Abstract
Curcumin, an active biphenolic molecule present in turmeric (Curcuma longa), has been reported to elicit plethora of health protective effects. The present study was carried out in vitro, in vivo and in silico to investigate the modulatory effects of curcumin on erythrocyte membrane Na+/K+-ATPase activity. In vitro curcumin (10−5 M to 10−8 M) was incubated with human erythrocytes membrane. In vivo curcumin (340 mg/kg b.w. and 170 mg/kg b.w.) was supplemented to wistar rats for 21 days. In silico, catalytic unit α of Na+/K+-ATPase (3b8e.pdb) protein was used as a receptor for the natural ligand ATP to study curcumin-mediated docking simulation using AutoDock4. The in vitro effect of curcumin on the Na+/K+-ATPase activity in human erythrocytes was biphasic. An inhibitory response was observed at 10−5 M (p < 0.001). An activation of the Na+/K+-ATPase activity was observed at 10−7 and 10−8 M (p < 0.001 and p < 0.01). In vivo, curcumin supplementation to rats increased the Na+/K+-ATPase activity at doses 340 mg/kg b.w. (p < 0.001) as well as at 170 mg/kg b.w., (p < 0.01). AutoDock4 docking simulation study showed that both ligands curcumin and ATP actively interacted with amino acids Glu214, Ser215, Glu216, Thr371, Asn377, Arg378, Met379, Arg438, Val440, Ala444, Lys451 and Asp586 at the catalytic cavity of Na+/K+-ATPase. ATP had more H bonding and hydrophobic interaction with active site amino acid residues compared to curcumin. These finding may explain some of the health beneficial properties of curcumin associated with deregulated Na+/K+-ATPase activity or ions homeostasis.
Abbreviations: CUR, curcumin; ATP, Adenosine Tri-Phosphate; RBCs, Red Blood Cells; RMS, root mean square
Keywords: Curcumin, Erythrocytes, Na+/K+ ATPase, In silico
Introduction
Curcumin (1E,6E)-1,7-Bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione, a natural biphenolic compound isolated from turmeric (Curcuma longa) has been reported to elicit a plethora of health protective effects [1]. Curcumin is lipophilic and therefore has the ability to localize between polar head and non-polar tail of lipid molecule in plasma membrane. This interaction influences the fluidity and thickness of the membrane. The localization between the membrane lipid bilayers significantly weakens the elasticity moduli of the bilayers [2], [3]. Insertion of curcumin into the membrane modifies the activity of several functionally unrelated membrane-bound proteins and related signalling cascade systems [4], [5].
Na+/K+-ATPase (EC 3.6.3.9) is a heterodimeric, trans-membrane, ubiquitously present protein that regulates neuronal signalling, ion homeostasis, substrate transportation and muscle contraction [6]. Besides its inotropic effects, Na+/K+-ATPase also acts as a signal transducer regulating many cellular events including those associated with tumour cell growth [7]. In view of its important cellular role, there is an increasing interest in the characterization of chemicals which may modulate this enzyme.
Na+/K+-ATPase (P-type ATPases) is made up of an active α unit (110 kDa) with 10 Trans membrane segments (TMS; αM1–αM10), sugar rich auxiliary β unit (55 kDa) and a hydrophobic single membrane crossing protein γ unit (12 kDa) for regulating ionic gradient across the cell membrane [8]. There are several isoforms of the binding units in the primary catalytic unit present in different tissues: α1 in nerves, kidney and lung, α2 in heart and skeletal muscle, α3 in the brain and α4 in testis and specifically in spermatozoa [9].
The membrane’s physical and biochemical properties are strongly regulated by lipid composition and redox status of the environment. Changes in membrane fluidity have been shown to modulate the activity of membrane bound receptors, enzymes and ion-exchangers [10], [11]. Na+/K+ ATPase activity is modulated by the surrounding microenvironment of lipids; thus, modifications in the membrane fluidity translate into effects on Na+/K+ ATPase activity. An altered Na+/K+-ATPase activity has been reported during late complications of diabetes mellitus such as nephropathy, neuropathy, retinopathy and in the development of diabetic vascular complications [12], [13], [14]. Elevation of intracellular sodium and potassium value was associated with reduced activity of erythrocyte Na+/K+-ATPase pump [15]. The present study was carried out in vitro, in vivo and in silico to investigate the modulatory effects of curcumin on ouabain-sensitive Na+/K+-ATPase from erythrocyte membrane of humans and rats. In addition, catalytic unit α of Na+/K+-ATPase (3b8e.pdb) protein has been used as a receptor for natural ligand adenosine triphosphate (ATP) and curcumin-mediated docking simulation using AutoDock4. The suitable docked conformation between receptor and ligands was predicted on the basis of cluster analysis.
Material and methods
Chemicals and instrument
Curcumin ((1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione) was purchased from Bio Basic Inc., Ontario, Canada (cat. # CB0346), and Imidazole, Ouabain, ATP, Bovine Serum Albumin (BSA) were purchased from Sigma Aldrich, India. Other chemicals of highest purity were purchased from Merck, India, and HIMEDIA Labs, India. Spectrophotometric measurements were performed on Shimadzu-UV-1800 (Japan) UV–VIS spectrophotometer.
Experimental study
Human erythrocyte
Human venous blood was collected in heparin from 26 healthy volunteers of both sexes between the 24 and 45 years of age by venipuncture. The human subjects were screened for diabetes mellitus, asthma, tuberculosis, or other major illness. None of the subjects were smokers or were taking any medication. All 26 selected subjects gave informed consent for the use of their blood samples for the research study. The protocol of study was in conformity with the guidelines of the Institutional Ethical Committee, University of Allahabad. The heparinized blood was centrifuged at 800g at 4 °C for 10 min. After the removal of plasma, buffy coat and top layer comprising approximately 15–20% of the packed cells, the remaining packed erythrocytes were washed twice with 10 mM phosphate buffered saline pH 7.4.
Animal
Male albino rats (Wistar strain) of 5–6 months weighing between 150 and 200 g were purchased from CDRI, Lucknow, India. Animals were housed in polypropylene cages at 24 ± 2 °C (6 rats per cage) and 12 h light:12 h dark cycles. Animals were fed with standard pellet diet obtained from Dayal Industries Limited, Lucknow, India, and had free access to drinking water. Rats were acclimated for one week before treatment. The protocol of study was in conformity with the guidelines of the Institutional Ethical Committee of University of Allahabad.
Twenty-four male Wistar rats were randomly divided into four groups (six rats/group). Group [I]: Control, receiving no treatment/supplementation. Group [II]: Experimental control, rats were supplemented with 0.9% NaCl solution through oral route. Group [III]: Curcumin-treated group (340 mg/kg b.w., saline) [16]. Group [IV]: Curcumin-treated group (170 mg/kg b.w., saline).
Curcumin and saline treatments were given through oral route for 21 days at fixed time 11.00 am to 12.00 pm to avoid circadian disturbance. At the end of treatment, rats were sacrificed under light anaesthesia. The blood was collected in heparinized tubes by heart puncture. The heparinized blood was centrifuged at 800g at 4 °C for 10 min. After the removal of plasma, buffy coat and upper 15–20% of the packed RBCs cells, the remaining RBCs were washed twice with 10 mM phosphate buffered saline pH 7.4.
Preparation of erythrocytes membrane
The erythrocyte membrane was isolated according to the method of Marchesi and Palade [17]. The erythrocyte membrane proteins were quantified according to the method of Lowry et al. [18].
Measurement of Na+/K+-ATPase activity
Na+/K+-ATPase activity was measured according to the method of Suhail and Rizvi [13]. The final assay mixture contained 0.5–1.2 mg membrane protein/mL, 20 mmol/L KCl, 140 mmol/L NaCl, 3 mmol/L MgCl2, 30 mmol/L imidazole (pH 7.24), with or without 5 × 10–4 mol/L ouabain and 6 mmol/L ATP. Assay mixture was incubated for 30 min at 37 °C and the reaction was stopped by the addition of 3.5 mL of a solution-A (0.5% ammonium molybdate, 0.5 mol/L H2SO4, and 2% sodium dodecyl sulphate). The amount of liberated phosphate (Pi) was estimated according to the method of Fiske and Subbarow [19]. In vitro experiment was carried out by adding curcumin (final concentration 10−5 M to 10−8 M) to the assay mixture and incubated for 30 min at 37 °C prior to enzyme assay. Na+/K+-ATPase activity was expressed as nmol pi released/mg protein per hour at 37 °C.
Statistical analysis
Statistical analysis was performed by the software GraphPad Prism 5 version 5.01. One way analysis of variance (ANOVA) was performed for multiple comparisons. P-values were evaluated by two tailed method. Statistical differences shown in Fig. 1 represent in vitro study (n = 26) on human erythrocytes, and Fig. 2 represents in vivo study (n = 6) on rat erythrocytes. p < 0.05 was considered to be significant. Values are represented as ±SD in graphs. Significance between experimental and control is represented by star (∗) in graph.
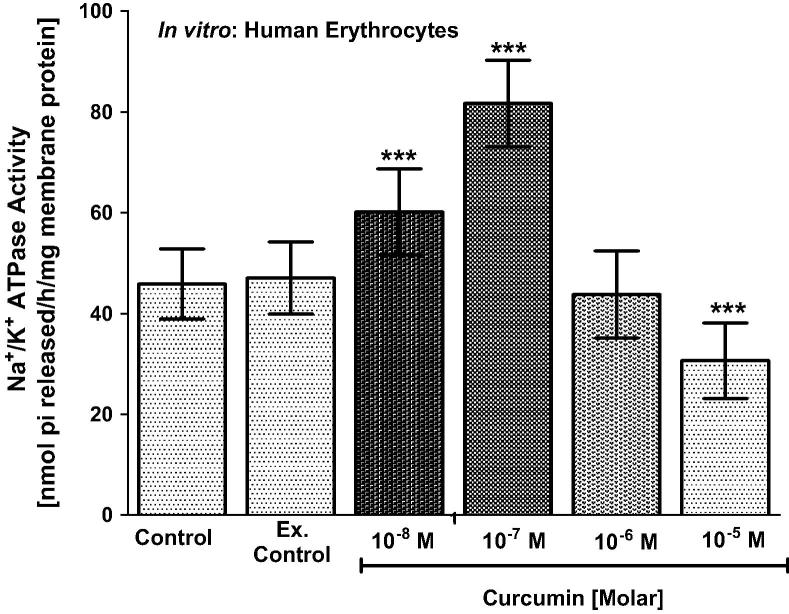
Fig. 1.
In vitro effect of curcumin (10−5 M to 10−8 M) on human erythrocyte’s membrane Na+/K+-ATPase activity (Ouabain-sensitive). Na+/K+-ATPase activity was expressed as nmol pi released/h/mg membrane protein at 37 °C. Values (n = 26) are means ± S.D.
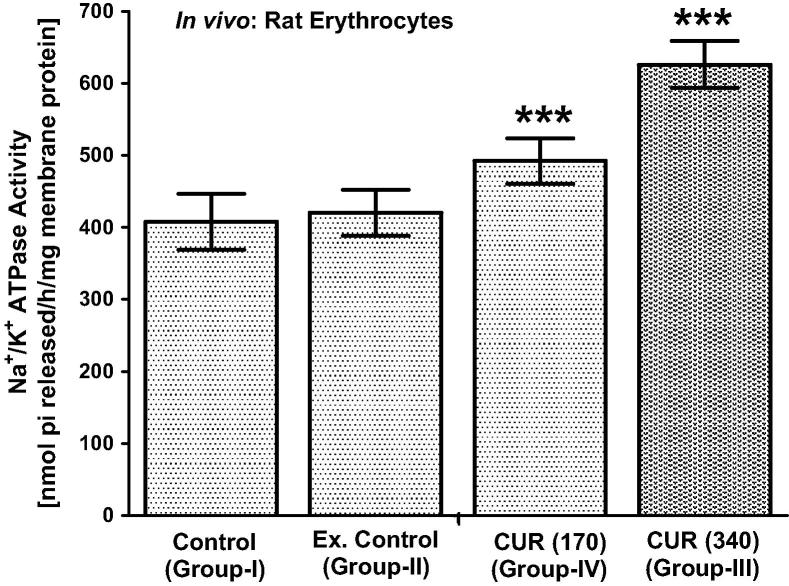
Fig. 2.
In vivo effect of curcumin (340 mg/kg b.w. and 170 mg/kg b.w. oral) on wistar albino rat’s erythrocyte membrane Na+/K+-ATPase activity (Ouabain-sensitive). Na+/K+-ATPase activity was expressed as nmol pi released/h/mg membrane protein at 37 °C. Values (n = 6) are means ± S.D.
Computational study
Selection of protein and ligand
A three-dimensional X-ray crystallized structure of Na+/K+-ATPase subunit α-1 protein complexed with 2 PC1 (2-diacyl-sn-glycero-3-phosphocholine) as a ligand and 6 Rb+ and 2 Mg++ together with 2 F4Mg−•− as a cofactor (PDB ID: 3B8E, Resolution = 3.50 Å, Chains: A, B, C, D, G, H) were downloaded from the Protein Data Bank [20], [21]. Pubchem compound database was used to retrieve the 3 dimensional structures of selected ligands (ATP and curcumin) and energy minimization was done using UCSF Chimera software (Developed by the Resource for Biocomputing, Visualization, and Informatics and can be downloaded from https://www.cgl.ucsf.edu/chimera/).
Proteins and ligands structure preparation
The selected target protein (PDB ID: 3B8E) and ligands (curcumin and ATP) were prepared as an input file for docking by using MGL (Molecular Graphics Laboratory) Tool 1.5.6 developed by The Scripps Research Institute for visualization and analysis of molecular structures [22], [23]. The co-crystallized heteroatoms and ligands were removed and final .pdbqs format file of receptor protein was used during docking simulation. On the basis of X-ray crystallized bound ligand MF4 (magnesium tetrafluoride), the active site was chosen for current docking study with curcumin and ATP [22].
Grid design and docking simulation
AutoGrid 4 was used to obtain the grid maps required prior to docking and AutoDock 4 for docking study [24]. AutoDock4 and AutoGrid4 tool have been developed by The Scripps Research Institute, U.S.A. which can be downloaded from http://autodock.scripps.edu/downloads. The user defined three dimensional grid covered the region of active or binding site of interest in the receptor and selected ligands and were limited to this search space during docking. A grid box size set on the basis of requirement (100 × 100 × 100 points with spacing of 0.375 Å) was used for the study. The binding constant or inhibition constant (kI) was directly calculated by AutoDock4 during the simulation for each conformation [22], [24]. The docked conformation of selected ligands with the receptor has been demonstrated in 2-dimensional page by using LIGPLOT version 4.5.3 (Developed by Wallace et al., 1995 and can be downloaded from www.ebi.ac.uk/thornton-srv/software/LIGPLOT/) [25].
Results
Fig. 1 shows that the effect of curcumin on the Na+/K+-ATPase activity in human erythrocytes was biphasic. An inhibitory response was observed at 10−5 M (P < 0.001) while an activation of the activity was observed at 10−7 M (P < 0.001) and 10−8 M (p < 0.001).
Fig. 2 shows that in vivo, curcumin (340 mg/kg b.w, oral) supplementation to rats significantly (P < 0.001) increased the Na+/K+-ATPase activity in erythrocyte membrane as compared with control and experimental control rats. Lower dose of curcumin supplementation (170 mg/kg b.w., oral) also significantly (P < 0.001) increased the Na+/K+-ATPase activity.
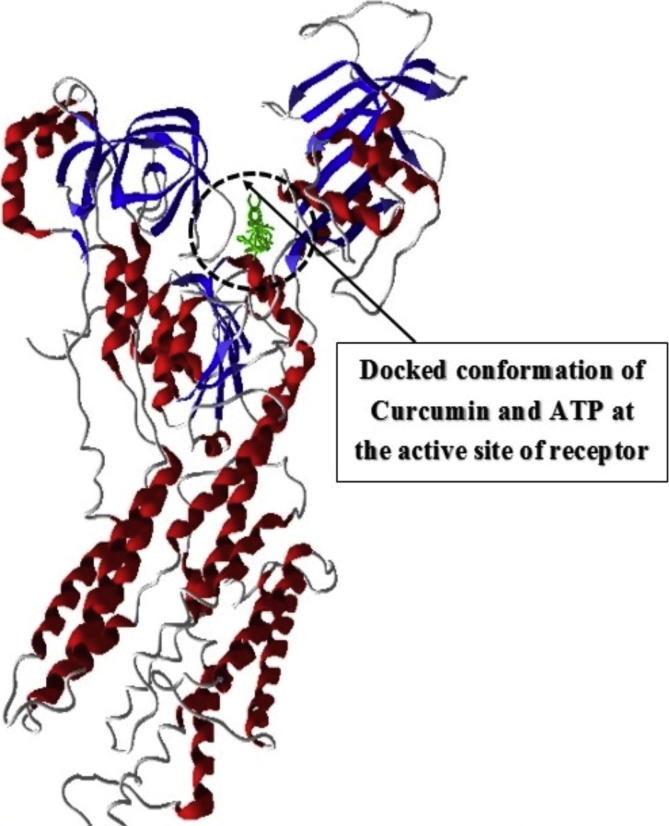
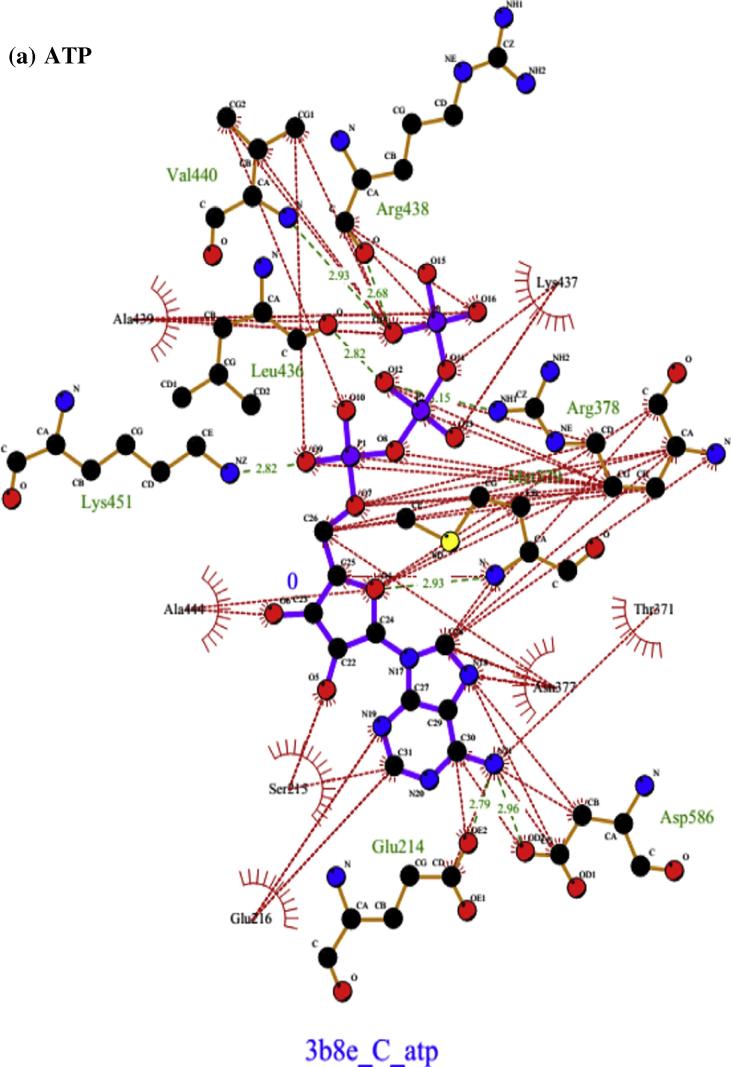
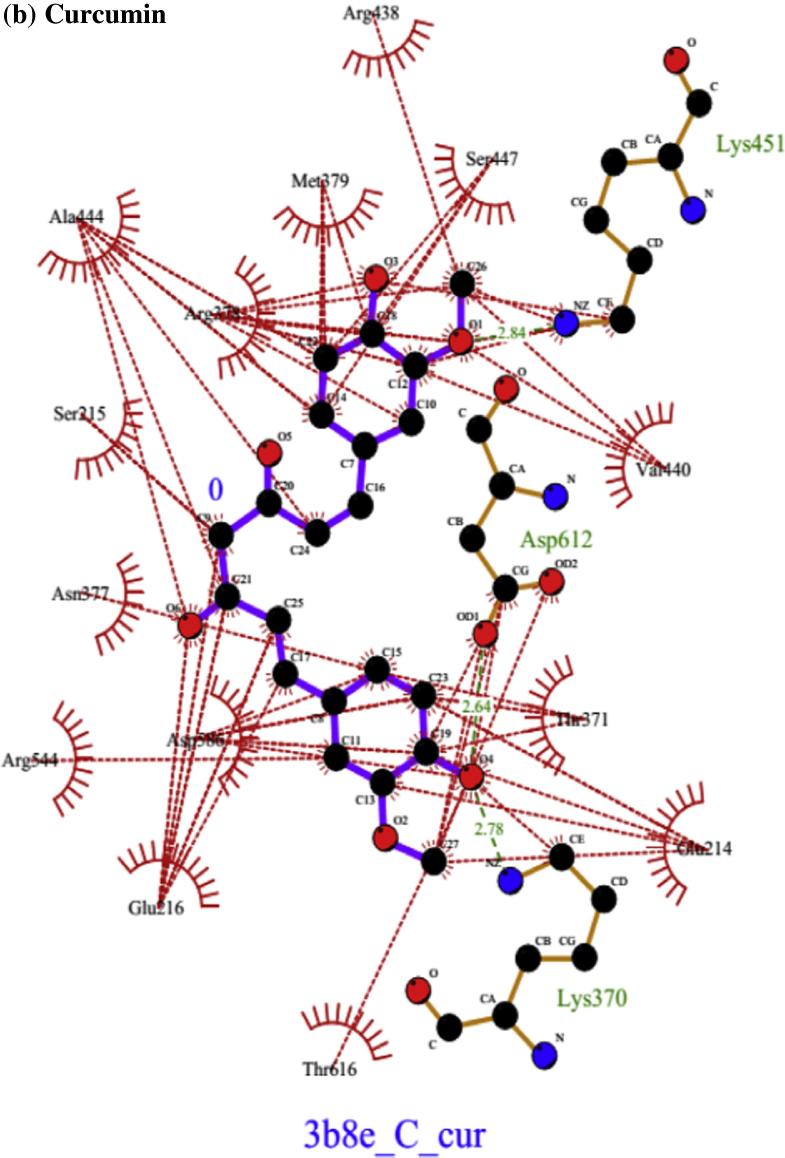
Fig. 3 shows the secondary structure (cartoon representation) of Na+/K+-ATPase subunit α-1 together with docked conformation of ligand ATP and curcumin. The AutoDock4 docking simulation results presented in Fig. 4(a) show that the active site residues at catalytic unit α of Na+/K+-ATPase (3b8e.pdb) Glu214, Ser215, Glu216, Thr371, Asn377, Arg378, Met379, Leu436, Lys437, Arg438, Ala439, Val440, Ala444, Lys451, Asp586 were actively involved in hydrogen bonding and hydrophobic interaction with natural ligand ATP. On the other hand Fig. 4(b) shows that curcumin interacted actively with Glu214, Ser215, Glu216, Lys370, Thr371, Asn377, Arg378, Met379, Arg438, Val440, Ala444, Ser447, Lys451, Arg544, Asp586, Asp612, Thr616 amino acids at the catalytic centre. Eight hydrogen bonds were formed with amino acids at active site by ATP. However, curcumin formed only three hydrogen bonds with different amino acids at active cavity. Table 1 shows that the binding energy of curcumin was relatively higher (−7.4 kcal) than ATP (−11.55 kcal).
Fig. 3.
Secondary structure (cartoon) representation at the active site of Na+/K+-ATPase subunit α-1 together with docked conformation of ligand ATP and curcumin.
Fig. 4.
Docked conformation of hydrogen bonding view and hydrophobic interaction of (a) ATP, (b) curcumin with amino acids of human Na+/K+-ATPase subunit α-1 protein (3b8e.pdb) at the active site cavity (hydrogen bonds as green dashed lines between the atoms involved and hydrophobic contacts as an arc with spokes radiating towards the ligand atoms).
Table 1.
Comparative docking simulation result of ligands (ATP and curcumin) with Na+/K+-ATPase protein (3b8e.pdb).
| Properties | ATP | Curcumin | |
|---|---|---|---|
| 1. | Binding energy (kcal/mol) | −11.55 | −7.4 |
| 2. | Ligand efficiency | −0.37 | −0.27 |
| 3. | Inhibition constant (kI) | 3.34e−009 | 3.75e−006 |
| 4. | Intermole energy (kcal/mol) | −16.034 | −10.39 |
| 5. | Torsional energy (kcal/mol) | 4.47 | 2.98 |
| 6. | No. of H-bond interactions | 8 | 3 |
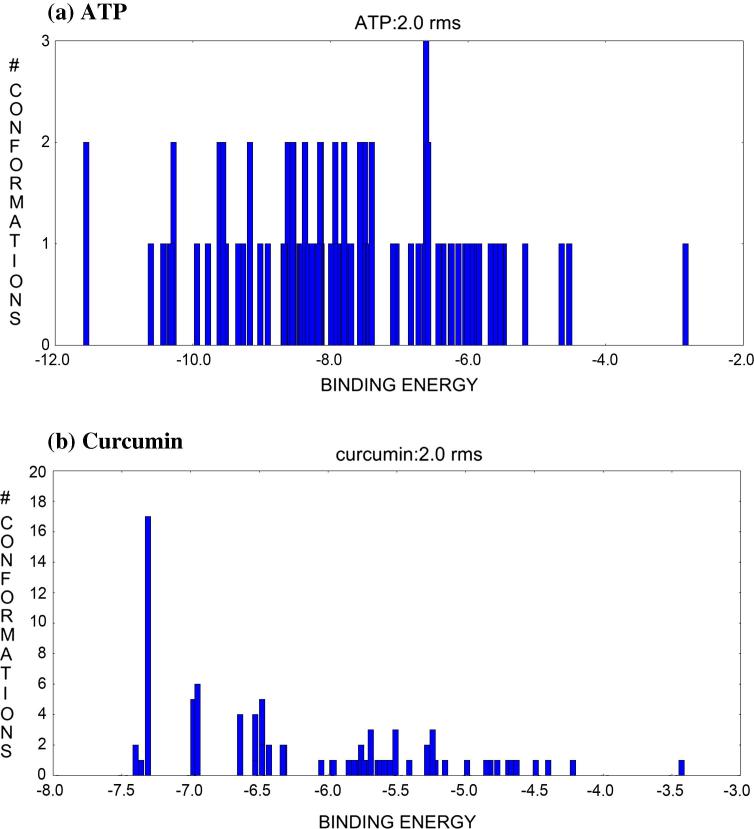
During docking simulation with cluster root mean square (RMS) = 2.0, different clusters were formed and each cluster except a single cluster (having 17 conformation) had 1–6 conformation of curcumin based cluster RMS value. ATP docking simulation showed several clusters with average conformation of 1–3. Fig. 5a and b shows all the conformations for clusters ranked according to binding energy.
Fig. 5.
Possible cluster between conformations and binding energy within range of root mean square (RMS) tolerance (a) ATP, (b) curcumin.
Discussion
Na+/K+-ATPase regulates different physiological and metabolic processes through regulating ion gradient across the cell membrane of excitable tissues [9], [26]. Several drugs modulate the Na+/K+-ATPase activity by binding at the active site, allosteric site, lipid microenvironment of the membrane or through a synergistic mechanism. Na+/K+-ATPase activity was reported to strongly synchronize with the fluidity of membrane and by drugs affecting fluidity [5], [27]. Na+/K+-ATPase activity inhibitory effect in the presence of high concentration of curcumin was reported by Mahmmoud (2005). This report showed that in shark rectal glands and pig kidney the EC50 of curcumin for Na+/K+-ATPase activity was 15.8 ± 1.12 μM and 5.24 ± 1.0 μM respectively [28]. Sharks contain higher cholesterol than pig kidney membranes; hence, the higher EC50 value of curcumin for Na+/K+-ATPase activity may be due to differences in membrane fluidity [29]. It was observed that curcumin interacted with trans-membrane domain(s) of the α-subunit of Na+/K+ ATPase protein and reduced the oligomycin inhibitory effects [16]. Oligomycin and curcumin have similar modulatory effects on the kinetic properties of the Na+/K+-ATPase [28]. We propose that higher concentrations of curcumin (10−5 M) caused down-regulation of Na+/K+-ATPase activity in human erythrocyte membrane by directly interacting at the active catalytic centre of the enzyme. However, lower concentrations of curcumin (10−7 M) modulated the membrane fluidity resulting in an increased activity of the enzyme.
Curcumin reduced the Na+/K+-ATPase activity in human blood mononuclear cells during initial days (up to 3 days of 7 days) of incubation, but prolonged incubation increased the Na+/K+-ATPase activity [30]. Curcumin supplementation to retinol deficient rats resulted in increased activity of brain microsomal membrane Na+/K+-ATPase activity mediated through improved cholesterol:phospholipid ratio [31]. In vitro we observed that curcumin at 10−7 M concentration increased the Na+/K+-ATPase activity maximally which decreased further on decreasing the curcumin concentration till 10−8 M. In vivo curcumin supplementation (340 mg/kg b.w. and 170 mg/kg b.w.) to rats increased the Na+/K+-ATPase activity in erythrocyte membrane thus corroborating the in vitro curcumin effects on Na+/K+-ATPase activity.
Minimum energy docked conformation after cluster analysis in MGL Tools and AutoDock4 docking simulation suggested that curcumin actively interacted to form hydrogen bond with amino acids viz., Lys370, Lys451 and Asp612 at the active site cavity of Na+/K+-ATPase α unit. Comparative binding analysis of curcumin and ATP shows that both ligands have common amino acids Glu214, Ser215, Glu216, Thr371, Asn377, Arg378, Met379, Arg438, Val440, Ala444, Lys451, Asp586 and among these Lys451 is the key residue for hydrogen bonding. Table 1 shows that ATP had more H bonding interactions compared to curcumin at the active site cavity of receptor protein.
In silico results show that curcumin has lower binding affinity due to less hydrogen bonding and hydrophobic interaction at the active site of Na+/K+ ATPase protein in comparison with natural ligand ATP. Down-regulating Na+/K+ ATPase activity effects of curcumin at higher concentration >10−6 M were hypothesized to be due to the interaction of curcumin with amino acids involved in active catalysis at α-subunit of Na+/K+ ATPase. However, increased Na+/K+ ATPase activity at lower concentration <10−7 M of curcumin is thought to be due to altering the membrane fluidity in favour of increased Na+/K+ ATPase activity in erythrocyte membrane.
Conclusions
The ion gradient and active membrane potential derived from Na+/K+-ATPase activity form the basis for a range of necessary cellular processes, especially Na+ and H+-dependent secondary transport systems. Curcumin binds and interacts at the active site cavity of Na+/K+-ATPase to down-regulate the enzyme activity at higher concentration (>10−6 M). However, curcumin surprisingly increased the enzyme activity at lower concentration (<10−7 M). In vivo oral supplementation of curcumin also increased Na+/K+-ATPase activity. The study concludes that curcumin has significant potential to modulate Na+/K+-ATPase activity dose dependently and the present findings may help to explain some of the biological effects of curcumin.
Conflict of Interest
The authors have declared no conflict of interest.
Acknowledgements
Prabhakar Singh acknowledges the support of Council of Scientific and Industrial Research (CSIR), New Delhi, India, for providing Senior Research Fellowship. Rajesh Kumar Kesharwani acknowledges the Indian Council of Medical Research (ICMR), New Delhi, India, for providing financial support as Senior Research Fellowship (SRF). Department of Biochemistry is also a recipient of FIST grant from Department of Science and Technology, Govt. of India.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Hung W.C., Chen F.Y., Lee C.C., Sun Y., Lee M.T., Huang H.W. Membrane-thinning effect of curcumin. Biophys. J. 2008;94:4331–4338. doi: 10.1529/biophysj.107.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kesharwani K.R., Misra K. Prediction of binding site for curcuminoids at human toposomerase II α protein; an in silico approach. Curr. Sci. 2011;101:1060–1065. [Google Scholar]
- 3.Ingolfsson H.I., Koeppe R.E., 2nd, Andersen O.S. Curcumin is a modulator of bilayer material properties. Biochemistry. 2007;46:10384–10391. doi: 10.1021/bi701013n. [DOI] [PubMed] [Google Scholar]
- 4.Singh P., Rizvi S.I. Curcumin activates erythrocyte membrane acetylcholinesterase. Lett. Drug Des. Discov. 2013;10:550–556. [Google Scholar]
- 5.Mahmmoud Y.A. Curcumin is a lipid dependent inhibitor of the Na, K-ATPase that likely interacts at the protein-lipid interface. Biochim. Biophys. Acta. 2011;1808:466–473. doi: 10.1016/j.bbamem.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Wang H.Y., O’Doherty G.A. Modulators of Na/K-ATPase: a patent review. Exp. Opin. Ther. Pat. 2012;22:587–605. doi: 10.1517/13543776.2012.690033. [DOI] [PubMed] [Google Scholar]
- 7.Rocafull M.A., Thomas L.E., del Castillo J.R. The second sodium pump: from the function to the gene. Pflugers Arch. 2012;463:755–777. doi: 10.1007/s00424-012-1101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suhail M. Na+, K+-ATPase: ubiquitous multifunctional transmembrane protein and its relevance to various pathophysiological conditions. J. Clin. Med. Res. 2010;2:1–17. doi: 10.4021/jocmr2010.02.263w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan J.H. Biochemistry of Na, K-ATPase. Annu. Rev. Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 10.Halliwell B., Gutteridge J.M.C. Oxygen free radicals and iron in relation to biology and medicine: some problems and concept. Arch. Biochem. Biophys. 1986;246:501–514. doi: 10.1016/0003-9861(86)90305-x. [DOI] [PubMed] [Google Scholar]
- 11.Maridonneau I., Barquet P., Garay R.P. Na+ and K+ transport damage induced by oxygen free radicals in human red cell membranes. J. Biol. Chem. 1983;258:3107–3113. [PubMed] [Google Scholar]
- 12.Jain S.K., Lim G. Lipoic acid decreases lipid peroxidation and protein glycosylation and increases (Na(+) + K(+)) and Ca(++)-ATPase activities in high glucose-treated human erythrocytes. Free Radic. Biol. Med. 2000;29:1122–1128. doi: 10.1016/s0891-5849(00)00410-x. [DOI] [PubMed] [Google Scholar]
- 13.Suhail M., Rizvi S.I. Red cell membrane (Na+ + K+)-ATPase in diabetes mellitus. Biochem. Biophys. Res. Commun. 1987;146:179–186. doi: 10.1016/0006-291x(87)90708-x. [DOI] [PubMed] [Google Scholar]
- 14.Jeffcoate S.L. Diabetes control and complications: the role of glycated haemoglobin, 25 years on. Diabet. Med. 2004;21:657–665. doi: 10.1046/j.1464-5491.2003.01065.x. [DOI] [PubMed] [Google Scholar]
- 15.Garay R., Adragna N., Canessa M., Tosteson D. Outward sodium and potassium co-transport in human red cells. J. Membr. Biol. 1981;62:169–174. doi: 10.1007/BF01998162. [DOI] [PubMed] [Google Scholar]
- 16.Marczylo T.H., Verschoyle R.D., Cooke D.N., Morazzoni P., Steward W.P., Gescher A.J. Comparison of systemic availability of curcumin with that of curcumin formulated with phosphatidylcholine. Cancer Chemother. Pharmacol. 2007;60:171–177. doi: 10.1007/s00280-006-0355-x. [DOI] [PubMed] [Google Scholar]
- 17.Marchesi V.T., Palade G.E. The localization of Mg–Na–K-activated adenosine triphosphatase on red cell ghost membranes. J. Cell. Biol. 1967;35:385–404. doi: 10.1083/jcb.35.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Fiske C., Subbarow Y. The colourimetric determination of phosphorus. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- 20.Morth J.P., Pedersen B.P., Toustrup-Jensen M.S., Sørensen T.L.M., Petersen J., Andersen J.P. Crystal structure of the sodium potassium pump. Nature. 2007;450:1043–1050. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H. The protein data bank. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 23.Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 24.Morris G.M., Goodsell D.S., Huey R., Olson A.J. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J. Comput. Aided Mol. Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 25.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Prot. Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi A., Reyes N., Artigas P., Gadsby D.C. The ion pathway through the opened Na(+), K(+)-ATPase pump. Nature. 2008;456:413–416. doi: 10.1038/nature07350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 28.Mahmmoud Y.A. Curcumin modulation of Na+, K+-ATPase: phosphoenzyme accumulation, decreased K+ occlusion, and inhibition of hydrolytic activity. Br. J. Pharmacol. 2005;145:236–245. doi: 10.1038/sj.bjp.0706185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornelius F., Turner N., Christensen H.R. Modulation of Na, K-ATPase by phospholipids and cholesterol. II. Steady-state and pre steady-state kinetics. Biochemistry. 2003;42(28):8541–8549. doi: 10.1021/bi034532e. [DOI] [PubMed] [Google Scholar]
- 30.Hari Cohly H.P., Rao M.R., Kanji V.K., Patlolla B., Taylor A., Wilson M.T. Effect of turmeric, turmerin and curcumin on Ca2+, Na+/K+ ATPases in concanavalin a-stimulated human blood mononuclear cells. Int. J. Mol. Sci. 2003;4:34–44. [Google Scholar]
- 31.Kaul S., Krishnakanth T.P. Effect of retinol deficiency and curcumin or turmeric feeding on brain Na(+)–K(+) adenosine triphosphatase activity. Mol. Cell. Biochem. 1994;137:101–107. doi: 10.1007/BF00944071. [DOI] [PubMed] [Google Scholar]