Abstract
Previous studies have hypothesized that diverse genetic causes of intellectual disability (ID) and autism spectrum disorders (ASDs) converge on common cellular pathways. Testing this hypothesis requires detailed phenotypic analyses of animal models with genetic mutations that accurately reflect those seen in the human condition (i.e., have structural validity) and which produce phenotypes that mirror ID/ASDs (i.e., have face validity). We show that SynGAP haploinsufficiency, which causes ID with co-occurring ASD in humans, mimics and occludes the synaptic pathophysiology associated with deletion of the Fmr1 gene. Syngap+/− and Fmr1−/y mice show increases in basal protein synthesis and metabotropic glutamate receptor (mGluR)-dependent long-term depression that, unlike in their wild-type controls, is independent of new protein synthesis. Basal levels of phosphorylated ERK1/2 are also elevated in Syngap+/− hippocampal slices. Super-resolution microscopy reveals that Syngap+/− and Fmr1−/y mice show nanoscale alterations in dendritic spine morphology that predict an increase in biochemical compartmentalization. Finally, increased basal protein synthesis is rescued by negative regulators of the mGlu subtype 5 receptor and the Ras–ERK1/2 pathway, indicating that therapeutic interventions for fragile X syndrome may benefit patients with SYNGAP1 haploinsufficiency.
SIGNIFICANCE STATEMENT As the genetics of intellectual disability (ID) and autism spectrum disorders (ASDs) are unraveled, a key issue is whether genetically divergent forms of these disorders converge on common biochemical/cellular pathways and hence may be amenable to common therapeutic interventions. This study compares the pathophysiology associated with the loss of fragile X mental retardation protein (FMRP) and haploinsufficiency of synaptic GTPase-activating protein (SynGAP), two prevalent monogenic forms of ID. We show that Syngap+/− mice phenocopy Fmr1−/y mice in the alterations in mGluR-dependent long-term depression, basal protein synthesis, and dendritic spine morphology. Deficits in basal protein synthesis can be rescued by pharmacological interventions that reduce the mGlu5 receptor–ERK1/2 signaling pathway, which also rescues the same deficit in Fmr1−/y mice. Our findings support the hypothesis that phenotypes associated with genetically diverse forms of ID/ASDs result from alterations in common cellular/biochemical pathways.
Keywords: fragile X syndrome, long-term depression, mGluR, neurodevelopmental disorder, STED, SynGAP
Introduction
Intellectual disability (ID) refers to a group of neurodevelopmental disorders characterized by an intelligence quotient (IQ) below 70, which affects ∼2–3% of the population (Huang, 2009). Large-scale genetic studies have shown that the genetic causes of ID are highly heterogeneous involving chromosomal abnormalities and single gene mutations, with several hundred ID-related genes identified (Krumm et al., 2014). Estimates of comorbidity indicate that ∼30% of people with ID also present with autism spectrum disorders (ASDs), suggesting significant genetic overlap between these two disorders. For example, one-third of all boys with fragile X syndrome (FXS) also meet the diagnostic criteria for autism (Wijetunge et al., 2013).
Although the genetic causes of ID/ASDs are highly diverse, previous studies suggest that there may be convergence at the level of biochemical pathways that are affected. For example, FXS and tuberous sclerosis complex (TSC), two monogenic causes of ID with overlapping clinical symptoms, appear to be on opposite ends of a common pathophysiological axis (Auerbach et al., 2011). Thus, it is possible that rare but highly penetrant genetic mutations (e.g., FXS and TSC) may give insight into the pathophysiology of a range of IDs/ASDs (Wijetunge et al., 2013).
A core physiological consequence of the loss of fragile X mental retardation protein (FMRP) is an increase in basal protein synthesis downstream of mGlu5 receptor activation (Qin et al., 2005; Dölen et al., 2007; Osterweil et al., 2010; Till et al., 2015). In the hippocampus, this change can be detected as an increase in metabotropic glutamate receptor (mGluR)-dependent long-term synaptic depression (LTD) that, unlike in wild type (WT), no longer requires new protein synthesis (Huber et al., 2002; Nosyreva and Huber, 2006). Reducing mGlu5 receptor signaling, either genetically or with negative allosteric modulators of mGlu5 receptors, rescues many of the phenotypes associated with the genetic deletion of FMRP (Dölen et al., 2007; Michalon et al., 2012; Michalon et al., 2014). In the absence of FMRP, the elevation in basal protein synthesis results from a hypersensitivity to ERK1/2-MAPK signaling, and inhibitors of this pathway also reverse cellular and behavioral effects resulting from the loss of FMRP (Osterweil et al., 2010, 2013).
In this study, we examine the pathophysiology in mice heterozygous for a null mutation in Syngap. Synaptic GTPase-activating protein (SynGAP) is a RasGAP encoded by a gene that gives rise to multiple isoforms, several of which have opposing effects on synaptic efficacy (McMahon et al., 2012). It is expressed throughout development with the highest levels observed in the forebrain at the peak of synaptogenesis (Porter et al., 2005; Barnett et al., 2006). In human patients, de novo SYNGAP1 mutations that result in a null allele lead to autosomal dominant nonsyndromic ID (NSID) and, in a minority of cases, ASDs (Hamdan et al., 2009, 2011; Berryer et al., 2013). Syngap heterozygous (Syngap+/−) mice also show a range of behavioral, physiological, and anatomical abnormalities throughout development (Komiyama et al., 2002; Vazquez et al., 2004; Barnett et al., 2006; Harlow et al., 2010; Clement et al., 2012, 2013). Fmr1−/y mice show a hypersensitivity to Ras–ERK1/2 signaling (Osterweil et al., 2010), whereas Syngap+/− mice show elevated ERK1/2 phosphorylation in the hippocampus (Komiyama et al., 2002; Ozkan et al., 2014). The involvement of the ERK1/2 pathway led us to hypothesize that these two genetic mutations, which cause human ID, may share a common hippocampal pathophysiology downstream of mGluR-dependent ERK1/2 activation.
Materials and Methods
Animals.
Syngap+/− mutant mice, originally generated by Komiyama et al. (2002), and Fmr1−/y mice were bred on a C57Black6JOla line (Harlan). For genetic occlusion experiments, Syngap+/− males were paired with Fmr1+/− females to obtain F1 male progeny with the following genotypes: WT (Syngap+/+/Fmr1+/y), Fmr1 KO (Syngap+/+/Fmr1−/y), Syngap heterozygous (Syngap+/−/Fmr1+/y), and double mutants (Syngap+/−/Fmr1−/y). All experiments were performed with WT littermates acting as controls and with the experimenter blind to genotype. All animal work was performed in accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986 under the authority of Project Licenses PPL 60/3631 and PPL 60/4290.
Hippocampal slice electrophysiology.
Horizontal hippocampal slices (400 μm) were prepared from postnatal day 25–32 (P25–32) animals. Slices were collected in carbogenated (95% oxygen, 5% CO2) ice-cold dissection buffer containing the following (in mm): 86 NaCl, 1.2 NaH2PO4, 25 KCl, 25 NaHCO3, 20 glucose, 75 sucrose, 0.5 CaCl2, and 7 MgCl2. Slices were incubated for 30 min at ∼30°C in artificial CSF (ACSF) containing the following (in mm): 124 NaCl, 1.2 NaH2PO4, 25 KCl, 25 NaHCO3, 20 glucose, 2 CaCl2, and 1 MgCl2, bubbled with 95% oxygen and 5% CO2. An incision was made through CA1–CA3 boundary, and slices were left to recover for a minimum of 1 h at room temperature (20−22°C) before any recordings were made. For electrophysiological recordings, slices were placed in a submersion chamber heated to 30°C and perfused with carbogenated ACSF containing picrotoxin (50 μm) at a rate of 4 ml/min.
Field EPSPs (fEPSPs) were recorded at Schaffer collateral/commissural inputs to CA1 pyramidal neurons using extracellular recording electrodes (1–3 MΩ) filled with ACSF and placed in the stratum radiatum layer of the CA1 area. Synaptic responses were evoked by applying single current pulses to the Schaffer collateral/commissural axons using a bipolar stimulating electrode. Stimuli (10–30 μA, 200 μs, delivered at a frequency of 0.033 Hz) were set to produce 30–60% of the maximal response amplitude.
Metabotropic glutamate receptor-dependent LTD (mGluR-LTD) was induced by acute application (5 min) of the group 1 mGluR agonist (R,S)-3,5-dihydroxyphenylglycine (DHPG; 50 μm) in the presence of the NMDAR antagonist d-AP5 (50 μm). For experiments involving anisomycin, slices were preincubated for 30 min in ACSF containing anisomycin (20 μm) before LTD induction, which remained present for the duration of the recording.
Electrophysiological traces were collected using WinLTP (University of Bristol, Bristol, UK) and exported to Microsoft Excel. The magnitude of LTD was calculated by dividing the average fEPSP slope from 40 to 60 min after DHPG application by the average fEPSP slope during the 20 min baseline before DHPG application. Statistical analysis was performed using GraphPad Prism routines (GraphPad Software). After genotyping, time-matched normalized data were averaged across experiments and expressed as means ± SE. Significant differences between the WT and mutant mice were determined using Student's t test or ANOVA with p ≤ 0.05.
Metabolic labeling.
Protein synthesis levels were measured following the protocol outlined by Osterweil et al. (2010). Briefly, 500 μm transverse hippocampal slices were prepared from either Syngap+/− or Fmr1−/y mice with WT littermates serving as interleaved controls. Dorsal hippocampal slices were left to recover for 4 h at 30°C in preoxygenated ACSF containing the following (in mm): 124 NaCl, 1.25 NaH2PO4, 3 KCl, 26 NaHCO3, 10 glucose, 2 CaCl2, 1 MgCl2. Slices were transferred to a chamber containing actinomycin D (25 μm) and vehicle, 2-chloro-4-((2,5-dimethyl-1-(4-(trifluoromethoxy)phenyl)-1H-imidazol-4-yl)ethynyl)pyridine (CTEP; 10 μm), 1,4-diamino-2,3-dicyano-1,4-bis (2-aminophenylthio)butadiene (U0126; 5 μm), or lovastatin (100 μm) for 30 min, and then transferred to a chamber with fresh ACSF containing 0.4 Mbq/ml 35S–Met/Cys protein labeling mix (PerkinElmer) and vehicle, CTEP (10 μm), U0126 (5 μm), or lovastatin (100 μm) for 45 min. After labeling, slices were frozen on dry ice and stored at −80°C. For processing, slices were homogenized in ice-cold homogenising buffer (10 mm HEPES, pH 7.4, 2 mm EDTA, 2 mm EGTA, 1% Triton X-100) with protease inhibitors (Roche) and phosphatase inhibitors (cocktails II and III; Sigma). Protein was precipitated with TCA (12.5% final), and total protein concentration was measured using a Bio-Rad protein assay. Samples were read using a scintillation counter, and data were expressed as the number of counts per minute per microgram of protein and normalized to the 35S–Met/Cys ACSF used for incubation.
Western blotting.
For Western blotting, metabolically labeled hippocampal homogenates were taken before TCA precipitation and boiled in Laemmli sample buffer, resolved on SDS polyacrylamide gels (Bio-Rad), transferred to nitrocellulose, and incubated overnight in primary antibodies for p-ERK1/2 (Thr202/Tyr204; Cell Signaling Technology), ERK1/2 (Cell Signaling Technology), and β-actin (Abcam). Blots were incubated with their respective fluorophore-conjugated secondary antibodies and imaged on an Odyssey imaging system (LiCor Biosciences).
Stimulated emission depletion microscopy and dendritic spine analysis.
Quantitative morphometric analysis of dendritic spines using stimulated emission depletion (STED) microscopy on dorsal hippocampal brain sections from perfusion-fixed P35 animals was performed as reported previously (Wijetunge et al., 2014). Spines on three to four apical oblique dendritic segments within stratum radiatum of CA1 were acquired per animal where each dendrite was from a different cell and analyzed blind to the genotype. Confocal microscopy analysis was performed as reported previously (Till et al., 2012), except that the cells were filled in lightly postfixed (4% paraformaldehyde) acute hippocampal slices from P26–P32 animals. Statistical analyses of spine morphology were performed as described previously (Wijetunge et al., 2014).
Results
Enhanced and protein synthesis independent mGluR-LTD in Syngap+/− hippocampal slices
To determine whether SynGAP haploinsufficiency results in a hippocampal pathophysiology similar to that observed in mouse models of FXS, we first examined whether the loss of a single copy of Syngap mimics the increase in mGluR-LTD reported in Fmr1−/y mice. The application of DHPG, a selective group 1 mGluR agonist, resulted in a significant increase in the magnitude of LTD in Syngap+/− hippocampal slices compared to WT controls (WT, 77 ± 3%, n = 18; Syngap+/−, 62 ± 5%, n = 15; p = 0.012). At this developmental time point (P25–P32), the expression of this form of LTD is dependent on the translation of mRNAs at local dendritic sites (Huber et al., 2000). Preincubation of hippocampal slices in the protein synthesis inhibitor anisomycin (20 μm) revealed that mGluR-LTD is no longer dependent on new protein synthesis in Syngap+/− mutants (64 ± 4%, n = 14; Fig. 1B). In contrast, anisomycin eliminated the persistent decrease in fEPSP slope following DHPG application in WT controls (91 ± 4%, n = 12). Furthermore, consistent with previous observations, and under our own experimental conditions, we also observed exaggerated mGluR-LTD that was independent of protein synthesis in Fmr1−/y mice (Fig. 1C,D; Huber et al., 2002; Nosyreva and Huber, 2006). Thus, these two ID-related mutations in Fmr1 and Syngap lead to similar alterations in protein synthesis-dependent mGluR-LTD in hippocampal CA1.
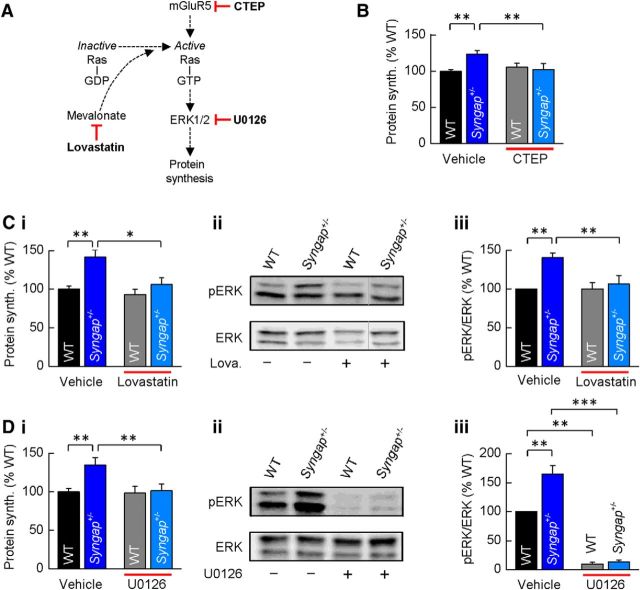
Figure 1.
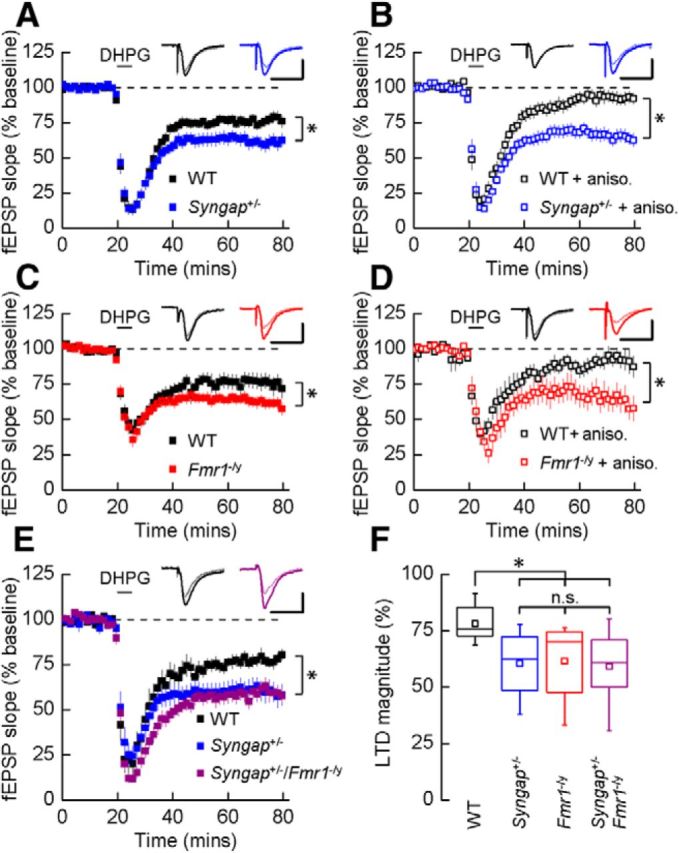
Syngap+/− phenocopies the hippocampal synaptic pathophysiology observed in Fmr1−/y mice. A, application of DHPG (50 μm) induced LTD that was significantly increased in Syngap+/− mice (62 ± 5%, n = 15; t test, p = 0.01) versus WT littermate controls (77 ± 3%, n = 18). Representative average fEPSPs before and after DHPG application are illustrated. B, In the presence of protein synthesis inhibitor anisomycin (aniso.; 20 μm), DHPG-induced LTD was not sustained in WT mice (91 ± 4%, n = 12), while remaining intact in Syngap+/− mice (64 ± 4%, n = 14). C, DHPG-induced mGluR-LTD was also significantly enhanced in Fmr1−/y mice (63 ± 4%, n = 17; t test, p = 0.04) versus WT littermate controls (77 ± 5%, n = 17). D, In the presence of anisomycin, mGluR-LTD remained at a similar magnitude in the Fmr1−/y (65 ± 7%, n = 9), whereas LTD could not be sustained in WT mice (91 ± 6%, n = 8). E, The magnitude of LTD was significantly enhanced in the Syngap+/−/Fmr1−/y double mutant mice (59 ± 4%, n = 12; ANOVA, p = 0.02) relative to WT littermate controls (78 ± 3%; n = 9). No significant differences were observed in LTD magnitude between the Syngap+/−/Fmr1−/y double mutant mice and either Syngap+/− (60 ± 4%, n = 10) or Fmr1−/y (62 ± 5%, n = 10; data not illustrated) single mutant mice. F, Summary of DHPG-induced LTD for each of the four genotypes generated from the Syngap+/− × Fmr1−/y cross. Box plots illustrate minima and maxima (whiskers), median (line), mean (square symbol), and interquartile range (box). Calibrations: A, D, 250 μV, 10 ms; B, C, 500 μV, 10 ms; E, 350 μV, 10 ms. n.s., Not significant. *p < 0.05.
Based on these LTD findings, we next wanted to examine whether mutations that cause FXS and SynGAP haploinsufficiency lead to the disruption of a common pathophysiological mechanism downstream of the mGlu5 receptor. To test this hypothesis, we assessed whether introducing the Syngap mutation into the Fmr1−/y mouse would occlude or exacerbate the LTD phenotype caused by the loss of FMRP. Figure 1, E and F, shows that in double mutant (Syngap+/−/Fmr1−/y) mice, the magnitude of mGluR-LTD is not significantly greater than that observed in either single mutant alone.
To confirm that the occlusion of LTD in the double mutants did not arise from a “floor effect,” mGluR-LTD was induced for a second time 1 h after the initial stimulation to determine whether the first application of DHPG led to a saturating level of LTD. In agreement with previous studies (Palmer et al., 1997; Schnabel et al., 1999; Huber et al., 2001), a subsequent application of DHPG (50 μm) significantly increased the magnitude of LTD relative to the first stimulation in slices prepared from all four genotypes (p < 0.05; Fig. 2A,B).
Figure 2.
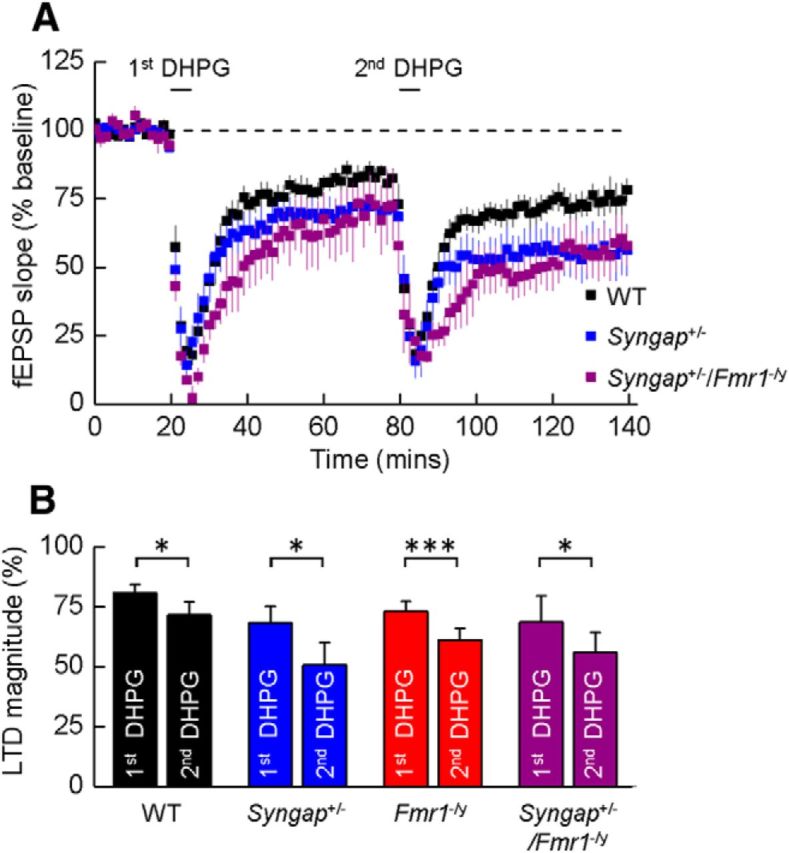
A single application of DHPG does not “saturate” LTD in Syngap+/−/Fmr1−/y double mutant mice. A, Mean time courses illustrating fEPSP slope in experiments on slices prepared from WT, Syngap+/−, and Syngap+/−/Fmr1−/y mice where two applications of DHPG (50 μm) were applied to determine whether the magnitude of LTD elicited by the first application could be augmented by the second. B, Bar graphs illustrating the magnitude of mGluR-mediated LTD produced by a first and then a second application of DHPG in slices prepared from WT (first, 81 ± 3%; second, 71 ± 5%; n = 8; p < 0.05, paired t test), Syngap+/− (first, 68 ± 7%; second, 51 ± 9%; n = 5; p < 0.05), Fmr1−/y (first, 73 ± 4%; second, 61 ± 5%; n = 3; p < 0.001), and Syngap+/−/Fmr1−/y (first, 69 ± 11%; second, 56 ± 8%; n = 4; p < 0.05) mice. *p < 0.05; ***p < 0.001.
Syngap+/− hippocampal slices have elevated basal protein synthesis and enhanced ERK1/2 activity
Previously, it was reported that exaggerated mGluR-LTD in Fmr1−/y mutants correlates with an increase in basal protein synthesis (Dölen et al., 2007). Therefore, we next examined protein synthesis rates in hippocampal slices from Syngap+/− mice employing a similar methodology to that described previously in Fmr1−/y mutants (Fig. 3A; Osterweil et al. 2010). Consistent with our electrophysiological findings, we found a significant increase in 35S-Met/Cys incorporation in Syngap+/− hippocampal slices (140 ± 12%, n = 9; Fig. 3B), indicating that SynGAP may function to suppress mRNA translation under steady-state conditions. Although the magnitude of this increase in basal protein synthesis was greater than that observed in Fmr1−/y versus WT slices (121 ± 7%; n = 7), our findings suggest that the proteins required for mGluR-LTD may be overtranslated in both the Fmr1+/− and Syngap+/− brain.
Figure 3.
Basal protein synthesis and phosphorylated ERK1/2 levels are elevated in the hippocampus of Syngap+/− mice. A, Schematic of the experimental timeline for 35[S]-Met/Cys metabolic labeling. B, Basal protein synthesis levels were significantly elevated in dorsal hippocampal slices from Syngap+/− versus WT mice (WT, 100 ± 3%; Syngap+/−, 140 ± 12%; t test, p = 0.0007; n = 9). Increased protein synthesis rates were also observed in the Fmr1−/y versus WT mice (WT, 100 ± 2%; Fmr1−/y, 121 ± 7%; t test, p = 0.01; n = 6). Ci, Cii, Basal activation state of ERK1/2 in hippocampal slices was significantly increased in Syngap+/− mice versus WT controls (phosphorylated/total ERK1/2, 137 ± 7%; n = 23; p = 1.5 × 10−5). D, Protein synthesis levels were significantly elevated in vehicle-treated dorsal hippocampal slices from Syngap+/− versus WT mice (WT, 100 ± 5%; Syngap+/−, 135 ± 7%; n = 6; ANOVA, p = 0.0038). DHPG treatment (100 μm, 5 min) significantly increased protein synthesis rates in WT slices, but did not further increase 35[S]-Met/Cys incorporation in Syngap+/− slices (WT DHPG, 138 ± 14%; Syngap+/− DHPG, 148 ± 15%; ANOVA, WT treatment, p = 0.0028; n = 6). Ei, Eii, Western blot analysis of vehicle/DHPG-treated hippocampal slices revealed DHPG significantly increases the phosphorylation status of ERK1/2 in both Syngap+/− and WT mice (phosphorylated/total ERK1/2, WT vehicle, 100%; Syngap+/− vehicle, 130 ± 6%; WT DHPG, 180 ± 16%; Syngap+/− DHPG, 173 ± 21%; n = 7; ANOVA, genotype × treatment, p < 0.05). *p < 0.05; **p < 0.01; ***p < 0.001.
In Fmr1−/y mice, increased protein synthesis rates are caused by a hypersensitive response to constitutive mGlu5–Ras–ERK1/2 signaling due the loss of the translational repression by FMRP (Osterweil et al., 2010). In the Syngap+/− hippocampus, both Ras and ERK1/2 activity are increased (Komiyama et al., 2002; Ozkan et al., 2014), leading us to hypothesize that elevated protein synthesis rates may arise from dysregulated ERK1/2 signaling due to the loss of Ras inactivation. To confirm that we observed a similar increase in ERK1/2 activity, phosphorylation levels were quantified in hippocampal slices from Syngap+/− mice and WT controls under identical conditions to which basal protein synthesis rates were found to be enhanced. In agreement with previous studies, ERK1/2 phosphorylation was significantly increased in Syngap+/− mutants under steady-state conditions in the absence of any detectable differences in total ERK1/2 protein (137 ± 7%, n = 23; p = 1.5 × 10−5; Fig. 3C).
It was shown previously that an mGluR-LTD-inducing stimulus increases protein synthesis rates in WT but not Fmr1−/y hippocampal slices (Osterweil et al., 2010). Thus, we measured 35S-Met/Cys incorporation following the acute application of DHPG (100 μm; 5 min) in Syngap+/− hippocampal slices. While DHPG stimulation significantly enhanced protein synthesis in WT slices, the same treatment failed to increase further the already exacerbated protein synthesis levels in Syngap+/− mutants (Fig. 3D), suggesting that mRNA translation downstream of group 1 mGluR activation is saturated under nonstimulated conditions. ERK1/2 activation is a critical step in group 1 mGluR-mediated protein synthesis governing the expression and maintenance of mGluR-LTD (Gallagher et al., 2004; Osterweil et al., 2010). In both WT and Syngap+/− hippocampal slices, DHPG induced a significant and robust increase in ERK1/2 phosphorylation (Fig. 3E). This result may suggest that exaggerated mGluR-LTD in Syngap+/− hippocampal slices could arise from the dysregulation of basal, rather than stimulated, ERK1/2 signaling.
Elevated basal protein synthesis in the Syngap+/− hippocampus can be ameliorated by negative modulators of mGlu5 receptor and Ras–ERK1/2 signaling
To test the hypothesis that excessive mGluR-mediated protein synthesis in Syngap+/− mice is a consequence of hyperactive Ras–ERK1/2 signaling, we examined the effects of inhibitors of this pathway on translational rates. CTEP, lovastatin, and U0126 selectively target and downregulate mGlu5 receptors, Ras, and ERK1/2 signaling, respectively (Fig. 4A), and have been shown previously to restore exaggerated levels of protein synthesis and/or functional deficits in Fmr1−/y mice (Osterweil et al., 2010, 2013; Michalon et al., 2012, 2014). We found that each of these compounds reduced basal protein synthesis in Syngap+/− hippocampal slices to levels not significantly different from WT controls (Fig. 4B,Ci,Di). Importantly, none of these compounds significantly altered basal protein synthesis in WT animals, suggesting that their effect is specific to mechanisms underlying the hyperactivation of ERK1/2 in Syngap+/− mice. To confirm that both lovastatin and U0126 elicited their effects on protein synthesis by downregulating ERK1/2 activity, we quantified the phosphorylation status of ERK1/2 in control and treated hippocampal slices from Syngap+/− and WT mice. Lovastatin significantly reduced ERK1/2 phosphorylation in Syngap+/− slices to WT levels without affecting ERK1/2 phosphorylation in WT treated slices (Fig. 4Cii,Ciii). In contrast, U0126 caused a dramatic reduction in ERK1/2 activity in both WT and Syngap+/− hippocampal slices (Fig. 4Dii,Diii). Together, these findings suggest that dysregulated ERK1/2 signaling leads to exaggerated protein synthesis rates in Syngap+/− hippocampus, and drugs that have been shown previously to ameliorate phenotypes associated with the loss of FMRP can restore translational rates in a mouse model of SYNGAP haploinsufficiency.
Figure 4.
Elevated protein synthesis in Syngap+/− hippocampus is corrected by inhibitors of mGluR5 and Ras–ERK1/2 signaling. A, schematic of the drug targets for CTEP, lovastatin, and U0126, each shown previously to reduce Ras–ERK1/2 activation and correct basal protein synthesis in Fmr1−/y mice. B, CTEP (10 μm) reverses elevated protein synthesis in Syngap+/− mice (WT, vehicle, 100 ± 3%; CTEP, 106 ± 5%; Syngap+/−, vehicle, 124 ± 5%; CTEP, 102 ± 8%; n = 9; ANOVA, genotype, p = 0.0007; KO treatment, p = 0.0014). Ci–Ciii, Lovastatin (100 μm) reverses elevated protein synthesis (Ci) in Syngap+/− mice (WT, vehicle, 100 ± 4%; lovastatin, 93 ± 7; Syngap+/−, vehicle, 142 ± 9%; lovastatin, 106 ± 8%; n = 7; ANOVA, genotype, p = 0.0061; KO treatment, p = 0.013). Example Western blot (Cii) and quantification of ERK1/2 activity (Ciii) in vehicle- and lovastatin-treated hippocampal slices revealed lovastatin significantly reduces phosphorylation levels of ERK1/2 in Syngap+/− mice (phosphorylated/total ERK1/2, WT, vehicle, 100%; Syngap+/−, vehicle, 140 ± 6%; WT, lovastatin, 100 ± 8%; Syngap+/−, lovastatin, 106 ± 11%; n = 10; ANOVA, genotype, *p = 0.0086; KO treatment, p = 0.0057). The gray line to the left of the lovastatin-treated Syngap+/− Western blot indicates that this lane was not adjacent to each of the other three lanes, but was run on the same gel, and hence was processed identically. D, U0126 (5 μm) reverses elevated protein synthesis (Di) in Syngap+/− mice (WT, vehicle, 100 ± 4%; U0126, 98 ± 8%; Syngap+/−, vehicle, 134 ± 9%; U0126, 102 ± 9%; n = 9; ANOVA, genotype, p = 0.003; KO treatment, p = 0.004). Example Western blot (Dii) and quantification (Diii) of ERK1/2 activity in vehicle- and U0126-treated hippocampal slices show U0126 abolishes ERK1/2 activity in both WT and Syngap+/− mice (phosphorylated/total ERK1/2, WT, vehicle, 100%; Syngap+/−, vehicle, 165 ± 15%; WT, lovastatin, 10 ± 3%; Syngap+/−, lovastatin, 13 ± 3%; n = 5; ANOVA, genotype, p = 0.0054; WT treatment, p = 0.005; KO treatment, p = 0.0002). *p < 0.05; **p < 0.01; ***p < 0.001.
Altered morphology of dendritic spines of CA1 pyramidal neurons in Syngap+/− mice
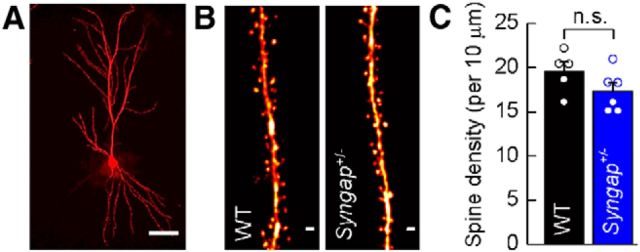
Next, we examined whether the electrophysiological and biochemical changes we observed in Syngap+/− mice are associated with density and morphological alterations in dendritic spines, which are the main site of excitatory input to CA1 pyramidal neurons. To determine whether the reduction in SynGAP expression affected the density of dendritic spines, we dye filled cells obtained from the “partner” hemispheres of brains that had been used for electrophysiological studies. Figure 5 illustrates that no difference in spine densities were observed in Syngap+/− mice and WT littermate controls (WT, 19.58 ± 1.03 spines/10 μm; Syngap+/−, 17.34 ± 0.94 spines/10 μm; p = 0.1430, two-tailed independent t test with Welch's correction).
Figure 5.
CA1 pyramidal neuron dendritic spine density in Syngap+/− mice. A, Composite image of a representative CA1 pyramidal neuron labeled by intracellular filling with Alexa Fluor 568 in lightly fixed section through CA1 of hippocampus. Scale bar, 50 μm. B, Representative composite images of apical oblique dendritic segments from WT and Syngap+/− mice. Scale bars, 1 μm. C, Mean spine density of apical dendrites for WT (n = 5) and Syngap+/− (n = 6) animals indicating no difference in dendritic spine density (WT, 19.58 ± 1.03 spines/10 μm; Syngap+/−, 17.34 ± 0.94 spines/10 μm; p = 0.1430, two-tailed independent t test with Welch's correction). n.s., Not significant.
To accurately resolve the fine details of spine morphology we used STED microscopy, quantifying the widths and lengths of spine heads and necks at the nanoscale (Fig. 6A). For spine head width, no difference in the distributions between Syngap+/− and WT neurons was observed, although spine neck lengths and widths showed significantly different distributions (Fig. 6B–D; WT, n = 393 spines; Syngap+/−, n = 424 spines; head width, p = 0.11; neck length, p = 0.005; neck width, p = 0.0008, Kolmogorov–Smirnov test). In agreement with our STED analysis, confocal analysis of dendritic spines in the animals from which electrophysiological analyses were performed (P25–P32) also indicates that there was no difference in the mean spine head width between genotypes (WT, 0.350 ± 0.01 μm; Syngap+/−, 0.360 ± 0.01 μm; p = 0.71, t test with Welch's correction; data not shown).
Figure 6.
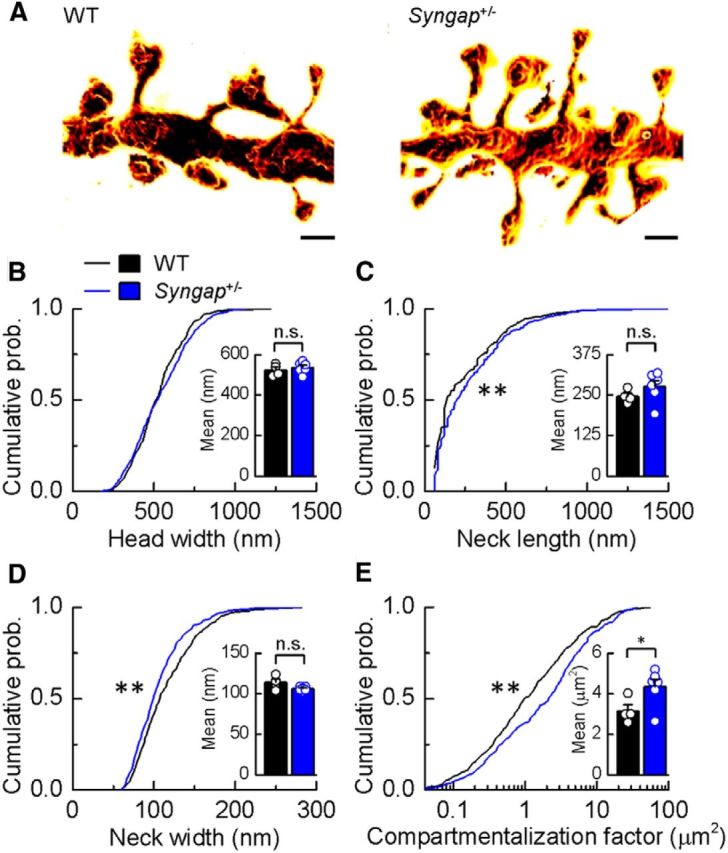
CA1 pyramidal neuron dendritic spine morphology in Syngap+/− mice. A, Surface-rendered STED images of apical dendritic segments from pyramidal neurons in the CA1 of the hippocampus of WT and Syngap+/− mice revealing morphological details that are invisible to conventional light microscopy. Scale bars, 500 nm. B–D, Kolmogorov–Smirnov tests of the cumulative frequency distributions show that the distribution profiles of spine head widths (B) are not significantly different between genotypes (p = 0.11), whereas distribution profiles of spine neck lengths and widths show that Syngap+/− mice (n = 424 spines) have significantly more spines with longer (C; p < 0.005) and narrower necks (D; p < 0.0008) compared to WT mice (n = 393 spines). The mean values for these parameters are, however, not significantly different (WT, 523 ± 14 nm, 246 ± 10 nm, 114 ± 4 nm, n = 4; Syngap+/−, 536 ± 11 nm, 276 ± 19 nm, 106 ± 1 nm, n = 6, for head width, neck length, and neck width, respectively). E, To predict the impact of these spine morphology changes on diffusional coupling, a morphological compartmentalization factor was calculated. The cumulative frequency distributions of the compartmentalization factor significantly differ between genotypes (Kolmogorov–Smirnov test, p < 0.0001), and this is also reflected in the mean values for each genotype (two-tailed independent t test with Welch's correction, p = 0.04; WT, 3.16 ± 0.30 μm2; Syngap+/−, 4.35 ± 0.37 μm2). n.s., Not significant; *p < 0.05; **p < 0.01.
To better understand the potential functional impact of these nanoscale morphological alterations on synapse function, we calculated the compartmentalization factor (defined as VL/A, where V is the head volume, L is the neck length, and A is the cross-sectional area of the spine neck), which serves as a measure of the degree of biochemical compartmentalization of spine synapses (Tønnesen et al., 2014). We found that the compartmentalization factor was substantially increased in Syngap+/− animals (Fig. 6E; Syngap+/−, n = 424 spines; WT, n = 393 spines; compartmentalization factor, p = 0.0001, Kolmogorov–Smirnov test; mean compartmentalization factor, WT, 3.16 ± 0.30 μm2, n = 4 animals; Syngap+/−, 4.35 ± 0.37 μm2, n = 6 animals; p = 0.04, two-tailed independent t test with Welch's correction), indicating that SynGAP expression influences the biochemical coupling of spines to their parent dendrites. Interestingly, in Fmr1−/y mice, the compartmentalization factor was also previously found to be increased relative to WT control animals (Wijetunge et al., 2014).
Discussion
De novo mutations in the SYNGAP1 gene result in moderate to severe ID, often with co-occurring epilepsy and/or ASDs that appear to be as prevalent as FXS (Berryer et al., 2013). However, unlike FXS, little is known about the underlying mechanisms by which SynGAP reduction causes these alterations in brain function. We now show that SynGAP haploinsufficiency in mice mimics and/or occludes the deficits in synaptic structure and physiology in hippocampal CA1 pyramidal neurons associated with the loss of FMRP. Furthermore, pharmacological interventions that reduce mGlu5 receptor signaling and rescue much of the pathophysiology associated with the loss of FMRP also rescue the protein synthesis deficits associated with loss of a single copy of Syngap. Together, these data suggest that these two prevalent monogenic causes of ID/ASDs share common and/or overlapping cellular mechanisms.
This is the first demonstration that Syngap+/− mice show altered mGlu5 receptor signaling and altered downstream protein synthesis. When considered in conjunction with previous studies that showed an increase in ERK1/2 activation following NMDA receptor activation (Vazquez et al., 2004; Rumbaugh et al., 2006), a decrease in NMDA receptor-dependent in LTP (Komiyama et al., 2002; Kim et al., 2003), and a block of the induction of NMDA receptor-dependent LTD (Carlisle et al., 2008), these data suggest that SynGAP regulates two distinct glutamate receptor dependent forms of synaptic plasticity. Intriguingly, we showed previously that the Syngap gene gives rise to a range of isoforms that are differentially regulated by synaptic activity and that have opposing effects on synaptic function (McMahon et al., 2012). For example, overexpression of SynGAP Aα1 results in a decrease in mEPSC frequency and amplitude, whereas SynGAP Cα2 increases mEPSC frequency and amplitude. SynGAP isoforms also demonstrate distinct activity- and age-dependent patterns of expression (McMahon et al., 2012). Therefore, the loss of a single functional allele of SynGAP may result in an imbalance of SynGAP isoforms as well as decreasing overall SynGAP levels. It remains to be determined whether the deficits seen in Syngap+/− mice result from a reduction in total SynGAP levels or whether an imbalance of SynGAP isoform expression is responsible for the altered cellular phenotypes associated with SynGAP haploinsufficiency.
Our findings are in good agreement with a previous report using acute and selective knockdown of the SynGAP α1 isoform in cortical cultures showing an ERK1/2-dependent increase in dendritic protein synthesis (Wang et al., 2013). Our data extend these findings by demonstrating that constitutive reduction of all SynGAP isoforms, a model that more closely reflects SYNGAP1 haploinsufficiency in humans (i.e., has excellent structural and face validity with the human condition), leads to an increase in ERK1/2 phosphorylation and basal protein synthesis. Furthermore, the latter can be corrected by inhibitors of the ERK1/2 signaling pathway. Importantly an inhibitor of ERK1/2 activator MEK1/2 (U0126) restored protein synthesis to wild-type levels, despite a dramatic reduction in ERK1/2 phosphorylation, indicating that only the exaggerated protein synthesis is regulated by ERK1/2. However, it seems likely that the dramatic reduction in ERK1/2 signaling could dramatically alter neuronal function and plasticity. In this respect, the finding that lovastatin restored WT levels of ERK1/2 phosphorylation and basal protein synthesis likely make it a far better candidate therapy for SYNGAP1 haploinsufficiency. Importantly, the selective effect on elevated protein synthesis in Syngap+/− mice is similar to that observed in Fmr1−/y mice, suggesting that dysregulation of mGlu5-ERK1/2-dependent protein synthesis may be a common feature of a subset of neurodevelopmental disorders (Osterweil et al., 2010, 2013; Tian et al., 2015). In summary, our findings suggest that targeting the exaggerated basal protein synthesis through modulation of the ERK1/2 pathway may provide a potential therapeutic strategy in the treatment of SYNGAP haploinsufficiency.
Using STED microscopy, we reveal nanoscale changes in spine morphology in Syngap+/− mice that indicate an increase in biochemical compartmentalization. These findings parallel those from the Fmr1−/y mice, where changes in spine neck and head parameters decrease the magnitude of the developmental change in biochemical compartmentalization that occurs in WT animals (Wijetunge et al., 2014). As we suggested previously for the Fmr1−/y mice (Wijetunge et al., 2014), we deem it unlikely that these subtle changes in spine morphology can, by themselves, fully account for the alterations in synaptic function and plasticity.
Previous findings from SynGAP-deficient mice suggest that changes in synaptic function are mirrored by changes in synaptic structure (Vazquez et al., 2004; Carlisle et al., 2008; Clement et al., 2012; but see McMahon et al. 2012). More specifically, SynGAP reduction resulted in an increase in dendritic spine head size (Vazquez et al., 2004; Carlisle et al., 2008; Clement et al., 2012). Our finding of no change in spine head width is at odds with those of Carlisle et al. (2008) and Clement et al. (2012), who examined hippocampal CA1 pyramidal neurons and hippocampal dentate gyrus cells, respectively. Although these differences may partly be due to differences in age and neuronal type that were examined [for example, Carlisle et al. (2008) examined animals up to 6 months of age, and we used animals up to 5 weeks of age], it is likely that methodological issues also explain some of the discrepancies. Both Clement et al. (2012) and Carlisle et al. (2008) categorized spines into “mushroom,” “thin,” and “stubby”; however, we demonstrated previously that spine shapes cover a wide continuum, and such categories are arbitrary (Tønnesen et al., 2014; Wijetunge et al., 2014). Carlisle et al. (2008) found small differences in the size of “mushroom” spines and no difference in “thin” or “stubby” spines. However, without such categorization, it is unclear whether the differences between genotype would have remained. Clement et al. (2012) found only modest increases in head diameter analyzed using cumulative distributions of spine head width. They also found an increase in the relative proportion of “mushroom” to “stubby” spines in Syngap+/− mice. However, the categorization of stubby spines may largely be a consequence of the limited resolution of regular confocal and two-photon microscopy (∼200 and 350 nm, respectively) to resolve short spine necks (Tønnesen et al., 2014). For these reasons, it is not appropriate to directly compare between these studies, and the apparent differences may be simply due to differences in methodological approach. Given the excellent resolution of the STED microscope (∼70 nm) and the good agreement of our findings of spine development with previous electron microscopic studies (Harris et al., 1992), we argue that our measurements are an accurate reflection of spine head morphology.
Previous studies in Fmr1−/y mice have found a preponderance of long thin spines and have correlated this with an increase in LTD (for review, see Wijetunge et al., 2013). We propose that our recent finding of an increase in spine head size (Wijetunge et al., 2014) is more suggestive of spines that have recently undergone potentiation and hence are pre-disposed to undergo LTD. Our findings that Syngap+/− mice have no change in head size but do show enhanced LTD indicate that these two parameters are not functionally interdependent. Notwithstanding, both models show an increase in biochemical compartmentalization, and future studies will be required to determine whether compartmentalization and excess LTD are causally linked. Finally, it should be noted that whereas an increase in head size in Fmr1−/y mice was generally thought to reflect a premature development of spines, we demonstrated previously (also using STED microscopy) that spine head size decreases with age in cortical layer 5 and hippocampal CA1 pyramidal neurons in WT mice. This implies that maintained large spine heads in Fmr1−/y mice actually reflects a developmental delay (Wijetunge et al., 2014).
In summary, our data demonstrate that two highly penetrant forms of ID/ASDs resulting from mutations in FMR1 and SYNGAP1 converge on common cellular processes. They are in agreement with studies of the neurofibromatosis 1 (NF1) gene, where mutations in NF1, encoding an RasGAP, also result in ID, and some of its associated cellular and behavioral phenotypes are alleviated by modulators of ERK1/2 signaling (Li et al., 2005). Furthermore, they support the hypothesis that alterations in synaptic plasticity resulting from dysregulation of new protein synthesis may be a common core pathophysiology underlying these disorders, and therapeutic strategies targeting modifiers of protein synthesis may be appropriate for treating genetically divergent forms of ID/ASD.
Footnotes
This work was supported by a Biotechnology and Biological Sciences Research Council studentship (S.A.B.), Medical Research Council Grants G0700967 (P.C.K.) and MR/K014137/1 (D.J.A.W.), the Patrick Wild Centre, the RS MacDonald Charitable Trust, a National Alliance for Research on Schizophrenia and Depression Young Investigator grant from the Brain and Behavior Research Foundation (L.S.W.), and France-BioImaging Grant ANR-10-INSB-04 (U.V.N.). We thank our colleagues in the Patrick Wild Centre for their constructive input during the course of this study.
The authors declare no competing financial interests.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License Creative Commons Attribution 4.0 International, which permits unrestricted use, distribution and reproduction in any medium provided that the original work is properly attributed.
References
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett MW, Watson RF, Vitalis T, Porter K, Komiyama NH, Stoney PN, Gillingwater TH, Grant SG, Kind PC. Synaptic Ras GTPase activating protein regulates pattern formation in the trigeminal system of mice. J Neurosci. 2006;26:1355–1365. doi: 10.1523/JNEUROSCI.3164-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryer MH, Hamdan FF, Klitten LL, Møller RS, Carmant L, Schwartzentruber J, Patry L, Dobrzeniecka S, Rochefort D, Neugnot-Cerioli M, Lacaille JC, Niu Z, Eng CM, Yang Y, Palardy S, Belhumeur C, Rouleau GA, Tommerup N, Immken L, Beauchamp MH, et al. Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum Mutat. 2013;34:385–394. doi: 10.1002/humu.22248. [DOI] [PubMed] [Google Scholar]
- Carlisle HJ, Manzerra P, Marcora E, Kennedy MB. SynGAP regulates steady-state and activity-dependent phosphorylation of cofilin. J Neurosci. 2008;28:13673–13683. doi: 10.1523/JNEUROSCI.4695-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, Almonte AG, Miller BH, Wiltgen BJ, Miller CA, Xu X, Rumbaugh G. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151:709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement JP, Ozkan ED, Aceti M, Miller CA, Rumbaugh G. SYNGAP1 links the maturation rate of excitatory synapses to the duration of critical-period synaptic plasticity. J Neurosci. 2013;33:10447–10452. doi: 10.1523/JNEUROSCI.0765-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SM, Daly CA, Bear MF, Huber KM. Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. J Neurosci. 2004;24:4859–4864. doi: 10.1523/JNEUROSCI.5407-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, Dobrzeniecka S, Côté M, Perreau-Linck E, Perreault-Linck E, Carmant L, D'Anjou G, Fombonne E, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Mouaffak F, Joober R, Mottron L, et al. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Daoud H, Piton A, Gauthier J, Dobrzeniecka S, Krebs MO, Joober R, Lacaille JC, Nadeau A, Milunsky JM, Wang Z, Carmant L, Mottron L, Beauchamp MH, Rouleau GA, Michaud JL. De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism. Biol Psychiatry. 2011;69:898–901. doi: 10.1016/j.biopsych.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Harlow EG, Till SM, Russell TA, Wijetunge LS, Kind P, Contractor A. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron. 2010;65:385–398. doi: 10.1016/j.neuron.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. SYNGAP: bridging the gap between genetic factors and autosomal non-syndromic mental retardation. Clin Genet. 2009;76:149–151. doi: 10.1111/j.1399-0004.2009.01247_3.x. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis–dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee HK, Takamiya K, Huganir RL. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J Neurosci. 2003;23:1119–1124. doi: 10.1523/JNEUROSCI.23-04-01119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O'Carroll CM, Martin SJ, Morris RG, O'Dell TJ, Grant SG. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, O'Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cui Y, Kushner SA, Brown RA, Jentsch JD, Frankland PW, Cannon TD, Silva AJ. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- McMahon AC, Barnett MW, O'Leary TS, Stoney PN, Collins MO, Papadia S, Choudhary JS, Komiyama NH, Grant SG, Hardingham GE, Wyllie DJ, Kind PC. SynGAP isoforms exert opposing effects on synaptic strength. Nat Commun. 2012;3:900. doi: 10.1038/ncomms1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, Jaeschke G, Bear MF, Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalon A, Bruns A, Risterucci C, Honer M, Ballard TM, Ozmen L, Jaeschke G, Wettstein JG, von Kienlin M, Künnecke B, Lindemann L. Chronic metabotropic glutamate receptor 5 inhibition corrects local alterations of brain activity and improves cognitive performance in fragile X mice. Biol Psychiatry. 2014;75:189–197. doi: 10.1016/j.biopsych.2013.05.038. [DOI] [PubMed] [Google Scholar]
- Nosyreva ED, Huber KM. Metabotropic receptor-dependent long-term depression persists in the absence of protein synthesis in the mouse model of fragile X syndrome. J Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil EK, Chuang SC, Chubykin AA, Sidorov M, Bianchi R, Wong RK, Bear MF. Lovastatin corrects excess protein synthesis and prevents epileptogenesis in a mouse model of fragile X syndrome. Neuron. 2013;77:243–250. doi: 10.1016/j.neuron.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan ED, Creson TK, Kramár EA, Rojas C, Seese RR, Babyan AH, Shi Y, Lucero R, Xu X, Noebels JL, Miller CA, Lynch G, Rumbaugh G. Reduced cognition in Syngap1 mutants is caused by isolated damage within developing forebrain excitatory neurons. Neuron. 2014;82:1317–1333. doi: 10.1016/j.neuron.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MJ, Irving AJ, Seabrook GR, Jane DE, Collingridge GL. The group I mGlu receptor agonist DHPG induces a novel form of LTD in the CA1 region of the hippocampus. Neuropharmacology. 1997;36:1517–1532. doi: 10.1016/S0028-3908(97)00181-0. [DOI] [PubMed] [Google Scholar]
- Porter K, Komiyama NH, Vitalis T, Kind PC, Grant SG. Differential expression of two NMDA receptor interacting proteins, PSD-95 and SynGAP during mouse development. Eur J Neurosci. 2005;21:351–362. doi: 10.1111/j.1460-9568.2005.03874.x. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc Natl Acad Sci U S A. 2006;103:4344–4351. doi: 10.1073/pnas.0600084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Kilpatrick IC, Collingridge GL. An investigation into signal transduction mechanisms involved in DHPG-induced LTD in the CA1 region of the hippocampus. Neuropharmacology. 1999;38:1585–1596. doi: 10.1016/S0028-3908(99)00062-3. [DOI] [PubMed] [Google Scholar]
- Tian D, Stoppel LJ, Heynen AJ, Lindemann L, Jaeschke G, Mills AA, Bear MF. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat Neurosci. 2015;18:182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till SM, Wijetunge LS, Seidel VG, Harlow E, Wright AK, Bagni C, Contractor A, Gillingwater TH, Kind PC. Altered maturation of the primary somatosensory cortex in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:2143–2156. doi: 10.1093/hmg/dds030. [DOI] [PubMed] [Google Scholar]
- Till SM, Asiminas A, Jackson AD, Katsanevaki D, Barnes SA, Osterweil EK, Bear MF, Chattarji S, Wood E, Wyllie DJ, Kind PC. Conserved hippocampal cellular pathophysiology but distinct behavioral deficits in a new rat model of FXS. Hum Mol Genet. 2015;24:5977–5984. doi: 10.1093/hmg/ddv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønnesen J, Katona G, Rózsa B, Nägerl UV. Spine neck plasticity regulates compartmentalization of synapses. Nat Neurosci. 2014;17:678–685. doi: 10.1038/nn.3682. [DOI] [PubMed] [Google Scholar]
- Vazquez LE, Chen HJ, Sokolova I, Knuesel I, Kennedy MB. SynGAP regulates spine formation. J Neurosci. 2004;24:8862–8872. doi: 10.1523/JNEUROSCI.3213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Held RG, Hall BJ. SynGAP regulates protein synthesis and homeostatic synaptic plasticity in developing cortical networks. PLoS One. 2013;8:e83941. doi: 10.1371/journal.pone.0083941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijetunge LS, Chattarji S, Wyllie DJ, Kind PC. Fragile X syndrome: from targets to treatments. Neuropharmacology. 2013;68:83–96. doi: 10.1016/j.neuropharm.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Wijetunge LS, Angibaud J, Frick A, Kind PC, Nägerl UV. Stimulated emission depletion (STED) microscopy reveals nanoscale defects in the developmental trajectory of dendritic spine morphogenesis in a mouse model of fragile X syndrome. J Neurosci. 2014;34:6405–6412. doi: 10.1523/JNEUROSCI.5302-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]