Abstract

Cyclotides are plant-derived mini proteins. They are genetically encoded as precursor proteins that become post-translationally modified to yield circular cystine-knotted molecules. Because of this structural topology cyclotides resist enzymatic degradation in biological fluids, and hence they are considered as promising lead molecules for pharmaceutical applications. Despite ongoing efforts to discover novel cyclotides and analyze their biodiversity, it is not clear how many individual peptides a single plant specimen can express. Therefore, we investigated the transcriptome and cyclotide peptidome of Viola tricolor. Transcriptome mining enabled the characterization of cyclotide precursor architecture and processing sites important for biosynthesis of mature peptides. The cyclotide peptidome was explored by mass spectrometry and bottom-up proteomics using the extracted peptide sequences as queries for database searching. In total 164 cyclotides were discovered by nucleic acid and peptide analysis in V. tricolor. Therefore, violaceous plants at a global scale may be the source to as many as 150 000 individual cyclotides. Encompassing the diversity of V. tricolor as a combinatorial library of bioactive peptides, this commercially available medicinal herb may be a suitable starting point for future bioactivity-guided screening studies.
Keywords: circular peptides, cystine-knot, transcriptome, peptidomics, Violaceae, ribosomally synthesized and post-translationally modified peptides, natural products, mass spectrometry, 1kp
Introduction
Cyclotides are gene-encoded cysteine-rich plant peptides. These peptides appear to be part of the plant defense system, based on observations that certain cyclotides exhibit insecticidal1 or antimicrobial activities.2 Their characteristic structure comprises a head-to-tail cyclized cystine-knot motif, which confers them great stability and resistance to enzymatic, thermal, and chemical degradation.3 Hence, cyclotides offer an interesting scaffold for peptide drug development,4 and they have been utilized as templates for grafting of bioactive epitopes onto the stabilized framework.5 Furthermore, cyclotides display a variety of intrinsic activities. For example, they are ligands of the human oxytocin and vasopressin V1a receptors, both members of the G protein-coupled receptor family,6 and they inhibit the activity of human prolyl oligopeptidase, a serine-protease involved in the processing of neuropeptides.7 These properties were discovered in bioassay-guided fractionation approaches using plant-derived peptide extracts as starting materials; needless to say, a robust and efficient analytical workflow is crucial to determine the molecular structure of each bioactive cyclotide. Therefore, advances in the discovery and identification of cyclotides are of great interest to provide novel peptide candidates for drug discovery8,9 and pharmacological applications.10
Cyclotide distribution within flowering plants has been explored extensively, and today members of the angiosperm families Cucurbitaceae, Fabaceae, Solanaceae, Poaceae, Rubiaceae, and Violaceae are known to express these circular peptides.11,12 All species of Violaceae examined hitherto are well-known as a rich source of cyclotides. The violets are the only plant family that ubiquitously contain cyclotides in every species sampled to date.11 Within the Violaceae ménage the species Viola tricolor L. is well-known as a traditional medicinal plant.13 In particular, herbal extracts of V. tricolor have been described to treat inflammatory diseases such as inflammatory atopic dermatitis or psoriasis.14,15 Although plants contain a number of different bioactive substances,16 a recent study highlighted that a cyclotide-enriched V. tricolor extract inhibited the proliferation of activated peripheral blood mononuclear cells.13 Earlier it was demonstrated that cyclotides isolated from V. tricolor are antiproliferative agents against cancer cell lines.17 Cyclotides have also been discovered in other members of Violaceae, including V. odorata L., V. yedoensis Mak., Melicitus ramiflorus J.R. & G. Forster, and Hybanthus floribundus Lindl. F.Muell., just to name a few.18−27 In addition, cyclotides isolated from species of Violaceae have been intensively investigated by mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy to provide structural data and insight into their stability and chemical folding.28−35 As a consequence of the cyclic nature of cyclotides and therefore, the lack of C- and N-termini, enzymatic processing is required to open the backbone ring. Given that an enzymatic digest of a peptide/protein-enriched plant extract yields thousands of peptides, the analysis of cyclotides in nonpurified preparations is extremely challenging, in particular due to the lack of appropriate nucleic acid/protein data sets available for proteomics analysis. To date, only laborious peptide purification combined with manual de novo sequencing appears to be suitable for precise sequence elucidation.21,36 More recently, next generation sequencing and bioinformatics have emerged as potential approaches for cyclotide discovery.36−39
In this study we combined transcriptome mining and MS to identify and characterize novel vitri cyclotides from V. tricolor. Transcriptome analysis provided new insights about the structure of cyclotide precursors, processing sites, and mature peptide sequences. Furthermore, we characterized the cyclotide peptidome by MALDI−TOF/TOF and LC–MS/MS. Systematic analysis and comparison of cyclotide identification at the nucleic acid level with application of different proteomics techniques provided the first comprehensive picture about cyclotide diversity and their post-translational modifications (PTM) in a single plant species. The outcome of this work may serve as a starting point for a routine high-throughput proteomics identification of cyclotides and other ribosomally synthesized and post-translationally modified natural peptide products.
Experimental Section
Chemicals
Acetonitrile (AcN), methanol (MeOH), dichlormethane, trifluoroacetic acid (TFA), and H2O were purchased as HPLC or LC-MS grade from Carl Roth (Karlsruhe, Germany). Dithiothreitol (DTT), α-cyano-hydroxy cinnamic acid, trypsin (sequencing grade), and iodoacetamide (IAA) were purchased as Bioultra grade from Sigma-Aldrich (Sigma-Aldrich, Vienna, Austria). Endoproteinase Glu C (sequencing grade) was purchased from New England Biolabs (Ipswich, MA).
Plant Material, RNA Extraction and Sequencing
V. tricolor was cultivated in a planting area near the entrance of Brywood, 16th avenue and NW 27th Terrace, Gainesville, Florida 32605 (coordinates 29°40′00.6″N 82°21′48.2″W). Fresh leaves of V. tricolor were identified by D.S. and colleagues and collected directly into liquid nitrogen. A voucher specimen (Soltis and Miles 2930) was deposited at the University of Florida Herbarium (FLAS). RNA was isolated using a Trizol/RNAqueous-Midi Kit (Ambion, Paisley, UK). Samples were sequenced as an indexed RNaseq library on an Illumina GA II machine (Illumina, San Diego, CA).40 Sequencing was paired-end (73 bp +75 bp) and produced a total of over 2.3 Gbp. The sequences were assembled into scaffolds using SOAPdenovo-Trans (Beijing Genomics Institute, China).41
Transcriptome Analysis of Cyclotide Precursor Proteins
Transcriptome data of V. tricolor were accessed via the 1kp-project (www.onekp.com). BLAST searches were conducted against these scaffolds. The V. tricolor data were accessed via tBLASTn using a set of published cyclotide precursor sequences as queries (www.cybase.org.au). Resulting hits and contig sequences were translated and annotated based on homology to known cyclotide precursors according to a similar methodology as described recently.36 Briefly, hits were collected and translated into their respective amino acid sequence (six-frame translation). Open reading frames were identified via manual sequence assignments. All transcriptome contig sequence IDs, nucleotide sequence data of precursors vitri 1 to vitri 99, as well as protein translations are listed in Supplementary Data S-1. Sequence alignments were prepared using ClustalW (www.ebi.ac.uk/Tools/msa/clustalw2/). Novel full-length precursor sequences are available in the GenBank database under the accession numbers KT203800–KT203810. Any ambiguous or erroneous assembly was analyzed by Transrate to identify mis-assembled contigs or assembly artifacts (http://www.stevekellylab.com/software/transrate).
Peptide Extraction
Dried V. tricolor total herbaceous plant material (Herba Violae Tricoloris plv.) was obtained from Dr. Kottas Pharma GmbH (Vienna, Austria). The peptide extraction was performed according to well-established protocols.13 Briefly, plant material was extracted in a dichloromethane/MeOH 1:1 (v/v) solution for 24 h. Afterward the addition of one-half volume of ddH2O allowed liquid–liquid phase extraction. The MeOH/water phase encompasses all moderately hydrophobic plant constituents such as peptides, whereas lipophilic plant pigments are enriched in the dichloromethane phase. The MeOH/water phase was further treated in batch by solid phase extraction. Zeoprep 60 Å, C18 irregular 40–63 μm material (Zeochem, Uetikon, Switzerland) was used according to the manufacturer’s recommendation with surface activation by MeOH, equilibration with 0.1% TFA and peptide elution with AcN/ddH2O/TFA 80/20/0.1% (v/v/v). The eluted extract was freeze-dried and will be referred to as “vitri extract”.
HPLC Fractionation
Fractionation of vitri extract was performed using a Dionex Ultimate 3000 HPLC system (Thermo Fisher Scientific, Waltham, MA) equipped with a binary pump, autosampler, multiwavelength detector, and a fraction collector. Chromeleon application software v. 6.80 was used to control the HPLC. Chromatography was performed using a preparative Dichrom Kromasil column (dichrom GmbH, Marl, Germany; 250 × 20 mm, spherical C18 modified particles, 10 μm, 110 Å). Mobile phase eluent A consisted of 0.1% TFA (aqueous) and mobile phase eluent B consisted of AcN/ddH2O/TFA 90/10/0.08% (v/v/v). The flow rate was set to 8 mL/min. Peptide separation on column was achieved with linear stepwise gradients (0′-5% B, 5′-5% B, 12.5′-20% B, 62.5′-70% B, 63.5′-99% B, 65′-99% B, 66′-5% B and 72′-5% B). Fractions of 40 mL (5 min) each, starting at 20% eluent B, were collected and lyophilized. The dried samples were kept at −20 °C until further analysis, e.g., mass deconvolution of cyclotide mass signals or automated MS/MS sequencing and database search of cyclotides for peptide identification in the proteomics workflow.
Sample Preparation for Mass Spectrometry
For MS-based analysis via MALDI−TOF or LC−MS vitri extract was dissolved in 0.1% TFA (aqueous) at a concentration of 3 mg/mL. For proteolytic digest the extract was dissolved in 0.1 M NH4HCO3 (pH 7.8) at a concentration of 3 mg/mL. Any insoluble material was removed by centrifugation at 12000g for 10 min, and the supernatant was used for further analysis. For proteomics analysis by MS/MS an aliquot of peptide solution (20–40 μL) was used for reduction, alkylation, and proteolytic digest. Briefly, disulfides were reduced with 20 mM dithiothreitol (DTT) for 1 h at 37 °C. Free thiols were carbamidomethylated using 100 mM iodoacetamide for 20 min at 23 °C in the darkness. This reaction was quenched with 1 μL of 100 mM DTT for 10 min at 37 °C. The reduced and alkylated samples were digested with trypsin (0.3 μg) or endoproteinase GluC (0.6 μg) for 12–16 h at 37 °C. Double digests were performed accordingly, starting with trypsin digestion overnight, followed by transient trypsin deactivation with DTT and addition of endoproteinase GluC. For all samples, proteolytic digests were stopped by adding TFA to a final concentration of 0.5% (v/v). The samples were stored at −20 °C until used for MS analysis.
Cyclotide Analysis by Mass Spectrometry
Vitri extracts and fractions were analyzed via MALDI-TOF to evaluate the samples for peaks in the cyclotide specific mass range of 2500–4000 Da. MALDI-TOF analysis was performed on a 4800 reflector TOF/TOF (time-of-flight) analyzer from ABSciex (Framingham, MA). Samples were mixed with a saturated matrix solution of α-cyano-hydroxy cinnamic acid dissolved in AcN/ddH2O/TFA 50/50/0.1% (v/v/v) at a ratio of 1:6, and 0.5 μL of this mixture was transferred onto the target plate to allow air-drying in the darkness. Mass accuracy was ensured by daily calibration using Peptide Mix 1 [(bradykinin fragment 8–39 (572.7 Da), angiotensin II (1046.2 Da), neurotensin (1672.9 Da), adrenocorticotropin fragment 18–39 (2465.7 Da) and bovine insulin chain B (3495.9 Da)] (Laserbiolabs, Sophia-Antipolis, France). MS spectra were recorded using the reflector in positive ionization mode in a spectral range between 2500 and 4500 Da. 2500–3000 shots per spot were recorded and accumulated. Laser intensity and amplifier adjustment were optimized for maximal signal intensity. Vitri extracts were evaluated via LC–MS scan and mass signal deconvolution of MS1 scans. MS scans were processed with deconvolution to evaluate mass signals in the cyclotide typical mass range of 2500–4000 Da. Peptide extract was analyzed via LC–MS on a QqTOF compact (Bruker Daltonics, Billerica, MA). Sample desalting and preconcentration were performed with a trap column, and peptides were separated with a PepMap Acclaim capillary column (Thermo Fisher Scientific; 150 × 0.3 mm, 2 μm 100 Å) using a Dionex Ultimate 3000 UPLC system. The mobile phase for the loading pump consisted of 0.1% TFA (aqueous); 0.1% formic acid (aqueous) and AcN/ddH2O/formic acid 80/20/0.08% (v/v/v) were used as mobile phases for chromatographic separation. The flow rates were set to 25 μL/min for the loading pump and 6 μL/min for the separation pump. The gradient for peptide separation was as follows: 0′-4% B, 10′-4% B, 120′-45% B, 125′-99% B, 132′-99% B, 135′-4% B and 150′-4% B. The LC QqTOF compact system was equipped with a captive nanoflow ionization source, and sample ionization was achieved in the positive electron ionization mode. Ion transfer parameters were kept constant for the duration of one run: prepulse storage 5 μs, collision energy transfer 9 eV, quadrupole energy 4 eV, funnel 1 400 Vpp, funnel 2 600 Vpp transfer time, and hexapole RF 400 Vpp. For collision energy RF and for ion transfer time stepping from 200 to 1500 Vpp and 30–150 μs were applied, respectively. External mass calibration was achieved via direct infusion of the low concentration tuning mix from Agilent Technologies (Santa Clara, CA). Furthermore, internal calibration via lock mass was applied for each analytical run using calibrant hexakis (1H, 1H, 4H-hexafluorobutyloxy)phosphazine (Agilent Technologies). Base peak chromatograms in a signal window of 800–2200 Da were processed with the “Compound Find” tool of the Data Analysis software 4.21 (Build 393) (Bruker Daltonics). The “Deconvolution” tool was applied to each identified compound using the following settings: deconvolution of masses between 2500–4000 Da, abundance cutoff 5%, maximum charge 4+, only consider [M + H]+ ions of proteins or peptides. Each deconvolution list was manually validated to exclude false-positive hits. Evidence by charge and/or isotope pattern was recorded for each signal. For MS/MS experiments of vitri extracts or vitri fractions F1–F9 the chromatographic conditions were identical as described for the mass signal deconvolution. Peptide sequencing and automated identification by subsequent database search were performed by MS/MS experiments on the QqTOF mass spectrometer using a bottom-up shotgun proteomics approach. The mass spectrometer was tuned and preoptimized to achieve best sequence coverage for precursors up to 2200 Da, as commonly observed from proteolysis of cyclotides. MS/MS sequencing was performed with a cycle time of 3 s and exclusion criteria to reduce triggering of background signals. Scan range for precursor recording was set to 50–2200 Da with a minimal precursor mass of 200 m/z. Peptides carrying charges from 2+ to 5+ were set to trigger MS/MS. High energy collision induced dissociation was performed under the regime of a set energy set. This set applies optimized collision energy for precursor signals based on the parameters: isolation m/z, isolation mass range width, and charge state of the ion. The set followed the manufacturer’s recommendations, and it was not further modified by the experimenter.
Database Construction and Database Search
The “Proteinscape” software package v. 3.1.5 474 (Build 20140711-1459) from Bruker Daltonics together with the Mascot algorithm (Matrixscience, MA) was used for data analysis and database search. The available cyclotide sequence database ERA42 was updated and customized with the most recent data set of circular peptide sequences (www.cybase.org.au) and from recent cyclotide discovery studies36,38 to yield in total 376 individual peptide sequences, including 55 novel precursors from the vitri transcriptome analysis of this study. Additionally, contaminants were implemented into the custom database, e.g., proteolytic fragments of keratins, bovine serum albumins and proteases. The publicly available contaminant database (http://maxquant.org/contaminants.zip) was provided by the Max Planck Institute of Biochemistry (Martinsried, Germany). The custom cyclotide FASTA database can be provided upon request. Peptide search parameters were set to peptide mass tolerance ±10 ppm, MS/MS tolerance ±0.05 Da, number of possible 13C atoms one, charge states 2+ – 5+ and considered up to one missed cleavage in the peptide search. Fixed modification was carbamidomethylation of cysteine side chains, and variable modifications included methionine oxidation, tryptophan modification to kynurenine and 3-hydroxykurenine, deamidation of asparagine and glutamine and ethylation as well as methylation of glutamate or aspartate residues, respectively. All peptides with more than one possible precursor were assigned to the rank one candidate. Identified proteins with a probability score >50 of two independent analyses of at least one digest (as described in the section Sample Preparation for Mass Spectrometry) were considered to be valid; this threshold score has been found by manual MS/MS validation to yield acceptable peptide assignment. Nevertheless, ion score cutoff was set to >15 to consider and exclude low quality peptides from multiple MS/MS triggering events. Cyclotides with a partial sequence and those giving rise to more than one candidate peptide were evaluated manually for assignment of one hit, and they were cross validated with the vitri extract for native peptide mass signals of the reported MS/MS precursor peak. The MS proteomics data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD002867 (project DOI: 10.6019/PXD002867).43
Results and Discussion
Since the discovery of “kalata” plant peptides in Oldenlandia affinis (R&S) DC. by Lorentz Gran44 in the 1970s and the appreciation of cyclotides as active substances in medical preparations for uterotonic purpose,6,44,45 these ultrastable peptides have been the focus of numerous investigations to delineate their biological diversity, structural arrangement, and phylogenetic distribution within the plant kingdom.6,11,36,46 With an estimated 50 000 cyclotides present in Rubiaceae alone, the current rate and performance of standard techniques of cyclotide discovery are clearly behind expectations, and it needs further optimization.47 So far laborious purification of peptides has been necessary to obtain accurate sequence information.36,37,39 Hence, not surprisingly, the number of novel cyclotides reported by peptide sequencing in single studies to date is 1839 and 24,42 respectively, and the overall number of cyclotides deposited in CyBase from >20 years of research is currently at 922.48 In the era of high-throughput nucleic acid sequencing, new tools have become available to examine cyclotide diversity on a genome/transcriptome level.36,37,39,47 Therefore, we aimed to discover as many cyclotides as possible expressed in a single plant species and to examine their molecular diversity by a combination of sequence analysis at the nucleic acid level and MS studies at the peptide level. In addition, we performed a state-of-the-art bottom-up proteomics approach using plant extracts as well as HPLC fractionated samples to identify the sequences and PTMs of many novel cyclotides present in V. tricolor and to establish a robust workflow for cyclotide analysis.
Analysis of Cyclotide Expression at Nucleic Acid Level
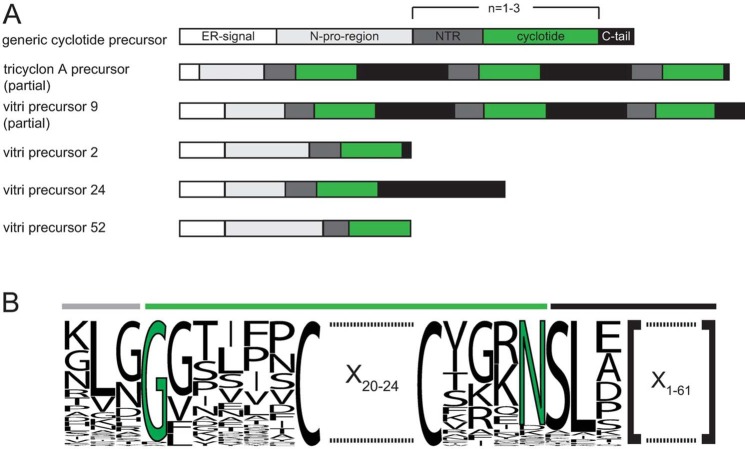
To date, only two precursor sequences have been reported for V. tricolor,12,49 and these were identified by PCR-based cDNA sequencing. Peptide extraction and MS analysis yielded another 14 mature cyclotides.17,50 Little is known about the variety of precursor sequences and how many cyclotides one plant is capable of expressing. Therefore, it is of interest to gain insight into the diversity of cyclotide precursors. V. tricolor transcriptome data were available via the 1kp-project (www.onekp.com) and identified by tBLASTn searches using known cyclotide precursor sequences as queries (www.cybase.org.au). Contigs from resulting homologous sequences have been translated and annotated based on published cyclotide precursors. This approach led to the identification of 98 unique precursor proteins encoding 108 peptide sequences (Supplementary Table S-1 and Supplementary Data S-1). These findings nearly double the number of precursors compared to a very recent report suggesting that a single Viola species contains 53 cyclotide precursor sequences.38 Most cyclotide precursors that have been discovered only contain one cyclotide domain, but it is well-known that precursor molecules may contain multiple cyclotide domains. In fact we identified eight precursor proteins with two cyclotide domains (vitri precursor 18, 22, 27, 42, 72, 85, 91, and 94) and one precursor, vitri precursor 9, encoding for three cyclotide domains (Figure 1A). Four cyclotide sequences (vitri 24/28, 36/37, 43/44 and 9a/53) have been found to be encoded by two different precursor proteins. Analysis of precursor architecture as well as putative processing sites provides further insight into cyclotide biosynthesis in V. tricolor (Figure 1B, Supplementary Data S-2) and may also help to distinguish cyclotides from other cysteine-rich plant peptides.38 The majority of identified precursor proteins, namely, 54 precursors contain an asparagine (or aspartic acid) at the C-terminus of the cyclotide domain followed by a highly conserved SL, or rarely SI, GL or AI in the positions P′1/P′2. Surprisingly, 10 precursors differ significantly in these common processing sites; vitri precursors 15, 16, 17, 19, 27a, 35, 40, 66, 76, and 79 lack the common asparagine (aspartic acid) in their cyclotide domain vital for cyclization (Supplementary Data S-1), and another seven peptides completely lack a C-terminal tail sequence altogether (vitri precursors 8, 25, 32, 33, 46, 51 and 52). Hence this raises the question of whether these peptides are being cyclized via different mechanisms or whether they represent acyclic homologues (Supplementary Table S-1, Supplementary Data S-1). The occurrence of linear cyclotides has infrequently been reported for species of the Rubiaceae, Violaceae, and Poaceae plant families.33,51,52 In fact, it was possible to confirm occurrence of acyclic cyclotide homologues in V. tricolor (see section MS/MS Peptide Sequencing of Cyclotides). These acyclic peptides not only lack the N-terminal glycine and/or the C-terminal asparagine, but the precursor proteins may also be significantly shortened.39 Besides containing unusual processing motifs, vitri precursors 24 and 28 exhibit a long C-tail region, which has not been observed previously in a cyclotide precursor. The biosynthesis of cyclotides is still not fully understood; however research has addressed the role of processing enzymes and the involvement of single residues at the processing sites within the peptide precursors (Supplementary Text S-1).33,53−57
Figure 1.
Viola tricolor cyclotide precursor analysis. (A) A generic cyclotide precursor is ribosomally synthesized and contains an ER signal, followed by a N-pro-region, N-terminal repeats (NTR), the cyclotide domain, and a C-terminal tail region. Cyclotide precursors may encode up to three peptide domains as it has been demonstrated for the V. tricolor tricyclon A precursor. Novel vitri precursors discovered by transcriptome mining contain one, two, or three cyclotide domains. Exemplarily shown are vitri precursors 2 and 9. As indicated each precursor domain may vary in length. For instance, vitri precursor 24 comprises an extended C-tail, whereas vitri precursor 52 completely lacks this domain. (B) Sequence comparison of vitri precursor processing site has been provided by a sequence logo. Three adjacent residues downstream of the C-terminus of the mature cyclotide domain in positions P′1, P′2, and P′3, and upstream of the N-terminus of the mature cyclotide domain in positions P1, P2, and P3 have been considered for being essential for precursor processing and cyclization in planta. Loop six comprises the highly conserved ligation sites, namely, glycine and asparagine or aspartic acid as highlighted in green.
These novel vitri precursor sequences encode up to 108 putative cyclotides of V. tricolor, and many of them exhibit novel or very rare features (Supplementary Table S-1). They have been discussed in the Supporting Information as extended discussion of the sequence information provided by the presented transcriptome analysis (Supplementary Figure S-1, Supplementary Text S-1). These new and rare sequence properties underline the structural plasticity of cyclotides, and they may be of future interest for studies utilizing cyclotides in peptide engineering and drug development.5,9
Although transcriptome data analysis revealed 108 peptide sequences, only a single known V. tricolor cyclotide (vitri D) has been identified at the transcript level. Moreover, the reported vitri precursor tricyclon precursor A could not be verified by transcriptome mining.12,49 It is known that cyclotide expression levels appear to be strongly determined by environmental conditions or seasonal changes.58 In fact, certain cyclotide precursors are exclusively expressed in certain plant tissues,59 and biotic and abiotic factors may influence cyclotide transcription.60,61 Most importantly, the major limitation of in silico sequence mining is the erroneous prediction of processing sites by homology, and hence certain cyclotides may be assigned alternatively (see footnotes Supplementary Table S-1). Only peptide analysis at the proteome level using MS will be able to verify and confirm the data obtained from transcriptome analysis.
Analysis of Cyclotide Expression at Proteome Level
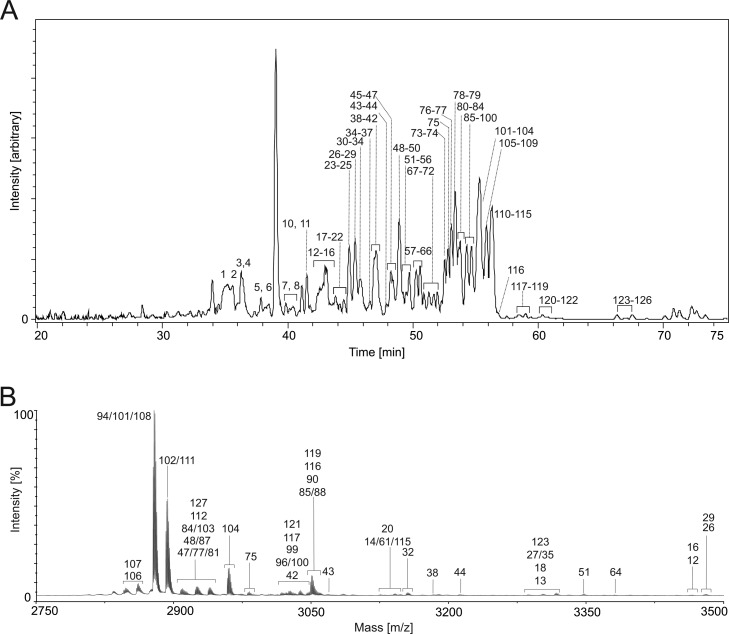
Having established a comprehensive view of the expression of cyclotide peptides at the nucleic acid level, we were interested in cyclotides present at the protein level. To elucidate the cyclotide proteome of V. tricolor MS was used to collect evidence of the expression of cyclotides in V. tricolor. Starting with powdered herb material, hydrophobic compounds were extracted and purified using well established protocols.7 This vitri extract was used as starting material for MS-based investigations. Capillary liquid chromatography coupled to a QqTOF mass spectrometer was used to record base peak chromatograms of all specimens in a signal range between 800–2200 Da (Figure 2A). LC–MS survey scans were evaluated for putative cyclotide masses using deconvolution tools (Supplementary Table S-2). LC–MS resolved 126 peptide signals by the detection of the isotopic pattern, charge state deconvolution, or both. Most notably, the LC–MS system takes advantage of its high resolution power by chromatographic separation and hence in total higher capacity in the scan mode. In contrast MALDI−TOF MS analysis of vitri plant extract using the 4800 MS analyzer suffers resolution problems due to multiple overlapping signals. This is why MALDI−MS only yielded 36 peptide signals (Figure 2B). On the other hand, cO28 was only detected by MALDI−MS, but not by LC–MS, which exemplifies sequence specific ionization differences. Among others, LC–MS revealed mass signals of vitri peptides kalata S, varv peptide D, E, F, H, He, Hm, vitri A, D, E, F, tricyclon A and cycloviolacin O2. Mass signals for vitri B, C and tricyclon B were not found with either technique. Obviously the LC–MS scan provided better resolution, which resulted in a higher number of identified cyclotide masses. Nevertheless, the combination of QqTOF and MALDI−TOF MS appeared to be more powerful for increasing the number of identified cyclotides. To gain deeper insight into cyclotide diversity of V. tricolor, in particular with regard to molecular structure and PTMs, we utilized MS-based peptide fragmentation and database searching to identify cyclotides by an automated proteomics workflow.
Figure 2.
Mass spectrometry analysis of Viola tricolor cyclotide extract. (A) Plant peptide extract from V. tricolor was analyzed via LC–MS. A base peak chromatogram of mass signals ranging from 800–2200 Da is shown. Cyclotide masses were found to carry +2, +3, and rarely +4 H-ions. Signals >5.0% intensity threshold were assigned to mass and charge deconvolution tool from Data Analysis software (Bruker Daltonics). The determined cyclotide mass signals were consecutively numbered by increasing retention times. (B) Accordingly a MALDI−TOF spectrum is shown between 2750 and 3500 Da. Mass signals with an intensity threshold of >0.5% are labeled. For cyclotide names, retention times and molecular weight (monoisotopic masses) refer to Supplementary Table S-2.
MS/MS Peptide Sequencing of Cyclotides
MS/MS fragmentation data of vitri extract were recorded with a capillary LC-QqTOF instrument and queried against the customized vitri database. For this purpose, the existing ERA database42 was updated to contain all vitri precursors or mature peptide sequences that were discovered by transcriptome mining (Supplementary Table S-1 and Supplementary Data S-1). The lack of a C- or N-terminus requires enzymatic processing for opening the cyclotide backbone, and linearization facilitates sufficient fragmentation using collision induced dissociation since intact cyclotides exhibit poor fragmentation.36 The most advanced strategy is to utilize a combination of multiple endopeptidases, i.e., trypsin and endoproteinase Glu C.21,36 This methodology resulted in multiple precursor peptides for one cyclotide with significant hits using the database analysis. Table 1 summarizes cyclotides identified by proteomics analysis of a vitri extract. Identification of a unique full or partial sequence in at least one of the digested samples with a Mascot score >50 was considered valid, and each cyclotide hit was further supported by peptide identification using MALDI−TOF or LC–MS scans. In total, 38 cyclotides were determined, 26 with full and 12 with partial sequence coverage (Table 1). Using this proteomics workflow, it was possible to confirm the expression of vitri cyclotides varv peptide A, D, E, F, H, vitri A, D, E, cycloviolacin O2 and tricyclon A.17,50 Five cyclotides (vitri 18b, 22a, 39, 42a, and 94b) were detected from the novel transcriptome data set, and tricyclon B as well as oak6 cyclotide 1 were verified at protein level for the first time.49,60 Further, this methodology yielded identification of the cyclotides cO13, cO22, and cO28, kalata B1, varv peptide B, C and G, vigno 4 and 5, viba 30 and 32, vodo M, vodo N.
Table 1. Identification and Discovery of Vitri Cyclotides by LC−MS/MS Peptide Sequencing of Viola tricolor Extract and Fractions.
| name | sequence | calc mass (monoiso.) [Da] | LC-MS | MALDI-MS | transcriptome | Mascot scored | full/partial sequence | PTMe |
|---|---|---|---|---|---|---|---|---|
| cycloviolacin O2 | GIPCGESCVWIPCISSAIGCSCKSKVCYRN | 3138.37 | x | x | 104.9 | partialb,c | Dea (NQ) | |
| cO4 | GIPCGESCVWIPCISSAIGCSCKNKVCYRN | 3165.38 | x | 113.3 | fullc | Dea (NQ) | ||
| cO13 | GIPCGESCVWIPCISAAIGCSCKSKVCYRN | 3122.37 | x | x | 91.6 | partialc | Dea (NQ) | |
| cO20 | GIPCGESCVWIPCLTSAIGCSCKSKVCYRD | 3153.37 | 90.2 | partiala | ||||
| cO22 | GLPICGETCVGGTCNTPGCTCSWPVCTRN | 2904.16 | x | x | 118.8 | fulla,b,c | Dea (NQ), Hkyn(W), | |
| cO22 (linear) | acyclo-GLPICGETCVGGTCNTPGCTCSWPVCTRN | 2922.16 | x | x | 93.7 | partialb | ||
| cO28 | GLPVCGETCVGGTCNTPGCSCSWPVCFRD | 2923.13 | x | 103.3 | fulla,b | Methyl (D) | ||
| chacur 1 | GLPVCGETCVGGTCNTPGCTCSWPICTRN | 2904.16 | x | x | 76.8 | partialc | Hkyn(W) | |
| vitri peptide 1 | GLIPCGESCVWIPCISSVIGCSCKSKVCYKN | 3251.48 | x | 58.2 | partialc | |||
| vitri peptide 2 | GSIPCGESCVWIPCISGIAGCSCSNKVCYLN | 3138.32 | x | x | x | 63.4 | partialc | |
| vitri peptide 3 | GSWPCGESCVYIPCITSIAGCECSKNVCYKN | 3289.35 | x | 86.8 | partialc | |||
| vitri peptide 4 | GTPCGESCIYVPCISAVFGCWCQSKVCYKD | 3221.32 | x | x | 74.3 | partiala | ||
| vitri peptide 8 (linear) | acyclo-PTPCGETCIWISCVTAAIGCYCHESICYR | 3172.31 | x | 91.4 | partialb,c | |||
| vitri peptide 9a/53 | acyclo-GTIFDCGETCLLGKCYTPGCSCGSWALCYGQN | 3343.11 | x | 106.4 | fulla,b,c | |||
| vitri peptide 14 | GSSCGETCEVFSCFITRCACIDGLCYRN | 3012.18 | x | x | 62.0 | partiala,c | ||
| vitri peptide 17 | GSDDQVACGESCAMTPCFMHVVGCVCSQKVCYR | 3488.37 | x | 81.5 | partialc | Ox (M) | ||
| vitri peptide 18a | GVPICGETCFQGTCNTPGCTCKWPICERN | 3092.25 | x | x | 121.1 | fulla,b,c | Dea (NQ), Methyl (D) | |
| vitri peptide 20 | GDLVPCGESCVYIPCLTTVLGCSCSENVCYRN | 3372.41 | x | x | x | 71.1 | partialb,c | |
| vitri peptide 21 | GGPLDCQETCTLSDRCYTKGCTCNWPICYKN | 3447.39 | x | 60.2 | partiala | |||
| vitri peptide 22a | GAPVCGETCFTGLCYSSGCSCIYPVCNRN | 2979.16 | x | x | x | 153.0 | fulla,b,c | Dea (NQ) |
| vitri peptide 22a (linear) | acyclo-GAPVCGETCFTGLCYSSGCSCIYPVCNRN | 2997.16 | x | 75.9 | partialb | |||
| vitri peptide 94b | GVAVCGETCTLGTCYTPGCSCDWPICKRN | 3012.22 | x | x | x | 139.0 | partialb,c | Hkyn(W), Methyl (D) |
| vitri peptide 23 | GLPTCGETCTLGTCYTPGCTCSWPLCTKN | 2985.19 | x | x | 53.8 | partialc | ||
| vitri peptide 24/28 | GEPVCGDSCVFFGCDDEGCTCGPWSLCYRN | 3194.14 | x | 57.9 | partialc | |||
| vitri peptide 27a | GAFTPCGETCLTGECHTEGCSCVGQTFCVKK | 3171.27 | x | 67.0 | partialb,c | Dea (NQ) | ||
| vitri peptide 29 | GVPSSDCLETCFGGKCNAHRCTCSQWPLCAKN | 3390.39 | x | 158.8 | partiala,c | |||
| vitri peptide 30 | GFACGETCIFTSCFITGCTCNSSLCFRN | 2960.15 | x | 86.6 | partialc | |||
| vitri peptide 36/37 | GGTIFSCGESCFQGTCYTKGCACGDWKLCYGEN | 3463.32 | x | 101.9 | partiala | Dea (NQ) | ||
| vitri peptide 38 | GDTCYETCFTGFCFIGGCKCDFPVCVKN | 3032.18 | x | 63.0 | fulla,c | |||
| vitri peptide 39 | GAPICGESCFTGTCYTVQCSCSWPVCTRN | 3048.18 | x | x | x | 207.0 | fulla,b,c | Dea (NQ) |
| vitri peptide 39 (linear) | acyclo-GAPICGESCFTGTCYTVQCSCSWPVCTRN | 3066.18 | x | n.a. | partialb | |||
| vitri peptide 24a | GGTIFNCGESCFQGTCYTKGCACGDWKLCYGEN | 3490.33 | x | 170.4 | partiala,b,c | Dea (NQ), Ethyl (E) | ||
| vitri peptide 50 | GDIPCGESCVYIPCITGVLGCSCSHNVCYYN | 3244.29 | x | 76.9 | fullc | Dea (NQ) | ||
| vitri peptide 18b | GSVFNCGETCVFGTCFTSGCSCVYRVCSKD | 3134.26 | x | x | 163.8 | fulla,b,c | Dea (NQ) | |
| kalata B1 | GLPVCGETCVGGTCNTPGCTCSWPVCTRN | 2890.14 | x | x | 161.7 | fulla,b,c | Dea (NQ), Hkyn(W), | |
| kalata B1 (linear) | acyclo-GLPVCGETCVGGTCNTPGCTCSWPVCTRN | 2908.14 | x | x | n.a. | partialb | ||
| kalata S | GLPVCGETCVGGTCNTPGCSCSWPVCTRN | 2876.13 | x | x | 307.0 | fulla,b,c | Dea (NQ), Hkyn(W), | |
| kalata S (linear) | acyclo-GLPVCGETCVGGTCNTPGCSCSWPVCTRN | 2894.13 | x | x | n.a. | partialb | ||
| mram 8 | GIPCGESCVFIPCLTSAIGCSCKSKVCYRN | 3113.37 | 100.2 | partiala,b | ||||
| oak6 cyclotide 1 | GLPVCGETCFGGTCNTPGCACDPWPVCTRN | 3033.18 | 75.4 | fulla | Hkyn(W), Dea (NQ) | |||
| tricyclon A | GGTIFDCGESCFLGTCYTKGCSCGEWKLCYGTN | 3478.35 | x | x | 331.3 | fulla,b,c | Dea (NQ) | |
| tricyclon A (linear) | acyclo-GTIFDCGESCFLGTCYTKGCSCGEWKLCYGTN | 3496.35 | x | n.a. | partialb | Dea (NQ) | ||
| tricyclon B | GGTIFDCGESCFLGTCYTKGCSCGEWKLCYGEN | 3506.35 | 200.0 | fulla,b,c | Dea (NQ), Ethyl (E) | |||
| vaby C | GLPVCGETCAGGRCNTPGCSCSWPVCTRN | 2903.15 | x | x | 122.6 | fulla,c | ||
| vaby E | GLPVCGETCFGGTCNTPGCSCDPWPVCTRN | 3049.17 | 91.9 | fulla | Dea (NQ) | |||
| varv peptide E/vigno 3 (linear) | acycloGLPLCGETCVGGTCNTPGCSCSWPVCTRN | 2908.15 | x | x | n.a. | partialb | ||
| varv B | GLPVCGETCFGGTCNTPGCSCDPWPMCSRN | 3067.13 | x | 229.0 | fulla,b,c | Me(Ox), Dea (NQ) | ||
| varv B (linear) | acyclo-GLPVCGETCFGGTCNTPGCSCDPWPMCSRN | 3085.13 | x | x | 77.4 | partialb | Ox (M) | |
| varv C | GVPICGETCVGGTCNTPGCSCSWPVCTRN | 2876.13 | x | x | 253.7 | fullb,c | Dea (NQ) | |
| varv C (linear) | acyclo-GVPICGETCVGGTCNTPGCSCSWPVCTRN | 2894.13 | x | n.a. | partialb | |||
| varv D | GLPICGETCVGGSCNTPGCSCSWPVCTRN | 2876.13 | x | x | 189 | fulla,b,c | Dea (NQ) | |
| varv D (linear) | acyclo-GLPICGETCVGGSCNTPGCSCSWPVCTRN | 2894.13 | x | 50.1 | partialb | Hkyn(W), | ||
| varv E/vigno 3 | GLPICGETCVGGTCNTPGCSCSWPVCTRN | 2890.14 | x | x | 251.3 | fulla,b,c | Dea (NQ), Hkyn(W), | |
| varv F | GVPICGETCTLGTCYTAGCSCSWPVCTRN | 2957.17 | x | x | 244.5 | fulla,b,c | Dea (NQ), Hkyn(W) | |
| varv F (linear) | acyclo-GVPICGETCTLGTCYTAGCSCSWPVCTRN | 2975.37 | x | n.a. | partialb | |||
| varv G | GVPVCGETCFGGTCNTPGCSCDPWPVCSRN | 3021.14 | x | x | 211.8 | fulla,b,c | Dea (NQ), Hkyn(W), | |
| varv G (linear) | acyclo-GVPVCGETCFGGTCNTPGCSCDPWPVCSRN | 3039.14 | x | n.a. | partialb | |||
| varv H | GLPVCGETCFGGTCNTPGCSCETWPVCSRN | 3053.17 | x | 151.1 | fulla,b,c | Dea (NQ), Ethyl (E), Methyl (D) | ||
| viba 11 | GIPCGESCVWIPCISGAIGCSCKSKVCYRN | 3108.36 | 60.8 | fullc | Dea (NQ) | |||
| viba 15 | GLPVCGETCVGGTCNTPGCACSWPVCTRN | 2880.13 | x | 99.7 | fullb,c | Hkyn(W), Dea (NQ) | ||
| viba 19 | GLPVCGETCFGGTCNTPGCSCEWPVCTRN | 2966.14 | 77.3 | partialb | Dea (NQ) | |||
| viba 30 | GPPVCGETCVGGTCNTPGCSCSWPVCTRN | 2860.10 | x | 191.0 | fulla,b,c | Dea (NQ), Kyn(W), | ||
| viba 30 (linear) | acyclo-GPPVCGETCVGGTCNTPGCSCSWPVCTRN | 2878.10 | x | x | n.a. | partialb | ||
| viba 32 | GLPVCGEACVGGTCNTPGCSCSWPVCTRN | 2846.12 | x | 110.9 | fulla,c | Dea (NQ), Ethyl (E) | ||
| viba 9 | GIPCGESCVWIPCISSAIGCSCKNKVCYRK | 3179.43 | x | x | 106.5 | fullc | ||
| vibi C | GLPVCGETCAFGSCYTPGCSCSWPVCTRN | 2973.15 | 52.8 | fulla | ||||
| vibi G | GTFPCGESCVFIPCLTSAIGCSCKSKVCYKN | 3220.40 | 74.1 | partialc | ||||
| vigno 10 | GTIPCGESCVWIPCISSVVGCSCKSKVCYKD | 3226.41 | 68.9 | partialc | Dea (NQ) | |||
| vigno 4 | GLPLCGETCVGGTCNTPACSCSWPVCTRN | 2904.16 | x | x | 141.7 | fullb,c | Dea (NQ), Hkyn(W), | |
| vigno 5 | GLPLCGETCVGGTCNTPGCSCGWPVCVRN | 2858.15 | x | x | 152.2 | fulla,b,c | ||
| vigno 5 (linear) | acyclo-GLPLCGETCVGGTCNTPGCSCGWPVCVRN | 2876.20 | x | x | 152.3 | fullb | ||
| vigno 6 | GIPCGESCVWIPCISSAIGCSCKGSKVCYRN | 3195.39 | x | 78.8 | partialc | |||
| vigno 7 | GTLPCGESCVWIPCISSVVGCSCKNKVCYKN | 3252.44 | 72.3 | partialc | Dea (NQ) | |||
| vigno 9 | GIPCGESCVWIPCISSALGCSCKSKVCYRN | 3138.37 | x | x | 90.8 | partialc | ||
| vitri A/cO3 | GIPCGESCVWIPCLTSAIGCSCKSKVCYRN | 3152.38 | x | x | 93.6 | partialb,c | Dea (NQ) | |
| vitri D | GLPVCGETCFTGSCYTPGCSCNWPVCNRN | 3043.16 | x | x | x | 69.6 | fulla,c | |
| vitri E | GLPVCGETCVGGTCNTPGCSCSWPVCFRN | 2922.15 | x | x | 192.9 | fulla,b,c | Dea (NQ) | |
| vitri E (linear) | acyclo-GLPVCGETCVGGTCNTPGCSCSWPVCFRN | 2940.13 | n.a. | partialb | ||||
| vitri F | GLTPCGESCVWIPCISSVVGCACKSKVCYKD | 3210.41 | x | 67.3 | partialc | |||
| vodo M | GAPICGESCFTGKCYTVQCSCSWPVCTRN | 3075.23 | x | 162.8 | fulla,b,c | |||
| vodo N | GLPVCGETCTLGKCYTAGCSCSWPVCYRN | 3046.24 | x | 201.4 | fulla,b,c | Dea (NQ) | ||
| vodo N (linear) | acyclo-GLPVCGETCTLGKCYTAGCSCSWPVCYRN | 3066.24 | n.a. | partialb |
Identified in trypsin digest.
Identified in endoproteinase GluC digest.
Identified in trypsin/endoproteinase GluC double digest.
Best score out of two independent analysis of each of the three digest experiments.
Modifications recorded in at least one peptide of assigned peptides of in total six independent analysis.
One advantage of using an automated proteomics workflow is the possibility to determine PTMs. In fact certain cyclotide modifications have been previously reported.39,42,50−52,62 It was possible to confirm many of them occur in cyclotides from V. tricolor. The most commonly observed modifications of vitri cyclotides have been illustrated, such as aspartate methylation, glutamate ethylation, tryptophan oxidation to kynurenine and 3-hydroxykynurenine, as well as deamidation of asparagine and glutamine (Figure 3A–D). It is noteworthy that some peptides differ only by one pivotal residue, for instance, vitri E (N29) and cycloviolacin O28 (D29). In this case deamidation of the N29 residue of vitri E would yield cycloviolacin O28 with a D29 residue. Judging from the MS/MS data, both cyclotides were found side-by-side in V. tricolor, but nevertheless it was necessary to manually reanalyze the automated assignments. Besides identification of common PTMs of amino acid side chains, proteomics is a powerful tool to determine cyclization of the N- and C-termini. Analysis of endoproteinase GluC digested samples revealed evidence for the presence of acyclic peptides in V. tricolor, namely, acyclo-kalata S, -tricyclon A, -vigno 5 -viba 30, -varv peptide C, D, and E and -viba 30. Here, the peptides from the C-terminal end to the first cleavage site of the protease served as indicative fragments (Table 1 and Figure 3E). The acyclic peptide species have been additionally confirmed by MS/MS analysis of reduced and alkylated samples. Since cyclized peptides emerge stabilized during collision-induced dissociation, only linear peptide species yielded MS/MS spectra with sufficient fragmentation for database searching.
Figure 3.
Evaluation of post-translational modifications in cyclotides identified by MS/MS sequencing and database analysis. Database searching of MS/MS spectra of cyclotides revealed a number of peptides with posttranslational modifications. The most abundant ones have been illustrated exemplarily: (A) Aspartate methylation in vitri peptide 94b; (B) tryptophan degradation product kynurenine in varv peptide E; (C) tryptophan oxidation to 3-hydroxy-tryptophan in kalata B1; and (D) glutamate ethylation and glutamine or asparagine deamidation in vitri peptide 42a. (E) The MS/MS fragment spectrum of a C-terminal fragment of endoproteinase GluC processed vitri peptide 22a has been shown to confirm the presence of acyclic cyclotides in V. tricolor. The major b- and y-ion series have been labeled in all spectra. Furthermore, peptide mass, cleavage protease, mass error, and Mascot score of the illustrated peptides have been provided.
Although cyclotide sequencing in nonfractionated vitri plant extract using the automated proteomics workflow yielded several known and novel cyclotides, many peptides lacked identification by sequence, as compared to analysis by molecular weight (Table 1). Thus, to maximize the number of identified cyclotides by sequence we introduced an HPLC-assisted fractionation approach with the aim to enrich cyclotides in a particular fraction and subsequently increase sensitivity of the proteomics workflow compared to the extract starting material.
Fractionation-Based Enrichment Approach
The vitri extract was subjected to preparative HPLC, and nine fractions were collected (Supplementary Figure S-2). Each fraction was evaluated for cyclotide masses by MALDI−MS (Supplementary Figure S-3). V. tricolor fraction F1 (vitri-F1) contained only minor amounts of cyclotide masses in the range of 2500–4500 Da, and vitri-F2 did not show any cyclotide-related signals. Instead, analysis of vitri-F3–F9 yielded significantly more molecular weight signals as compared to MALDI−TOF MS of nonfractionated vitri extract. According to previous studies, fractionation has been confirmed as an efficient tool for enrichment and increased identification of cyclotides.7,63 The digested samples of each HPLC fraction were applied to LC–MS/MS sequencing. The results of the proteomics study of the vitri fractions have been summarized in Table 1. As compared to analysis of the vitri extract, overall 82 peptides have been validated by proteomics of the fractionated samples. Most notable, 20 novel vitri peptides (1, 2, 3, 4, 8, 9a/53, 14, 17, 18a, 20, 21, 23, 24/28, 27a, 29, 30, 36/37, 38, 42a, 50) have been unambiguously characterized by MS/MS sequencing. In addition, vitri F, cycloviolacin O4 and O20, chacur 1, vaby C, E, viba 9, 11, 15, 19 vibi C, G, and vigno 6, 7, 9, and 10 have all been validated and identified by the proteomics approach. Moreover, an additional 11 acyclic peptides have been found as compared to the analysis of the vitri extract; these were acyclo-kalata B1, -varv peptide B, -F, -G, -vitri E, -vodo N, -vitri peptide 8, 9a/53, 22a, 39 and -cycloviolacin O22. Most acyclic peptides were found alongside the cyclic species, except for vitri 8 of which only the acyclic peptide has been identified. In fact, vitri precursor 8 encoding for vitri 8 is missing the essential C-tail for successful cyclization in planta. In summary, sample enrichment by HPLC fractionation is very effective to enhance the number of total cyclotides identified by about 2-fold. Using proteomics provided evidence for 82 cyclotides present at the protein level.
Comparison of Cyclotide Discovery Approaches
Cyclotide analysis is associated with challenges regardless if they are studied at the nucleic acid or proteome level. To highlight the benefits and drawbacks of each method, it may be useful to compare the results, i.e. the number of identified cyclotides, from transcriptome mining, mass deconvolution, and MS/MS sequencing (Figure 4A). Analyzing cyclotides at the transcript level, by mass signals deconvolution or by MS/MS sequencing 108, 127, and 82 cyclotides were detected, respectively. Essentially, 54 cyclotides were identified by MS/MS sequencing as well as mass deconvolution, 31 by MS/MS as well as transcriptome analysis, and 17 were found by both mass deconvolution and transcriptome mining; 11 peptides were identified by all three methods. After all, 28% of 108 vitri cyclotides identified by transcriptome mining were confirmed by MS/MS or molecular weight; 50% of cyclotides were assigned from partial sequences, 2% constitute cyclotide-like peptides, and only 20% of all full-length cyclotides determined by transcriptome mining remained unverified by MS (Figure 4B). Considering full-length precursor molecules, 46% of all predicted peptides were found by MS/MS sequencing. Importantly, 12 out of 14 previously published vitri peptides have been identified by MS/MS sequence, which confirms the effectiveness of our approach, considering that we worked with plant extracts and fractions but avoided using purified cyclotides for proteomics analysis.
Figure 4.
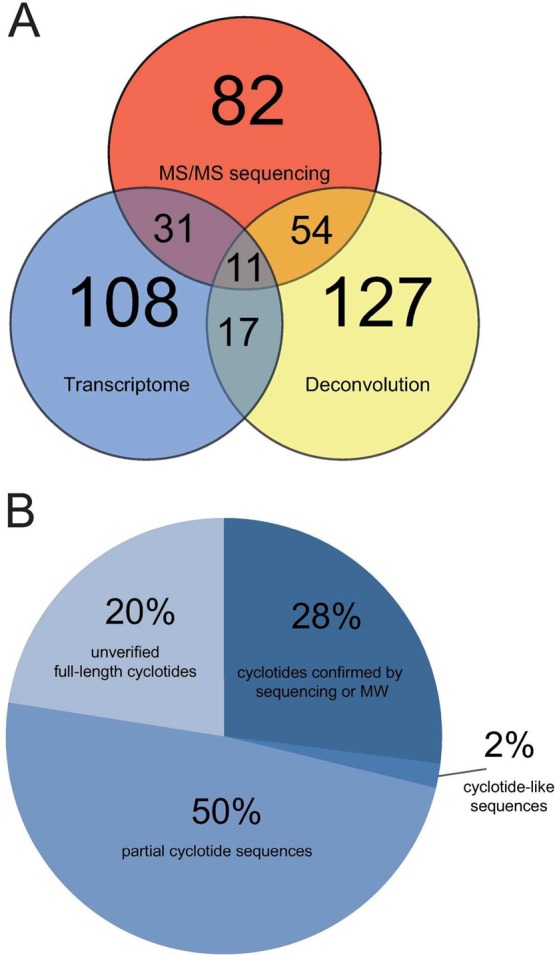
Comparative analysis of vitri cyclotides identified by transcriptome mining, mass deconvolution, and MS/MS sequencing. (A) An overview of vitri peptides discovered in this study is summarized by a Venn diagram. The number of identified cyclotides by each approach (transcriptome mining, mass deconvolution or MS/MS sequencing) is indicated by the numbers in each of the three interwheeling circles. The overlapping regions represent the numbers of identified cyclotides by combination of two or all three approaches. (B) A pie chart illustrates coverage for confirmation of predicted vitri cyclotides by MS. All predicted 108 vitri cyclotides identified by transcriptome mining were grouped in cohorts: (i) confirmed by sequence or molecular weight (28%), (ii) partial sequences from transcriptome mining (50%), (iii) cyclotide-like molecules (2%) and unverified full-length cyclotides (20%).
Clearly, analyzing cyclotides by molecular weight only is the most error-prone method for unambiguous characterization of cyclotides. Nevertheless, combined with transcriptome analysis, a simple MS run may provide useful information about the diversity of cyclotides in a given plant extract. Similarly, transcriptome mining by itself resulted in the identification of many novel cyclotides, but the sequences of many vitri precursors contained uncommon C- or N-terminal processing sites, such as the missing of the conserved asparagine or aspartic acid residues or complete lack of C-tail motifs. In combination with MS however, these powerful tools were able to verify some of these peptides as acyclic cyclotide homologues (Supplementary Table S-1). Obviously MS/MS sequencing using an automated proteomics workflow appears to be the most effective approach for identifying and characterizing novel cyclotides from V. tricolor, since it is capable of providing evidence not only for the presence of a cyclotide and further yields detailed information sequence and PTMs. Thus, we are confident that our results may provide a proof-of-principle and motivation for continued efforts in cyclotide discovery in many plant families using a combined transcriptome and proteome analysis workflow.
Conclusion
The presented study combined cyclotide peptide analysis at both the nucleic acid and the proteome level to obtain insights into the diversity of circular cysteine-rich peptides of V. tricolor. It provides a comprehensive view of the transcription and on the translation of plant cyclotides and to our knowledge the largest study to date in the field of cyclotide discovery and characterization. Our results may provide a guideline for future cyclotide discovery projects combining transcriptome and proteome analysis. Here we were able to shed light to the unexplored diversity of plant-derived cyclotides within the Violaceae family, which comprises up to 31 genera and approximately 1000 species.64 Previously 10 cyclotides have been estimated as a realistic number of peptides to be expressed per Viola species.24 A more recent study determined that a single species may express up to 53 peptides.38 To date, 161 individual cyclotides are known in Violaceae, and in total 922 cyclotides are published online in CyBase (www.cybase.org.au). Our finding that V. tricolor contains at least 164 different cyclotides significantly augments the number of cyclotides present in Violaceae. Thus, we determined that violaceous plants may indeed exhibit a much higher cyclotide diversity than earlier anticipated24 and contain up to >150 000 individual cyclotides. Similarly, other plant families such as Rubiaceae may contain 150 000 cyclotides or more, determined by the number of cyclotides per species and multiplied by the estimated number of species known to express cyclotides.12 This simple calculation does not account for any peptide that may be ubiquitously expressed in many species. In addition other plant families of Cucurbitaceae, Fabaceae, Poaceae and Solanaceae also contain cyclotides. Encompassing the diversity of V. tricolor as a natural library of bioactive peptides, this commercially available medicinal herb may be a suitable starting point for future bioactivity-guided screening studies, and the presented methodology may provide a guideline for cyclotide analytics combining transcriptome and proteome analysis.
Acknowledgments
The authors would like to thank M. Gold-Binder and J.H. Leebens-Mack for technical assistance. Further we thank C. Gründemann for providing a photograph of Viola tricolor, courtesy of Weleda AG, Schwäbisch-Gmünd, Germany.
Glossary
Abbreviations
- Vitri
Viola tricolor
- CCK
cyclic cystine-knot
- MS
mass spectrometry
- FLAS
University of Florida Herbarium
- BLAST
Basic Local Assignment Search Tool
- NMR
nuclear magnetic resonance
- MS
mass spectrometry
- PTM
post-translational modification
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00681.
Supplementary Figure S-1. Loop lengths of Viola tricolor cyclotides. Supplementary Text S-1. Extended Discussion. Supplementary Figure S-2. Fractionation of cyclotide extract by preparative HPLC. Supplementary Figure S-3. MALDI−TOF MS analysis of vitri extract and fractions 1–9. Supplementary Data S-1. Novel cyclotide precursor sequences from Viola tricolor. Supplementary Data S-2. Alignment of mature cyclotides and processing sites. Supplementary Table S-2. LC−MS deconvolution and MALDI−TOF MS of vitri extract (PDF)
Supplementary Table S-1. Viola tricolor cyclotides discovered by transcriptome mining (XLSX)
This work was financially supported by the Austrian Science Fund (FWF P-24743). C.W.G. is an ARC Future Fellow. J.K. received a UQ Postdoctoral Research Fellowship. The 1000 Plants (1kp) initiative, led by G.K-S.W., is funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures (AITF), Innovates Centres of Research Excellence (iCORE), Musea Ventures, BGI-Shenzhen and China National Genebank (CNGB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Gruber C. W.; Cemazar M.; Anderson M. A.; Craik D. J. Insecticidal plant cyclotides and related cystine knot toxins. Toxicon 2007, 49, 561–75 10.1016/j.toxicon.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Tam J. P.; Lu Y. A.; Yang J. L.; Chiu K. W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 8913–8 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgrave M. L.; Craik D. J. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry 2004, 43, 5965–75 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- Northfield S. E.; Wang C. K.; Schroeder C. I.; Durek T.; Kan M. W.; Swedberg J. E.; Craik D. J. Disulfide-rich macrocyclic peptides as templates in drug design. Eur. J. Med. Chem. 2014, 77, 248–57 10.1016/j.ejmech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Wang C. K.; Gruber C. W.; Cemazar M.; Siatskas C.; Tagore P.; Payne N.; Sun G.; Wang S.; Bernard C. C.; Craik D. J. Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chem. Biol. 2014, 9, 156–63 10.1021/cb400548s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehbach J.; O’Brien M.; Muttenthaler M.; Miazzo M.; Akcan M.; Elliott A. G.; Daly N. L.; Harvey P. J.; Arrowsmith S.; Gunasekera S.; Smith T. J.; Wray S.; Goransson U.; Dawson P. E.; Craik D. J.; Freissmuth M.; Gruber C. W. Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 21183–88 10.1073/pnas.1311183110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinger R.; Koehbach J.; Puigpinos A.; Clark R. J.; Tarrago T.; Giralt E.; Gruber C. W. Inhibition of Human Prolyl Oligopeptidase Activity by the Cyclotide Psysol 2 Isolated from Psychotria solitudinum. J. Nat. Prod. 2015, 78, 1073–82 10.1021/np501061t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C. I.; Swedberg J. E.; Craik D. J. Recent progress towards pharmaceutical applications of disulfide-rich cyclic peptides. Curr. Protein Pept. Sci. 2013, 14, 532–42 10.2174/13892037113149990069. [DOI] [PubMed] [Google Scholar]
- Thell K.; Hellinger R.; Schabbauer G.; Gruber C. W. Immunosuppressive peptides and their therapeutic applications. Drug Discovery Today 2014, 19, 645–53 10.1016/j.drudis.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundemann C.; Koehbach J.; Huber R.; Gruber C. W. Do Plant Cyclotides Have Potential As Immunosuppressant Peptides?. J. Nat. Prod. 2012, 75, 167–174 10.1021/np200722w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman R.; Gunasekera S.; Stromstedt A. A.; Goransson U. Chemistry and biology of cyclotides: circular plant peptides outside the box. J. Nat. Prod. 2014, 77, 724–36 10.1021/np401055j. [DOI] [PubMed] [Google Scholar]
- Gruber C. W.; Elliott A. G.; Ireland D. C.; Delprete P. G.; Dessein S.; Goransson U.; Trabi M.; Wang C. K.; Kinghorn A. B.; Robbrecht E.; Craik D. J. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 2008, 20, 2471–83 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinger R.; Koehbach J.; Fedchuk H.; Sauer B.; Huber R.; Gruber C. W.; Grundemann C. Immunosuppressive activity of an aqueous Viola tricolor herbal extract. J. Ethnopharmacol. 2014, 151, 299–306 10.1016/j.jep.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovekorn W.; Tepe A.; Danesch U. A randomized, double-blind, vehicle-controlled, half-side comparison with a herbal ointment containing Mahonia aquifolium, Viola tricolor and Centella asiatica for the treatment of mild-to-moderate atopic dermatitis. Int. J. Clin. Pharmacol. Ther. 2007, 45, 583–91 10.5414/CPP45583. [DOI] [PubMed] [Google Scholar]
- Reuter J.; Merfort I.; Schempp C. M. Botanicals in dermatology: an evidence-based review. Am. J. Clin. Dermatol. 2010, 11, 247–67 10.2165/11533220-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Toiu A.; Muntean E.; Oniga I.; Vostinaru O.; Tamas M. Pharmacognostic research on Viola tricolor L. (Violaceae). Planta Med. 2009, 113, 264–7 10.1055/s-0029-1234429. [DOI] [PubMed] [Google Scholar]
- Svangard E.; Goransson U.; Hocaoglu Z.; Gullbo J.; Larsson R.; Claeson P.; Bohlin L. Cytotoxic cyclotides from Viola tricolor. J. Nat. Prod. 2004, 67, 144–7 10.1021/np030101l. [DOI] [PubMed] [Google Scholar]
- Broussalis A. M.; Goransson U.; Coussio J. D.; Ferraro G.; Martino V.; Claeson P. First cyclotide from Hybanthus (Violaceae). Phytochemistry 2001, 58, 47–51 10.1016/S0031-9422(01)00173-X. [DOI] [PubMed] [Google Scholar]
- Burman R.; Gruber C. W.; Rizzardi K.; Herrmann A.; Craik D. J.; Gupta M. P.; Goransson U. Cyclotide proteins and precursors from the genus Gloeospermum: filling a blank spot in the cyclotide map of Violaceae. Phytochemistry 2010, 71, 13–20 10.1016/j.phytochem.2009.09.023. [DOI] [PubMed] [Google Scholar]
- Goransson U.; Luijendijk T.; Johansson S.; Bohlin L.; Claeson P. Seven novel macrocyclic polypeptides from Viola arvensis. J. Nat. Prod. 1999, 62, 283–6 10.1021/np9803878. [DOI] [PubMed] [Google Scholar]
- Hashempour H.; Koehbach J.; Daly N. L.; Ghassempour A.; Gruber C. W. Characterizing circular peptides in mixtures: sequence fragment assembly of cyclotides from a violet plant by MALDI-TOF/TOF mass spectrometry. Amino Acids 2013, 44, 581–95 10.1007/s00726-012-1376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A.; Burman R.; Mylne J. S.; Karlsson G.; Gullbo J.; Craik D. J.; Clark R. J.; Goransson U. The alpine violet, Viola biflora, is a rich source of cyclotides with potent cytotoxicity. Phytochemistry 2008, 69, 939–52 10.1016/j.phytochem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Ireland D. C.; Colgrave M. L.; Craik D. J. A novel suite of cyclotides from Viola odorata: sequence variation and the implications for structure, function and stability. Biochem. J. 2006, 400, 1–12 10.1042/BJ20060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen S. M.; Sando L.; Ireland D. C.; Colgrave M. L.; Bharathi R.; Goransson U.; Craik D. J. A continent of plant defense peptide diversity: cyclotides in Australian Hybanthus (Violaceae). Plant Cell 2005, 17, 3176–3189 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabi M.; Mylne J. S.; Sando L.; Craik D. J. Circular proteins from Melicytus (Violaceae) refine the conserved protein and gene architecture of cyclotides. Org. Biomol. Chem. 2009, 7, 2378–88 10.1039/b823020j. [DOI] [PubMed] [Google Scholar]
- Wang C. K.; Colgrave M. L.; Gustafson K. R.; Ireland D. C.; Goransson U.; Craik D. J. Anti-HIV cyclotides from the Chinese medicinal herb Viola yedoensis. J. Nat. Prod. 2008, 71, 47–52 10.1021/np070393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeshak M. Y.; Burman R.; Asres K.; Goransson U. Cyclotides from an extreme habitat: characterization of cyclic peptides from Viola abyssinica of the Ethiopian highlands. J. Nat. Prod. 2011, 74, 727–31 10.1021/np100790f. [DOI] [PubMed] [Google Scholar]
- Chen B.; Colgrave M. L.; Wang C.; Craik D. J. Cycloviolacin H4, a hydrophobic cyclotide from Viola hederaceae. J. Nat. Prod. 2006, 69, 23–8 10.1021/np050317i. [DOI] [PubMed] [Google Scholar]
- Gerlach S. L.; Yeshak M.; Goransson U.; Roy U.; Izadpanah R.; Mondal D. Cycloviolacin O2 (CyO2) suppresses productive infection and augments the antiviral efficacy of nelfinavir in HIV-1 infected monocytic cells. Biopolymers 2013, 100, 471–9 10.1002/bip.22325. [DOI] [PubMed] [Google Scholar]
- Goransson U.; Herrmann A.; Burman R.; Haugaard-Jonsson L. M.; Rosengren K. J. The conserved glu in the cyclotide cycloviolacin O2 has a key structural role. ChemBioChem 2009, 10, 2354–60 10.1002/cbic.200900342. [DOI] [PubMed] [Google Scholar]
- Goransson U.; Svangard E.; Claeson P.; Bohlin L. Novel strategies for isolation and characterization of cyclotides: the discovery of bioactive macrocyclic plant polypeptides in the Violaceae. Curr. Protein Pept. Sci. 2004, 5, 317–29 10.2174/1389203043379495. [DOI] [PubMed] [Google Scholar]
- Herrmann A.; Svangard E.; Claeson P.; Gullbo J.; Bohlin L.; Goransson U. Key role of glutamic acid for the cytotoxic activity of the cyclotide cycloviolacin O2. Cell. Mol. Life Sci. 2006, 63, 235–45 10.1007/s00018-005-5486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland D. C.; Colgrave M. L.; Nguyencong P.; Daly N. L.; Craik D. J. Discovery and characterization of a linear cyclotide from Viola odorata: Implications for the processing of circular proteins. J. Mol. Biol. 2006, 357, 1522–1535 10.1016/j.jmb.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Leta Aboye T.; Clark R. J.; Craik D. J.; Goransson U. Ultra-stable peptide scaffolds for protein engineering-synthesis and folding of the circular cystine knotted cyclotide cycloviolacin O2. ChemBioChem 2008, 9, 103–13 10.1002/cbic.200700357. [DOI] [PubMed] [Google Scholar]
- Svangard E.; Burman R.; Gunasekera S.; Lovborg H.; Gullbo J.; Goransson U. Mechanism of action of cytotoxic cyclotides: cycloviolacin O2 disrupts lipid membranes. J. Nat. Prod. 2007, 70, 643–7 10.1021/np070007v. [DOI] [PubMed] [Google Scholar]
- Koehbach J.; Attah A. F.; Berger A.; Hellinger R.; Kutchan T. M.; Carpenter E. J.; Rolf M.; Sonibare M. A.; Moody J. O.; Wong G. K.; Dessein S.; Greger H.; Gruber C. W. Cyclotide discovery in Gentianales revisited-identification and characterization of cyclic cystine-knot peptides and their phylogenetic distribution in Rubiaceae plants. Biopolymers 2013, 100, 438–52 10.1002/bip.22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth A. G.; Colgrave M. L.; Lyons R. E.; Daly N. L.; Craik D. J. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 10127–32 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Li J.; Huang Z.; Yang B.; Zhang X.; Li D.; Craik D. J.; Baker A. J.; Shu W.; Liao B. Transcriptomic screening for cyclotides and other cysteine-rich proteins in the metallophyte Viola baoshanensis. J. Plant Physiol. 2015, 178, 17–26 10.1016/j.jplph.2015.01.017. [DOI] [PubMed] [Google Scholar]
- Nguyen G. K.; Lim W. H.; Nguyen P. Q.; Tam J. P. Novel cyclotides and uncyclotides with highly shortened precursors from Chassalia chartacea and effects of methionine oxidation on bioactivities. J. Biol. Chem. 2012, 287, 17598–607 10.1074/jbc.M111.338970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordon-Thaden I. E.; Chanderbali A. S.; Gitzendanner M. A.; Soltis D. E. Modified CTAB and TRIzol protocols improve RNA extraction from chemically complex Embryophyta. Appl. Plant Sci. 2015, 3, 1400105. 10.3732/apps.1400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Wu G.; Tang J.; Luo R.; Patterson J.; Liu S.; Huang W.; He G.; Gu S.; Li S.; Zhou X.; Lam T. W.; Li Y.; Xu X.; Wong G. K.; Wang J. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–6 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- Colgrave M. L.; Poth A. G.; Kaas Q.; Craik D. J. A new ″era″ for cyclotide sequencing. Biopolymers 2010, 94, 592–601 10.1002/bip.21400. [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A.; Cote R. G.; Csordas A.; Dianes J. A.; Fabregat A.; Foster J. M.; Griss J.; Alpi E.; Birim M.; Contell J.; O’Kelly G.; Schoenegger A.; Ovelleiro D.; Perez-Riverol Y.; Reisinger F.; Rios D.; Wang R.; Hermjakob H. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013, 41, D1063–9 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gran L. Oxytocic principles of Oldenlandia affinis. Lloydia 1973, 36, 174–8. [PubMed] [Google Scholar]
- Gran L. On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol. Toxicol. 1973, 33, 400–8 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Wang C. K.; Hu S. H.; Martin J. L.; Sjogren T.; Hajdu J.; Bohlin L.; Claeson P.; Goransson U.; Rosengren K. J.; Tang J.; Tan N. H.; Craik D. J. Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold. J. Biol. Chem. 2009, 284, 10672–83 10.1074/jbc.M900021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber C. W. Global cyclotide adventure: a journey dedicated to the discovery of circular peptides from flowering plants. Biopolymers 2010, 94, 565–72 10.1002/bip.21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach S. L.; Goransson U.; Kaas Q.; Craik D. J.; Mondal D.; Gruber C. W. A systematic approach to document cyclotide distribution in plant species from genomic, transcriptomic, and peptidomic analysis. Biopolymers 2013, 100, 433–7 10.1002/bip.22258. [DOI] [PubMed] [Google Scholar]
- Mulvenna J. P.; Sando L.; Craik D. J. Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure 2005, 13, 691–701 10.1016/j.str.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Tang J.; Wang C. K.; Pan X.; Yan H.; Zeng G.; Xu W.; He W.; Daly N. L.; Craik D. J.; Tan N. Isolation and characterization of cytotoxic cyclotides from Viola tricolor. Peptides 2010, 31, 1434–40 10.1016/j.peptides.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Nguyen G. K.; Lian Y.; Pang E. W.; Nguyen P. Q.; Tran T. D.; Tam J. P. Discovery of linear cyclotides in monocot plant Panicum laxum of Poaceae family provides new insights into evolution and distribution of cyclotides in plants. J. Biol. Chem. 2013, 288, 3370–80 10.1074/jbc.M112.415356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plan M. R.; Goransson U.; Clark R. J.; Daly N. L.; Colgrave M. L.; Craik D. J. The cyclotide fingerprint in oldenlandia affinis: elucidation of chemically modified, linear and novel macrocyclic peptides. ChemBioChem 2007, 8, 1001–11 10.1002/cbic.200700097. [DOI] [PubMed] [Google Scholar]
- Barbeta B. L.; Marshall A. T.; Gillon A. D.; Craik D. J.; Anderson M. A. Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 1221–5 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J.; Malik U. Cyclotide biosynthesis. Curr. Opin. Chem. Biol. 2013, 17, 546–54 10.1016/j.cbpa.2013.05.033. [DOI] [PubMed] [Google Scholar]
- Gillon A. D.; Saska I.; Jennings C. V.; Guarino R. F.; Craik D. J.; Anderson M. A. Biosynthesis of circular proteins in plants. Plant J. 2008, 53, 505–15 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- Gruber C. W.; Cemazar M.; Clark R. J.; Horibe T.; Renda R. F.; Anderson M. A.; Craik D. J. A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 2007, 282, 20435–46 10.1074/jbc.M700018200. [DOI] [PubMed] [Google Scholar]
- Saska I.; Gillon A. D.; Hatsugai N.; Dietzgen R. G.; Hara-Nishimura I.; Anderson M. A.; Craik D. J. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 2007, 282, 29721–8 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- Trabi M.; Svangard E.; Herrmann A.; Goransson U.; Claeson P.; Craik D. J.; Bohlin L. Variations in cyclotide expression in viola species. J. Nat. Prod. 2004, 67, 806–10 10.1021/np034068e. [DOI] [PubMed] [Google Scholar]
- Trabi M.; Craik D. J. Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell 2004, 16, 2204–2216 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J. S.; Wang C. K.; van der Weerden N. L.; Craik D. J. Cyclotides are a component of the innate defense of Oldenlandia affinis. Biopolymers 2010, 94, 635–46 10.1002/bip.21419. [DOI] [PubMed] [Google Scholar]
- Seydel P.; Gruber C. W.; Craik D. J.; Dornenburg H. Formation of cyclotides and variations in cyclotide expression in Oldenlandia affinis suspension cultures. Appl. Microbiol. Biotechnol. 2007, 77, 275–84 10.1007/s00253-007-1159-6. [DOI] [PubMed] [Google Scholar]
- Conlan B. F.; Colgrave M. L.; Gillon A. D.; Guarino R.; Craik D. J.; Anderson M. A. Insights into processing and cyclization events associated with biosynthesis of the cyclic Peptide kalata B1. J. Biol. Chem. 2012, 287, 28037–46 10.1074/jbc.M112.347823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biass D.; Violette A.; Hulo N.; Lisacek F.; Favreau P.; Stocklin R. Uncovering intense protein diversification in a cone snail venom gland using an integrative venomics approach. J. Proteome Res. 2015, 14, 628–38 10.1021/pr500583u. [DOI] [PubMed] [Google Scholar]
- Hekking W. H. A. Studies on Neotropical Violaceae. II. Arrangement of Leaves, Inflorescences and Branchlets in Neotropical Rinorea. Flora 1988, 180, 345–376. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.