditional remedy for the treatment of piles, headache, stomach ache, influenza, snake bites, curing dysentery, diarrhea and cancer [12]. Although l-asparaginase production from these selected plants is not known, we draw conclusions that because of the anticancer properties of the plants and that fungus are eukaryotes; there is a possibility that l-asparaginase endophytes may be isolated. Many other discoveries of endophytes with active biomolecules were made from various medicinal plants irrespective of the properties found in the plants [5], [6], [7]. In addition, l-asparaginase is an important anticancer molecule which is often sourced from microorganisms, and we were hoping to achieve similar discoveries as the taxol-producing T. andrenae [4].
To date, endophyte studies for P. bleo and M. koenigii are limited, although isolation of Campylospora chaetocladia as an endophyte in M. koenigii [13] has been reported. Similarly, endophyte isolation has not been attempted for O. diffusa although this plant has been used in China for many years to treat hepatitis, sore throat and malignant tumors of the liver, lung and stomach [14]. On the contrary, C. citratus is the only plant selected which has a more extensive endophyte profiling. This plant, used to treat fever, infection, headaches as well as having antitumor and anti-inflammatory properties, are host to a variety of endophytes such as Aspergillus sp., Cladosporium sp., Gleocladium roseum, Macrophoma sp., Penicillium notatum and Trichoderma viride [15]. However, l-asparaginase producing endophytes from this plant have not been reported as well.
This study was conducted to explore the potential of isolating and harnessing the potential of l-asparaginase-producing endophytes from the four Malaysian medicinal plants (P. bleo, M. koenigii, O. diffusa and C. citratus) with anticancer properties. Endophyte diversity was established and their l-asparaginase production is reported here.
Material and methods
Isolation and culture establishment
Endophytes were isolated from fresh leaves and stems of anticancer plants; C. citratus, M. koenigii, O. diffusa and P. bleo. These plants were sampled randomly from a housing garden at Bandar Tun Hussein Onn, Selangor, Malaysia. The plant tissues were first left under running tap water for 15 min. The leaf tissues were then cut into 2 cm × 2 cm segments whereas the stem tissues were cut to a length of 2 cm each. The tissues were subjected to triple sterilization technique, beginning with 40% household bleach for 5 min, then in 50%, 70%, 90% and 100% ethanol for 2 min each immersion, and finally a quick rinse in sterilized distilled water. This procedure was performed and repeated twice on the same batch of plant tissues [16]. Surface-sterilized tissue segments were then injured and seeded onto Potato Dextrose Agar (PDA, Merck) supplemented with Rose bengal (0.033 g L−1) and chloramphenicol (0.05 g L−1), to select for fungal endophytes [8]. Controls were prepared by seeding non-injured surface-sterilized tissue segments onto PDA plates. The absence of mycelial growth indicated effective surface sterilization while mycelial growth from injured tissues was identified as endophytes. All PDA plates were incubated at 28 ± 2 °C for 7–14 days. The fungal colonies were subsequently transferred to fresh PDA plates to establish pure cultures.
Screening of l-asparaginase-producing endophytes
A 5-mm fungal mycelial plug was inoculated onto Modified Czapex Dox’s (McDox) agar [agar powder (20.0 g L−1), glucose (2.0 g L−1), l-asparagine (10.0 g L−1), KH2PO4 (1.52 g L−1), KCl (0.52 g L−1), MgSO4·7H2O (0.52 g L−1), CuNO3·3H2O (0.001 g L−1), ZnSO4·7H2O (0.001 g L−1), FeSO4·7H2O (0.001 g L−1)] supplemented with l-asparagine (10.0 g L−1) and 0.3 mL of 2.5% phenol red dye (indicator) [17]. Controls were prepared by inoculating mycelial plugs on McDox agar without asparagine. Triplicates for each isolate were prepared. All the plates were incubated at 28 ± 2 °C. After 5 days of incubation, the diameter of the pink zone was measured [8].
l-Asparaginase activities in selected endophytes
Selected fungal cultures were first established in 30 mL of McDox broth and incubated for 5 days at 120 rpm. After incubation, 100 μL of broth (crude enzyme) was pipetted into 2 mL microcentrifuge tubes. To this, 100 μL of Tris HCl (pH 7.2), 200 μL of 0.04 M asparagine and 100 μL of sterile distilled water (SDW) were added. The mixture was incubated at 37 ± 2 °C for 1 h. After incubation, 100 μL of 1.5 M Trichloroacetic Acid (TCA) was then added to stop the enzymatic reaction. This was followed by pipetting 100 μL of the mixture into fresh tubes containing 750 μL SDW and 300 μL of Nessler’s reagent [18] and incubated at 28 ± 2 °C for 20 min, after which the absorbance of the samples was measured at 450 nm. One unit of asparaginase is expressed as the amount of enzyme that catalyzes the formation of 1 μmol of ammonia per minute at 37 ± 2 °C [18].
Identification of l-asparaginase producing endophytes
Selected endophytes were first established in Potato Dextrose Broth (PDB) and incubated for 5 days at 28 ± 2 °C. Genomic DNA was extracted from 200 mg wet weight of fresh mycelium [19]. The mycelium was ground in liquid nitrogen. The genomic DNA (gDNA) was extracted using the GF-1 Plant DNA Extraction Kit-50 preps, as described by the manufacturer (Vivantis® Technologies, California, United States of America). The genomic DNA was amplified using the universal primer ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [32]. The PCR was performed on the reaction mixture (50 μL) which consisted of 5 μL genomic DNA (∼100 ng), 5 μL of forward (ITS1) and reverse (ITS4) primers each (10 μM), 25 μL GoTaq @ Green Master Mix 2x (Promega, Malaysia), and 10 μL nuclease free water (Promega, Malaysia). Amplification process was initiated by pre-heating at 95 °C for 1 min, followed by 34 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 40 s, extension at 72 °C for 90 s and a final extension at 72 °C for 5 min. Amplifications were performed on a MyCycler Thermocycler (Bio-Rad). The products of the PCR reaction were then examined by electrophoresis using 1% (w/v) agarose gel, stained with gel red (Biotium®) and visualized with a UV transilluminator. Subsequently, the PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega®) and outsourced to 1st Base® Techniques for sequencing. The sequence results were compared to the database from National Center for Biotechnology Information, USA (NCBI) using BLAST search (http://blast.ncbi.nlm.nih.gov/BLAST.cgi).
Statistical analysis
The experiment was conducted in a Complete Randomized Design (CRD) with triplicates for each parameter assessed. The data were statistically analyzed using the software Statistical Package for the Social Sciences (SPSS) version 20.0. One-way ANOVA with Tukey’s Studentized Range Test (HSD(0.05)) were applied to analyze all the data collected. Differences were considered significant at p < 0.05.
Results
Isolation of endophytes
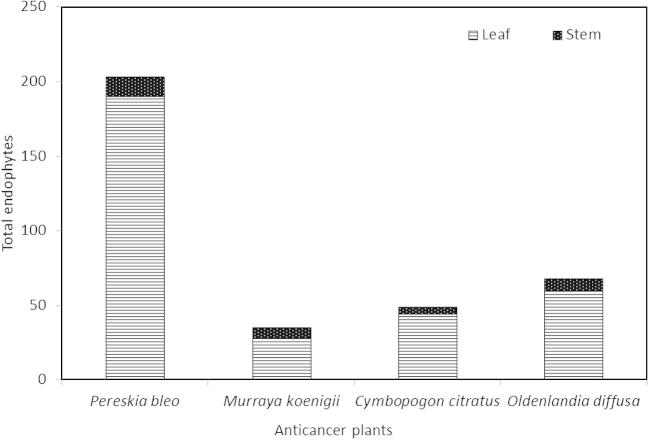
Endophytes were found in all four anticancer plant species with a total of 355 endophytes isolated from the 699 tissue segments used. Endophytes were mostly recovered from P. bleo (203 isolates), followed by O. diffusa (68 isolates), C. citratus (49 isolates) and M. koenigii (35 isolates) (Fig. 1). It was also observed that for all four plants, a higher number of endophytes were recovered from leaf tissues than in stem tissues (Fig. 1). This was most evident in P. bleo where 190 isolates were found in leaf tissues compared to only 13 isolates from stem tissues (Fig. 1). Due to the large number of endophytes, the isolates were further grouped into 89 morphotypes with each morphotype comprising of isolates with similar morphological and cultural characteristics. P. bleo have the most morphotypes (40 morphotypes), followed by O. diffusa (25 morphotypes), C. citratus (14 morphotypes) and M. koenigii (10 morphotypes). For all subsequent tests, morphotypes were used.
Fig. 1.
Total number of fungal endophytes isolated from leaf and stem tissues of respective anticancer plants.
Detecting l-asparaginase producing endophytes and their activities
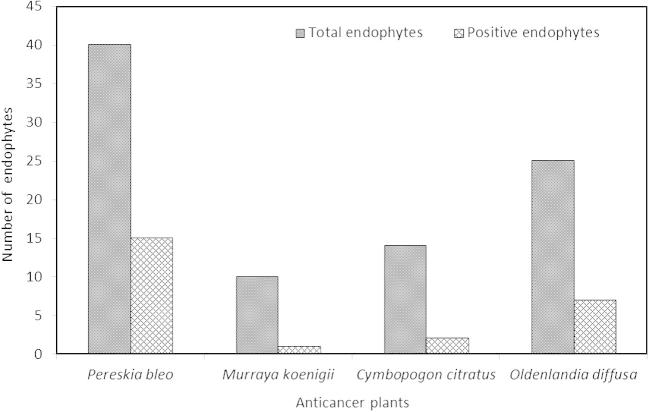
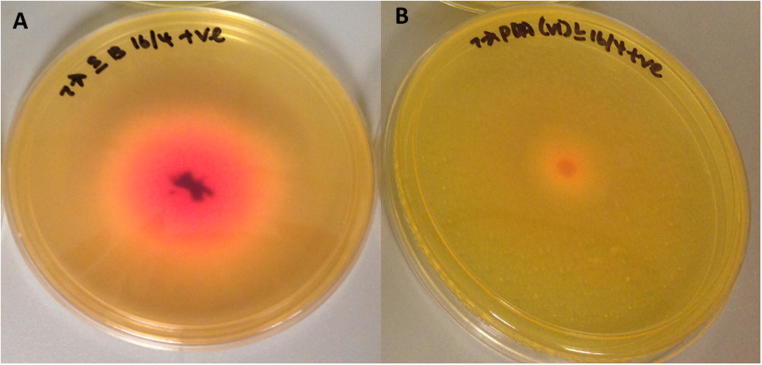
Of the 89 endophytes (morphotypes), only 25 morphotypes were l-asparaginase-producing endophytes (Fig. 2). For these positive isolates, formation of pink zone was evident as a result of l-asparaginase produced by the endophytes, which hydrolyzes asparagine into aspartic acid and ammonia, converting the phenol red dye indicator from yellow (acidic condition) to pink (alkaline condition) (Fig. 3). Each morphotype produced different sizes of pink zone diameter, ranging from 0.4 to 3.7 cm with a median of 1.2 cm.
Fig. 2.
Comparison between total morphotypes against positive l-asparaginase-producing fungal endophytes in selected anticancer plants (P. bleo, M. koenigii, C. citratus and O. diffusa).
Fig. 3.
l-Asparaginase producing fungal endophytes was distinguished based on the formation of pink zone (A) as compared to endophytes without l-asparaginase activity (B). Cultures were 5-day old cultures.
The positive endophytes produced asparaginase activities ranging from 0.0069 to 0.025 μM−1 mL−1 min−1 (Table 1). Eighteen of these endophytes were considered to have relatively high asparaginase activity (0.010–0.025 μM−1 mL−1 min−1) whereas 7 endophytes (PBL1, PBL2, PBL4, PBL6, PBS2, PBL9, CCL2) have low asparaginase activity (0.0069–0.0098 μM−1 mL−1 min−1) (Table 1). Isolates PBS1, PBL13, CCL1, ODL4 and MKS1, from various plants, were among the 18 endophytes with high l-asparaginase activities. The endophyte with significantly highest l-asparaginase activity was ODL4 from O. diffusa (0.025 μM−1 mL−1 min−1), followed by CCL1 (0.023 μM−1 mL−1 min−1), PBL13 (0.019 μM−1 mL−1 min−1), PBS1 (0.018 μM−1 mL−1 min−1) and MKS1 (0.013 μM−1 mL−1 min−1). The least l-asparaginase activity was produced by PBS2 from P. bleo (0.0069 μM−1 mL−1 min−1) (Table 1). Most of the strong l-asparaginase producers were isolated from O. diffusa (5 isolates), C. citratus (2 isolates) and M. koenigii (1 isolate). P. bleo host both strong and weak-producers of l-asparaginase, with 10 and 6 isolates, respectively (Table 1). This suggested that there was no trend in host-specificity with l-asparaginase producers.
Table 1.
Asparaginase activities (μM−1 mL−1 min−1) of the 25 fungal endophytes isolated from Leaf (L) and Stem (S) tissues of Pereskia bleo (PB), Cymbopogon citratus (CC), Oldenlandia diffusa (OD) and Murraya koenigii (MK). Mean and standard deviation are included. Means with the same letters are not significantly different as determined by Tukey’s Studentized Range Test (HSD0.05) (Tukey grouping).
| Plant | Endophyte isolate | Mean asparaginase activity (μM−1 mL−1 min−1) | Standard deviation | Tukey grouping |
|---|---|---|---|---|
| Pereskia bleo | PBL1 | 0.08 | 0.0013 | bc |
| PBL2 | 0.098 | 0.0038 | abc | |
| PBL3 | 0.015 | 0.0032 | abc | |
| PBS1 | 0.018 | 0.0038 | abc | |
| PBL4 | 0.009 | 0.0005 | bc | |
| PBL5 | 0.015 | 0.0042 | abc | |
| PBL6 | 0.008 | 0.0012 | bc | |
| PBL7 | 0.011 | 0.0011 | abc | |
| PBL8 | 0.01 | 0.0005 | abc | |
| PBS2 | 0.007 | 0.0016 | c | |
| PBL9 | 0.009 | 0.0006 | bc | |
| PBL10 | 0.016 | 0.002 | abc | |
| PBL11 | 0.012 | 0.0004 | abc | |
| PBL12 | 0.017 | 0.0003 | abc | |
| PBS3 | 0.013 | 0.0015 | abc | |
| PBL13 | 0.021 | 0.0191 | abc | |
| Oldenlandia diffusa | ODL1 | 0.013 | 0.0013 | abc |
| ODL2 | 0.017 | 0.0028 | abc | |
| ODL3 | 0.014 | 0.0043 | abc | |
| ODL4 | 0.0246 | 0.0053 | a | |
| ODL5 | 0.012 | 0.0023 | abc | |
| Cymbopogon citratus | CCL1 | 0.023 | 0.0073 | abc |
| CCL2 | 0.009 | 0.0095 | bc | |
| CCL3 | 0.017 | 0.0009 | abc | |
| Murraya koenigii | MKS1 | 0.013 | 0.0015 | abc |
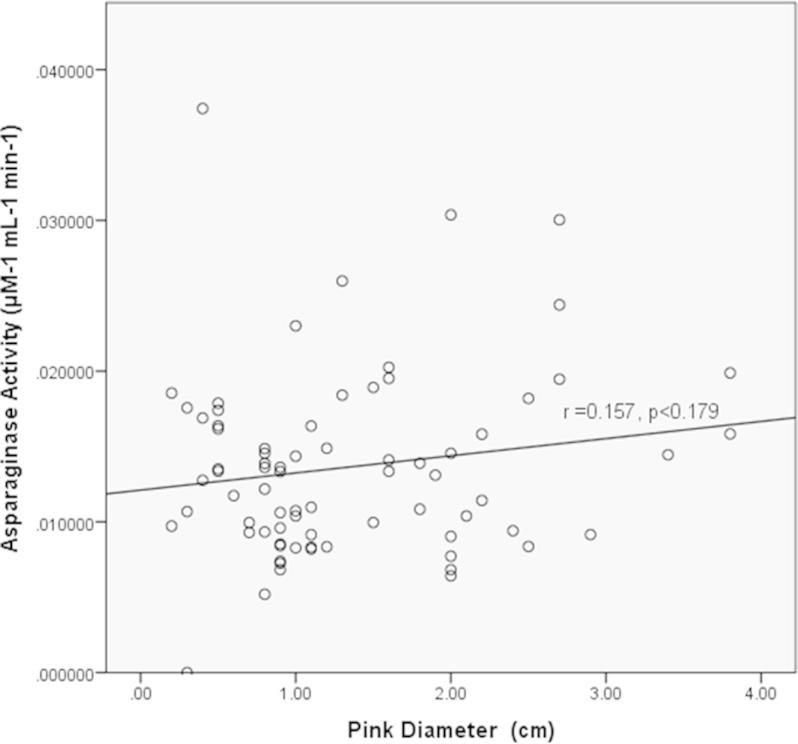
In addition, results from both the qualitative (pink zone diameter) and quantitative (l-asparaginase activities) assays were used to perform a simple regression analysis to study the correlation between the two methods used. This regression analysis has a weak correlation coefficient (r) of 0.157, indicating a weak relationship between pink zone diameters on agar plates with asparaginase activities determined quantitatively. In addition, the value of p < 0.179 indicated that the coefficient is not significantly different from 0. These results complied with the observation where 11 of the 25 positive endophytes showed high asparaginase activities which did not correlate to the diameter size of the pink zones. ODL4 which recorded the highest asparaginase activity (0.025 μM−1 mL−1 min−1), have only 2.7 cm diameter, compared to CCL3 with a 3.7 cm diameter but only 0.017 μM−1 mL−1 min−1 l-asparaginase activity.
Identification of l-asparaginase producing endophytes
Identification was performed based on sequencing results of the 23 selected endophytes with search sequences referred to the NCBI database. Nineteen of the l-asparaginase producing endophytes were identified as probable species of Colletotrichum (12 morphotypes), Fusarium (4 morphotypes), Penicillium (2 morphotypes) and Phoma (1 morphotype). Another four endophytes were identified only to the probable phylum and class of Ascomycota (2 morphotypes) and Dothideomycetes (2 morphotypes), respectively (Table 2).
Table 2.
Identification of selected fungal endophytes isolated from P. bleo, M. koenigii, C. citratus and O. diffusa to the most probable species based on similarities of ITS sequences to the NCBI database.
| Isolate | GenBank accession number | Closest related species | No of base pairs analyzed | Percentage of similarity (%) |
|---|---|---|---|---|
| PBL1 | N/A | |||
| PBL2 | KM104576 | Colletotrichum siamense F272 | 464 | 99 |
| PBL3 | KM104578 | Penicillium simplicissimum strain KUC 5153 | 578 | 100 |
| PBS1 | KM104575 | Dothiodeomycetes sp. P15E6 | 477 | 99 |
| PBL4 | KM104582 | Colletotrichum sp. NK29 | 459 | 99 |
| PBL5 | KM104583 | Colletotrichum sp. NK29 | 390 | 100 |
| PBL6 | KM104585 | Colletotrichum gloeosporioides strain M2.1 | 440 | 99 |
| PBL7 | N/A | |||
| PBL8 | KM104586 | Colletotrichum gloeosporioides | 445 | 99 |
| PBS2 | KM104587 | Colletotrichum gloeosporioides UFMGCB | 441 | 99 |
| PBL9 | KM104588 | Colletotrichum sp. NK22 | 479 | 100 |
| PBL10 | KM104591 | Colletotrichum sp. NK29 | 446 | 99 |
| PBL11 | KM104592 | Fusarium proliferatum | 558 | 99 |
| PBL12 | KM104594 | Colletotrichum gloeosporioides isolates Cglo TIN03 | 524 | 99 |
| PBS3 | KM104596 | Colletotrichum sp. NK29 | 480 | 99 |
| PBL13 | KM104597 | Penicillium simplicissimum KUC5153 | 559 | 100 |
| ODL1 | KM104580 | Fusarium verticillioides strain jb111 | 372 | 100 |
| ODL2 | KM104589 | Ascomycota sp. AR-2010 | 516 | 100 |
| ODL3 | KM104590 | Colletotrichum sp. GM414 | 464 | 99 |
| ODL4 | KM104579 | Ascomycota sp. | 480 | 99 |
| ODL5 | KM104595 | Colletotrichum sp. NK29 | 472 | 99 |
| CCL1 | KM104577 | Dothideomycetes sp. P15E6 | 465 | 98 |
| CCL2 | KM104581 | Fusarium proliferatum | 420 | 98 |
| CCL3 | KM104593 | Phoma sp. | 544 | 98 |
| MKS1 | KM104584 | Fusarium oxysporum isolate h13 | 386 | 99 |
Note: N/A indicated that cultures perished during study due to mite infection.
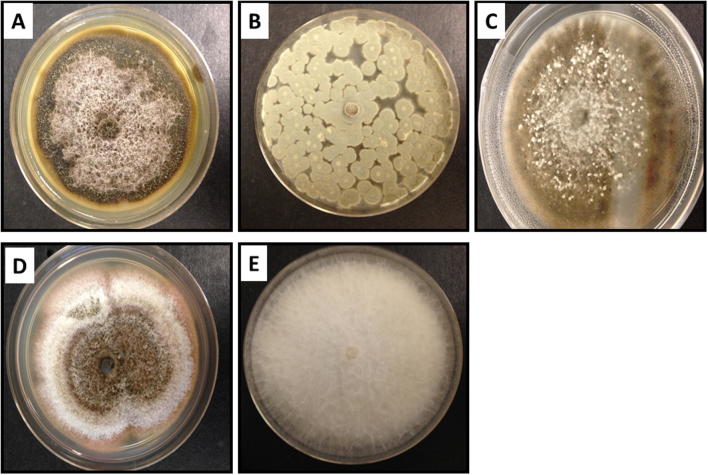
The morphological and cultural appearance of the five most active producers of l-asparaginase is represented in Fig. 4. Isolate PBL13 identified as Penicillium simplicissimum have green powdery spores, typical of Penicillium species; isolates PBS1 and CCL1 have slight discrepancies in morphological appearance despite being identified as Dothideomycetes sp. which may be attributed to species variability. MKS1 was identified as Fusarium sp. which has white fluffy mycelium. ODL4 was identified as Ascomycota sp. (Fig. 4).
Fig. 4.
Cultural characteristics of the five fungal endophytes with strong l-asparaginase activities, which include (A) PBS1 (Dothiodeomycetes sp.), (B) PBL13 (Penicillium simplicissimum), (C) ODL4 (Ascomycota sp.), (D) CCL1 (Dothideomycetes sp.) and (E) MKS1 (Fusarium oxysporum).
Discussion
The selected Malaysian plants with anticancer properties (P. bleo, M. koenigii, C. citratus and O. diffusa) were found to host endophytes with l-asparaginase activities, an essential tumor-controlling enzyme. This is the first reporting of l-asparaginase fungal endophytes for C. citratus, P. bleo, M. koenigii and O. diffusa, although l-asparaginase–producing bacteria have been isolated from C. citratus [20], [21] prior to this study. Our results supported the hypothesis that medicinal plants are potential hosts of endophytes with medicinal properties. Although we did not confirm in this study if this was a result of co-evolution of host plants with endophytes, many studies have observed similar trends and the hypothesis remained strong as postulated by [8]. We also observed that the number of endophytes isolated from each of the four medicinal plants varies. We did not vary or increase the sampling size as typically, beneficial endophytes can be recovered from few plant samples irrespective of seasonal change [35]. Several factors may influence endophyte distribution and colonization frequency, with rainfall and age of host plant as the two most common factors. Rainfall enhances endophyte dispersal and colonization [22], while older plants have larger organs to host more endophytes [23]. We do not rule out the possible influence of these two factors although we did not ascertain the age of the four plants sampled nor measured the rainfall. In addition the distinct numbers of endophytes residing in leaf and stem tissues observed in P. bleo, M. koenigii, C. citratus and O. diffusa was consistent with observations in other important medicinal plants where frequency of endophytes was highest in leaves, followed by stems and roots [24]. Leaves are generally preferred by endophytes as leaves have greater surface area [23], is nutrient-rich and thin-walled to allow endophytic colonization [25]. These factors coincide with the nature of leaves and stem of P. bleo, M. koenigii, C. citratus and O. diffusa, hence the higher number of endophytes from leaves.
l-Asparaginase producing endophytes were detected from the plate assay by the formation of pink zone around the colony. The reaction of l-asparaginase that hydrolyzed l-asparagine into ammonia increased the pH of the media which was in turn detected by phenol red, a pH indicator. Isolates incapable of expressing l-asparaginase enzymes have agar media which remained yellow in color. The correlation coefficient (r) used to explain the relationship between pink zone diameter and asparaginase activity [33], revealed that there is no significant relationship between both variables. This was also observed in numerous other studies [33]. Therefore, plate assays are often accompanied by quantitative assays using positive isolates from initial plate assay to select isolates with most potential for further studies.
Endophytes positive for l-asparaginase activity were predominantly of the genus Colletotrichum, followed by species of Fusarium, Penicillium and Phoma [26]. This result could be reflective of the endophyte diversity in the four plants tested as the teleomorph and anamorph states of ascomycetes typically dominate endophyte populations especially in leaves [27]. The genus Colletotrichum, can be found in many plants including legumes, grasses, vegetables and medicinal plants such as Justicia gendarussa and Taxus mairei [28]. Similarly, endophytic species of Fusarium [29], Phoma and Penicillium, are equally ubiquitous in distribution, in both tropical and temperate climates [30]. Although literatures on diversity of endophytes are available, documentation of their l-asparaginase production is lacking. To date, Colletotrichum, Fusarium, Penicillium and Phoma spp. are more well-known as producers of a variety of bioactive compounds [27], [28] such as alkaloids and antibiotics [31]. Hence, from this study, we add l-asparaginase production to the list, particularly for Fusarium oxysporum (MKS1) and P. simplicissimum (PBL13) from the Malaysian plants.
The l-asparaginase activities of endophytes from this study were however, inferior to levels produced by some known fungi of the Fusarium and Penicillium sp. For example, l-asparaginase activities by a Fusarium sp. were 0.08–3.14 unit mL−1 [32], while a Penicillium sp. can produce as high as 3.75 unit mL−1 [33]. These levels were significantly higher than the 0.013 and 0.019 μM−1 mL−1 min−1 produced by MKS1 (F. oxysporum) and PBL13 (P. simplicissimum) in this study, respectively. We presume the difference in amount of l-asparaginase produced may be attributed to several factors, which include culture media used and the different species of endophytes tested although of the same genus [32]. Nevertheless, this could only be further validated by performing western blot analysis to determine the relative expression patterns and characterization of l-asparaginase from fungal endophytes in comparison to existing bacteria-derived l-asparaginase [35]. The bioactivity of the l-asparaginases from various endophytes against various cancer cell lines can also we further tested to determine their application as anticancer agents [35]. Thus, we recommend these analyses for future explorations.
In our study, three endophytes were identified to only the phylum (Ascomycota) and class level (Dothideomycetes), probably due to the non-specificity of the universal primers (ITS1 and ITS4) used. Consequently, the l-asparaginase activities of ODL4 (Phylum Ascomycota), PBS1 (Class Dothideomycetes) and CCL1 (Class Dothideomycetes) cannot be compared with other studies. In future, this can be improved using genus–species specific primers. We also observed that the percentage of endophytes with l-asparaginase production was 28% of the total number isolated, with 17%, 8%, 2% and 1% for P. bleo, O. diffusa, C. citratus and M. koenigii. This percentage of l-asparaginase producing endophytes was higher than the 18% recovered from the anticancer plant Ocimum sanctum L. [34], but was relatively lower than the 35% recovered from Thai medicinal plants (Adenanthera microsperma, Betula alnoides, Cassia alata, Eupatorium odoratum, Hiptage benghalensis, Houttuynia cordata and Stemona tuberosa) [8]. Clearly, the choice of host plants is important to increase the chances of isolating more l-asparaginase producing endophytes.
Conclusions
To conclude, Malaysian anticancer plants (P. bleo, M. koenigii, C. citratus and O. diffusa) have shown potential to host l-asparaginase producing endophytes. Of this, P. bleo hosted the highest number of l-asparaginase producing endophytes, mostly in leaf tissues than stem tissues. There is no correlation between quantitative and qualitative estimation of asparaginase activity. The endophytes with good activities were ODL4 (Ascomycota), PBS1 (Dothideomycetes), PBL13 (P. simplicissimum), CCL1 (Dothideomycetes) and MKS1 (F. oxysporum), isolated from various host plants. These endophytes can be cultured in the laboratory and l-asparaginase produced can be harvested for development as anticancer compound.
Conflict of Interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
The authors express their gratitude to Monash University Malaysia for the funding (BCHH-SS-10-01-2012) and facilities to complete the project.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jare.2014.07.005.
Appendix A. Supplementary material
Supplementary Fig. 1.
Sequence similarity blast search.
References
- 1.Nelson K.E., Nelson B.J. Genetic diversity of microbial endophytes and their biotechnical applications. In: Strobels G., editor. Genomics applications for the developing world, advances in microbial ecology. Springer Science + Business Media; New York: 2012. pp. 249–262. [Google Scholar]
- 2.Patil M.P., Patil R.H., Mahjeshwari V.L. A novel and sensitive agar plug assay for screening of asparagine-producing endophytic fungi from Aegle marmelos. Acta Biol Szeged. 2012;56:175–177. [Google Scholar]
- 3.Mehanni M.M., Safwat M.S.A. Endophytes of medicinal plants. Acta Hort (ISHS) 2010;854A:31–39. [Google Scholar]
- 4.Harper J.K., Arif A.M., Ford E.J. Pestacin: a 1,3-dihydro isobenzofuran from Pestalotiopsis microspora possessing antioxidant and antimycotic activities. Tetrahedron. 2003;59:2471–2476. [Google Scholar]
- 5.Gutierrez R.M.P., Gonzalez A.M.N., Ramirez A.M. Compounds derived from endophytes: a review of phytochemistry and pharmacology. Curr Med Chem. 2012;19:2992–3030. doi: 10.2174/092986712800672111. [DOI] [PubMed] [Google Scholar]
- 6.Cassady J.M., Chan K.K., Floss H.G., Leistner E. Recent developments in the maytansinoid antitumor agents. Chem Pharm Bull. 2004;52:1–26. doi: 10.1248/cpb.52.1. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J., Li C., Wang W., Zhao C., Luo M., Mu F. Hypocrea lixii, novel endophytic fungi producing anticancer agent cajanol, isolated from pigeon pea (Cajanus cajan [L]. Mill sp.) J Appl Microb. 2013;115:102–113. doi: 10.1111/jam.12195. [DOI] [PubMed] [Google Scholar]
- 8.Theantana T., Hyde K.D., Lumyong S. Asparaginase production by endophytic fungi from Thai medicinal plants: cytotoxicity properties. Int J Integr Biol. 2009;7:1–8. [Google Scholar]
- 9.Verma N., Kumar K., Kaur G., Anand S. l-Asparaginase: a promising chemotherapeutic agent. Crit Rev Biotechnol. 2007;27:45–62. doi: 10.1080/07388550601173926. [DOI] [PubMed] [Google Scholar]
- 10.McCredie J.A., Inch W.R., Kruuv J., Watson T.A. The rate of tumor growth in animals. Growth. 1965;29:331. [Google Scholar]
- 11.Goh K.L. millennium ed. Advanco Press; Malaysia: 2007. Malaysian herbaceous plants. p. 142 [in Chinese] [Google Scholar]
- 12.Kong Y.C., Ng K.H., But P.P., Yu Q.L.S.X., Zhang H.T., Cheng K.F. Sources of the anti-implantation alkaloid yuehchukene in the genes Murraya. J Ethnopharmacol. 1986;15:195–200. doi: 10.1016/0378-8741(86)90155-8. [DOI] [PubMed] [Google Scholar]
- 13.Sati S.C., Belwal M. Aquatic hyphomycetes as endophytes of riparian plant roots. Mycologia. 2005;97:45–49. doi: 10.3852/mycologia.97.1.45. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez J.A., Astudillo L., Hirschmann G.S. Oleanolic acid promotes healing of acetic acid-induced chronic gastric lesions in rats. Pharmacol Res. 2003;48:291–294. doi: 10.1016/s1043-6618(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 15.Krishnanmurthy Y.L., Hemalatha T.V. Isolation of endophytic fungi from some grasses. J Mycol Plant Pathol. 2003;33:305–306. [Google Scholar]
- 16.Ting A.S.Y., Meon S., Kadir J., Radu S., Singh G. Endophytic microorganisms as potential growth promoters of banana. Biocontrol. 2008;53:541–553. [Google Scholar]
- 17.Kumar N.S.M., Ramasamy R., Manonmani H.K. Production and optimization of l-asparaginase from Cladosporium sp. using agricultural residues in solid state fermentation. Ind Crop Prod. 2013;43:150–158. [Google Scholar]
- 18.Tan M.L., Sulaiman S.F., Najimuddin N., Samian M.R., Tengku Muhammad T.S. Methanolic extract of Pereskia bleo (Kunth) DC. (Cactaceae) induces apoptosis in breast carcinoma, T47-D cell line. J Ethnopharmacol. 2005;96:287–294. doi: 10.1016/j.jep.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Diaz P.L., Hennell J.R., Sucher N.J. Genomic DNA extraction and barcoding of endophytic fungi. In: Sucher N.J., Hennell J.R., Carles M.C., editors. Plant DNA fingerprinting and barcoding: methods and protocols, methods in molecular biology. Springer Science + Business Media, LLC; New York: 2012. pp. 171–179. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura C.T., Wilkinson R., Woodruff K. Pancreatitis and parotitis following therapy with l-asparaginase. Int Ped. 2012;14:25–27. [Google Scholar]
- 21.Khamna S., Yokota A., Lumyong S. l-Asparaginase production by actinomycetes isolated from Thai medicinal plant rhizosphere soils. Int J Integr Biol. 2012;6:22–26. [Google Scholar]
- 22.Wilson D., Faeth S.H. Do fungal endophytes result in selection for leaf miner ovipositional preference. Ecology. 2001;82:1097–1111. [Google Scholar]
- 23.Chareprasert S., Piapukiew J., Thienhirun S., Whalley A.J.S., Sihanonth P. Endophytic fungi of teak leaves Tectona grandis L. and rain tree leaves Samanea saman Merr. World J Microb Biot. 2006;22:481–486. [Google Scholar]
- 24.Verma V.C., Gond S.K., Kumar A., Kharwar R.N. Endophytic mycoflora from leaf, bark and stem of Azadirachata indica A. Juss (Neem) from Varanasi (India) Microb Ecol. 2007;54:119–125. doi: 10.1007/s00248-006-9179-9. [DOI] [PubMed] [Google Scholar]
- 25.Huang W.Y., Cai Y.Z., Hyde K.D., Corke H. Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fung Divers. 2008;33:61–75. [Google Scholar]
- 26.Thirunavukkarasu N., Suryanarayanan T.S., Murali T.S., Ravishankar J.P., Gummadi S.N. l-Asparaginase from marine derived fungal endophytes of seaweeds. Mycosphere. 2011;2:147–155. [Google Scholar]
- 27.Gangadevi V., Sethumeenai S., Yogeswari S., Rani G. Screening endophytic fungi isolated from a medicinal plant, Acalypha indica L. for antibacterial activity. Indian J Sci Tech. 2008;1:1–6. [Google Scholar]
- 28.Senthil R.K., Joon H.H., Kim H.J. In vitro screening of taxol, an anticancer drug produced by the fungus, Colletotrichum capsici. Eng Life Sci. 2011;11:254–271. [Google Scholar]
- 29.Bottalico A., Perrone G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur J Plant Pathol. 2002;108:611–624. [Google Scholar]
- 30.Aweskamp M.M., Verkley G.J.M., Gruyter J.D., TurcoGroenewald E.J.Z., Crous P.W. Development of taxon-specific sequence characterized amplified region (SCAR) markers based on actin sequences and DNA amplification fingerprinting (DAF): a case study in the Phoma exigua species complex. Mol Plant Pathol. 2009;10:403–414. doi: 10.1111/j.1364-3703.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumara P.M., Zuehlke S., Priti V., Ramesha B.T., Shweta S., Ravikanth G. Fusarium proliferatum, an endophytic fungus from Dysoxylum binectariferum Hook. F, produces rohitukine, a chromane alkaloid possessing anti-cancer activity. A Van Leeuw. 2012;101:323–329. doi: 10.1007/s10482-011-9638-2. [DOI] [PubMed] [Google Scholar]
- 32.Nakahama K., Imada A., Igarasi S. Formation of l-asparaginase by Fusarium species. J Gen Microbiol. 1973;75:269–273. doi: 10.1099/00221287-75-2-269. [DOI] [PubMed] [Google Scholar]
- 33.Soniyamby A.R., Lalitha S., Praveesh B.V., Priyadarshini V. Isolation, production and antitumour activity of l-asparaginase of Penicillium sp. Int J Microbiol. 2011;2:38–42. [Google Scholar]
- 34.Pradeep S.M., Mahmood R., Jagadeesh K.S. Screening and characterization of l-asparaginase producing microorganisms from tulsi. J Agric Sci. 2010;23:660–661. [Google Scholar]
- 35.Mahajan R.V., Kumar V., Rajendran V., Saran S., Ghosh P.C., Saxena R.K. Purification and characterization of a novel and robust l-asparaginase having low-glutaminase activity from Bacillus licheniformis: in vitro evaluation of anti-cancerous properties. PLoS ONE. 2014;9:e99037. doi: 10.1371/journal.pone.0099037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence similarity blast search.