Abstract
Activation of hepatic stellate cells (HSCs) by transforming growth factor-β1 (TGF-β1) initiates HBV-associated fibrogenesis. The mechanism of TGF-β1 modulating HSC activation is not fully uncovered. We hypothesized a positive feedback signaling loop of TGF-β1-CD147 promoting liver fibrogenesis by activation of HSCs. Human HSC cell line LX-2 and spontaneous liver fibrosis model derived from HBV transgenic mice were used to evaluate the activation of molecules in the signaling loop. Wound healing and cell contraction assay were performed to detect the CD147-overexpressed HSC migration and contraction. The transcriptional regulation of CD147 by TGF-β1/Smad4 was determined using dual-luciferase reporter assay and chromatin immunoprecipitation. We found that a positive reciprocal regulation between TGF-β1 and CD147 mediated HSC activation. CD147 over-expression promoted HSC migration and accelerated TGF-β1-induced cell contraction. Phosphorylation of Smad2 and Smad3 in cooperation with Smad4 mediated the TGF-β1-regulated CD147 expression. Smad4 activated the transcription by direct interaction with CD147 promoter. Meanwhile, CD147 modulated the activated phenotype of HSCs through the ERK1/2 and Sp1 which up-regulated α-SMA, collagen I, and TGF-β1 synthesis. These findings indicate that TGF-β1-CD147 loop plays a key role in regulating the HSC activation and combination of TGF-β receptor inhibitor and anti-CD147 antibody might be promised to reverse fibrogenesis.
Liver fibrosis results from chronic liver injury during a long-term wound-healing response, which causes increasing excessive accumulation of extracellular matrix (ECM) proteins and eventually leads to fibrogenesis and later cirrhosis1. The hepatic stellate cells (HSCs) are the main ECM-producing cells during this process, and they activate and differentiate from quiescent vitamin A-storing cells into proliferative myofibroblasts in response to fibrogenic liver injury. Activated HSCs express many ECM proteins including collagen type I, α-smooth muscle actin (α-SMA), transforming growth factor-β1 (TGF-β1), matrix metalloproteinase (MMP), and tissue inhibitors of metalloproteinases, which contributes to liver fibrosis2. Clinical studies suggest that hepatitis B virus (HBV) chronic infection is the most important cause of liver cirrhosis and hepatocellular carcinoma (HCC) in human patients3. TGF-β1 is considered a key mediator of liver fibrogenesis and detected in HBV-related liver fibrogenesis4,5. The TGF-β1 gene is transcriptionally activated by hepatitis B virus X protein (HBx) which is one of HBV encoded-proteins through the Egr-1 binding sites6.
Liver-damage-induced levels of active TGF-β1 mediate HSC transdifferentiation through the canonical Smad signaling pathway involving TGF-β receptor-mediated phosphorylation of Smad2 and Smad3 (p-Smad2/3) to enhance collagen synthesis7. The p-Smad2/3 form complexes with Smad4, which are translocated to the nucleus to regulate the transcription of certain genes. Putative target genes of Smad4 are screened by promoter-wide analysis in human epithelial cells8. However, the target genes transcriptionally regulated by Smad4 in HSCs are unknown.
Our previous study and others′ reveal that a glycosylated transmembrane protein, CD147 presents on HSCs9,10. CD147 expression in HSCs is elevated by TGF-β1 stimulation9, but the regulating mechanism is not uncovered. In this study, we hypothesized a direct role of TGF-β1 in the development of liver fibrosis by the activation of HSCs through TGF-β1-CD147 signaling loop. We here showed that TGF-β1 was released from hepatocytes which was transfected by HBx, and exerted on HSC activation by directly transcriptional regulation of CD147 through TGF-β1/Smad4 signaling pathway. Over-expression of CD147 was positively feedback on TGF-β1 expression via the ERK1/2/Sp1 transduction. The TGF-β1-CD147 loop contributed to HBV-associated liver fibrosis progression.
Results
A positive reciprocal regulation between TGF-β1 and CD147 in HSC activation
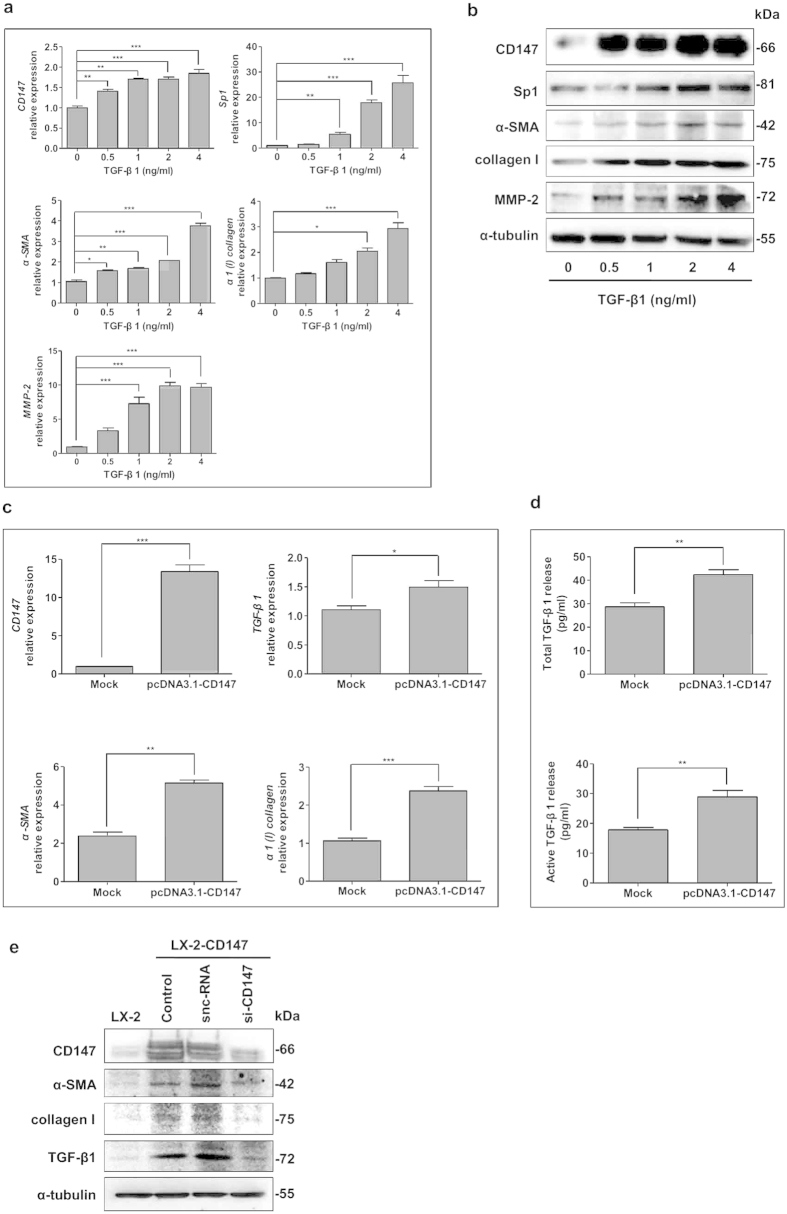
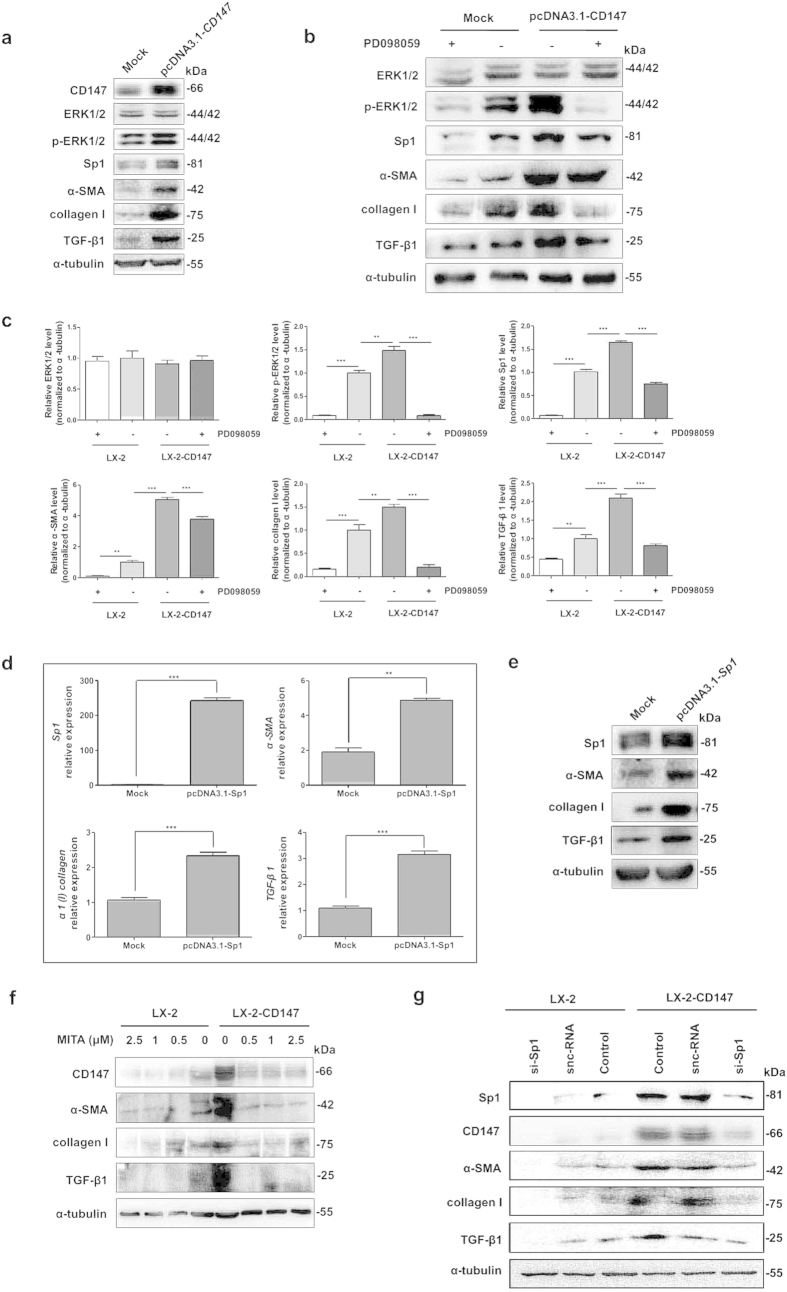
It is found that HSCs exposed to conditioned medium from HBx-expressing hepatocytes show increased expression of TGF-β111,12. We confirmed that the ectopic expression of HBx in L02 cells (named L02-HBx) significantly induced the elevation of total and active TGF-β1 levels compared with controls (Supplemental Fig. 1a,b). Strikingly, we observed that CD147 was significantly increased in LX-2 cells either incubation with L02-HBx conditioned medium or co-cultured with L02-HBx cells. This up-regulation was inhibited with a selective antagonist of TGF-β1 type I receptor SB431542 (Sigma, St Louis, MO, USA), which demonstrated that TGF-β1 signaling transduction was involved in CD147 expression by a paracrine way (Supplemental Fig. 1c). We then evaluated the levels of CD147 and fibrosis-related genes in response to different doses of TGF-β1 in LX-2 cells. The mRNA and protein levels of CD147, α-smooth muscle actin (α-SMA), α1(I) collagen, and MMP-2 were significantly up-regulated with TGF-β1 stimulation in dose-dependent manners. A transcription factor Sp1 was also markedly increased by TGF-β1 (Fig. 1a,b). Meanwhile, Real-time RT-PCR analysis showed that the transfection of CD147 gene in LX-2 cells induced the increased mRNA expressions of TGF-β1, α-SMA, and α1(I) collagen (Fig. 1c). Also, both total and active forms of TGF-β1 were up-regulated by CD147 over-expression as detected by enzyme-linked immunosorbent assay (ELISA) (Fig. 1d). As endogenous level of CD147 was very low in quiescent LX-2 cells, thus we generated a LX-2-CD147 cell line that stably expressed CD147 by puromycin selection. Then we knocked down CD147 by treating LX-2-CD147 cells with a small interference RNA specific to CD147 (si-CD147). As shown in Fig. 1e, the elevated expression of CD147 in LX-2-CD147 cells was depressed by si-CD147 efficiently, which was coupled with down-regulation of α-SMA, collagen I, and TGF-β1. Collectively, our data suggested that HBx was capable of inducing the secretion of TGF-β1 in hepatocytes that resulted in paracrine activation of HSCs by CD147-TGF-β1 positive feedback loop transduction.
Figure 1. A positive reciprocal regulation between TGF-β1 and CD147 in HSC activation.
(a) Real-time RT-PCR and (b) western blot analysis of CD147, Sp1, α-SMA, collagen I, and MMP-2 stimulated with various doses of TGF-β1 for 24 hours. (c) Real-time RT-PCR analysis of CD147, TGF-β1, α-SMA, and α1(I) collagen in LX-2 cells transiently transfected with plasmid pcDNA3.1-CD147 at 24 hours. (d) ELISA detection of total and active forms of TGF-β1 in supernatant of LX-2-transfected pcDNA3.1-CD147 cells. Cells were transfected with pcDNA3.1(+) as a mock control. (e) Western blot analysis of CD147, α-SMA, collagen I, and TGF-β1 in LX-2-CD147 cells treated with siRNA targeting CD147 (si-CD147). snc-RNA was used as a control. *P < 0.05, **P < 0.01, ***P < 0.001.
Over-expression of CD147 promoted HSC transdifferentiation
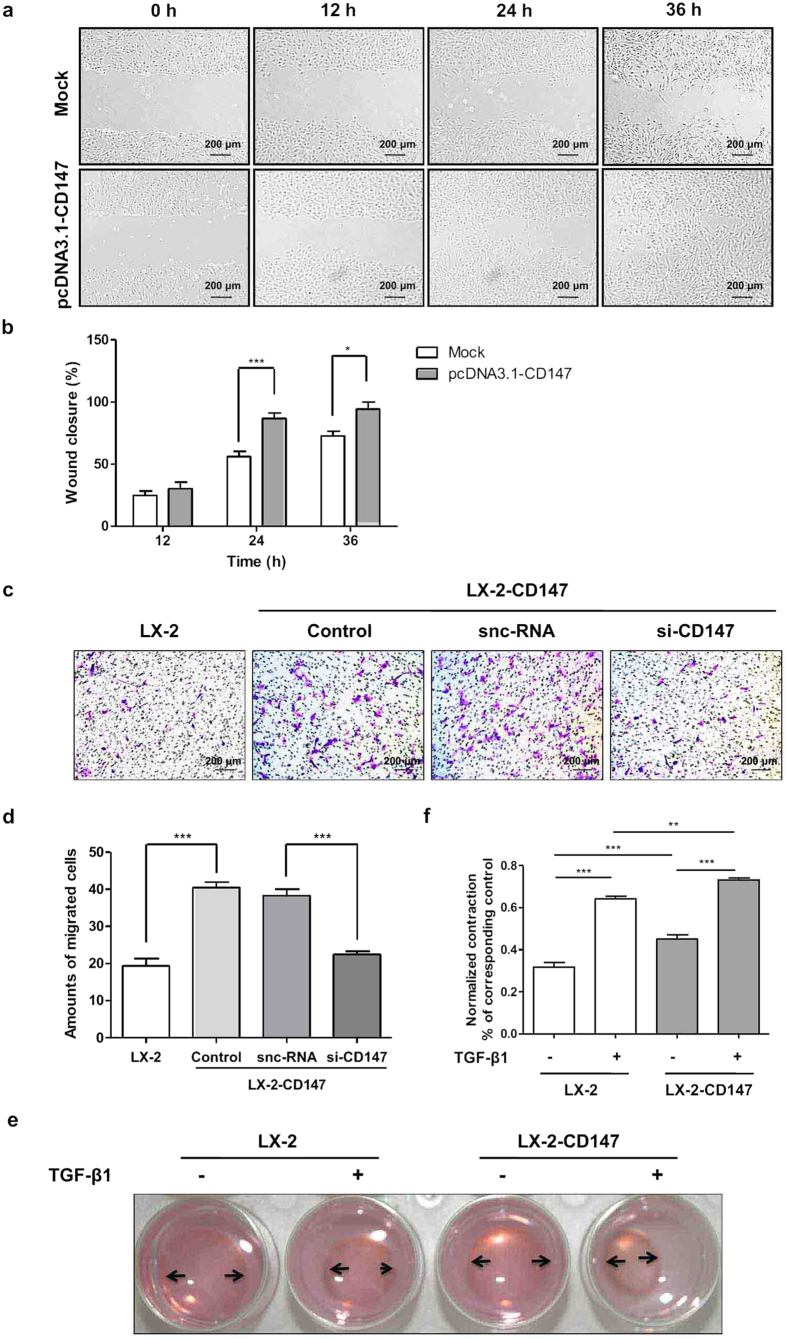
As shown in Fig. 2a,b, transient over-expression of CD147 promoted LX-2 cell migration as detected by wound healing assay. Moreover, the transwell migration assay confirmed that the migration of LX-2 cells was accelerated by CD147 up-regulation, whereas, knockdown of CD147 by si-CD147 significantly decreased the migration of LX-2-CD147 cells compared with that of silencer-negative control siRNA (snc-RNA, Fig. 2c,d). With LX-2-CD147 cell model, collagen-based cell contraction assay showed that CD147 promoted HSC contraction and even accelerated TGF-β1-induced cell contraction (Fig. 2e,f). These results indicated that CD147 cooperated with TGF-β1 in HSC transdifferentiation.
Figure 2. Over-expression of CD147 accelerated HSC migration and contraction.
(a) Representative phase contrast image and (b) quantitative analysis of wound healing assay in LX-2-transfected pcDNA3.1-CD147 cells and mock controls. (c) Representative phase contrast image and (d) quantitative analysis of transwell migration assay in LX-2 and LX-2-CD147 cells which were treated with siRNA targeting CD147 (si-CD147). snc-RNA was used as a control. (e) Representative phase contrast image and (f) quantitative analysis of collagen-based cell contraction in LX-2-CD147 and LX-2 cells with or without TGF-β1 stimulation for 24 hours. *P < 0.05, **P < 0.01, ***P < 0.001.
Smads signaling was involved in TGF-β1-induced CD147 expression
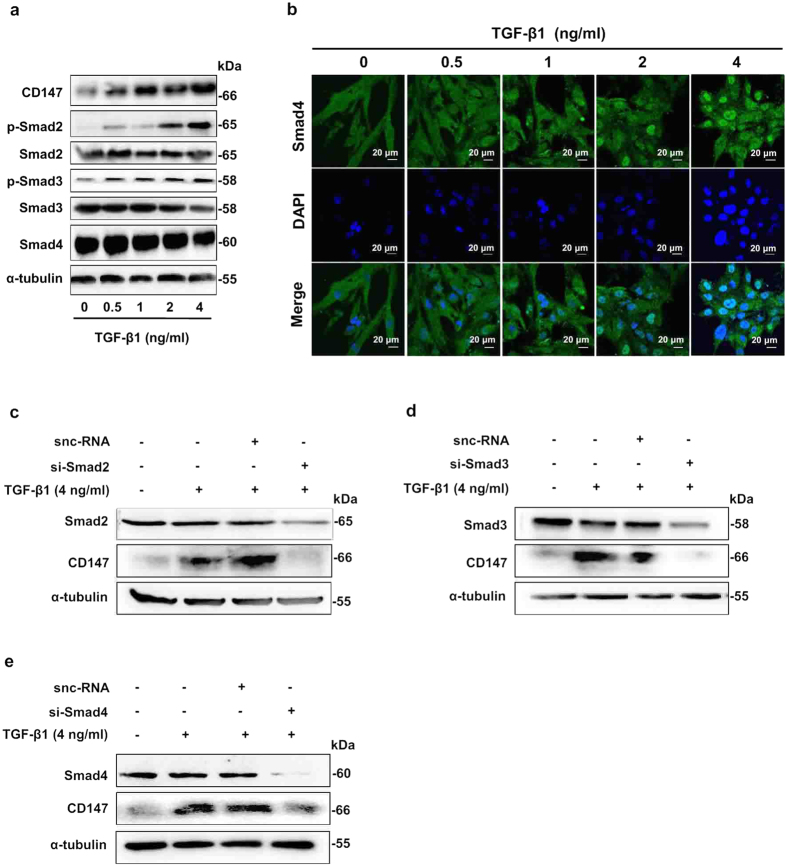
TGF-β1/Smads signaling is demonstrated to participate in the fibrogenic response by activating HSCs5,7. Therefore, we determined whether this canonical Smad-dependent pathway also had a role in TGF-β1-driven CD147 expression. As shown in Fig. 3a, after treated with TGF-β1, the protein levels of Smad2, Smad3, and Smad4 showed no changes in LX-2 cells. However the phosphorylations of Smad2 (p-Smad2) and Smad3 (p-Smad3) were elevated in a TGF-β1-dose-dependent manner. Nuclear translocation of Smad4 and its direct binding to target gene promoter are necessary for TGF-β1 signaling. To understand the molecular mechanism of TGF-β1 regulation of CD147 expression, we determined whether Smad4 translocated to the nucleus from cytoplasm with immunofluorescence. As shown in Fig. 3b, Smad4 was distributed in the cytoplasm of LX-2 cells. However, it aggregated to nucleus in the presence of TGF-β1. In this process, CD147 expression was up-regulated significantly in TGF-β1-treated cells compared with that in control cells (Fig. 3a). Then specific small interfering RNAs targeting Smad2 (si-Smad2), Smad3 (si-Smad3), and Smad4 (si-Smad4) were synthesized to validate the Smad proteins regulating CD147 expression. When LX-2 cells were treated with si-Smad2, si-Smad3, and si-Smad4 respectively, the corresponding Smad expressions were silenced, coupled with the fact that the TGF-β1-upregulated CD147 was depressed (Fig. 3c–e). Collectively, these findings suggested that TGF-β1 induced expression of CD147 in HSCs through activation of Smad signaling by the cooperation of Smad2, Smad3, and Smad4.
Figure 3. Smads signaling was involved in TGF-β1-induced CD147 expression.
(a) Western blot analysis of CD147, p-Smad2, Smad2, p-Smad3, Smad3, and Smad4 expressions in LX-2 cells stimulated with various doses of TGF-β1 for 24 hours. (b) Immunofluorescent staining of Smad4 in LX-2 cells stimulated with TGF-β1. Cell nuclei were stained with DAPI. Western blot analysis of CD147 expression in LX-2 cells which were transfected with siRNA targeting Smad2 (c), Smad3 (d), and Smad4 (e) in the presence of 4 ng/ml TGF-β1 for 24 hours. snc-RNA was used as control.
Specific binding of Smad4 to the CD147 promoter
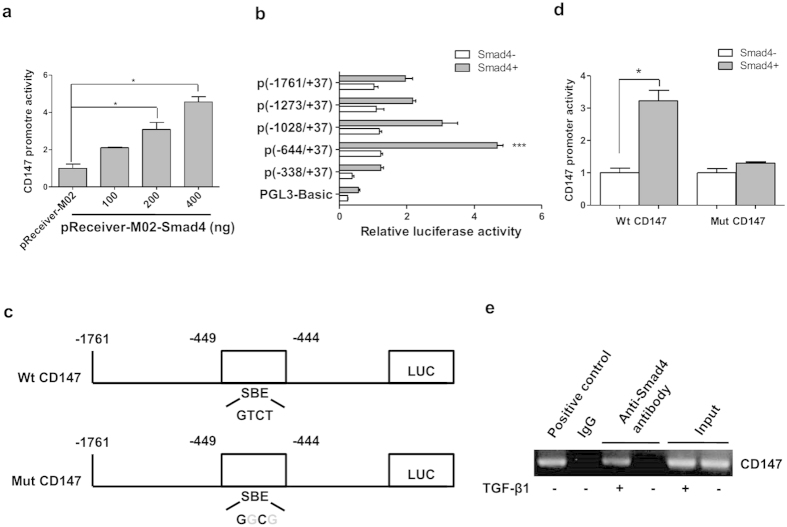
Previous reports have revealed an indispensable role of Smad4 in TGF-β1-induced expression of a subset of target genes, and it can directly bind to an 8-bp palindromic sequence in the promoter which is called Smad-binding element (SBE)13. We thus investigated the transcriptional regulation of Smad4 on CD147 expression using a dual-luciferase reporter assay system. As shown in Fig. 4a, co-transfection of pReceiver-M02-Smad4 and pGL3-Basic vector containing Luc gene under the control of human CD147 promoter (−1761/+37) fragment into LX-2 cells led to a Smad4 dose-dependent increase in luciferase activity. This result indicates that Smad4 has an inducible effect on the activity of the CD147 promoter, which directly regulates the transcriptional expression of CD147.
Figure 4. Specific binding of Smad4 to CD147 promoter.
(a) LX-2 cells were co-tranfected with pGL3-Basic vector containing human CD147 promoter (−1761/+37) and pReceiver-M02-Smad4, CD147 promoter activity was detected by dual-luciferase reporter assay. (b) LX-2 cells were co-transfected with different length of CD147 promoters and pReceiver-M02-Smad4, CD147 promoter activity was detected by dual-luciferase reporter assay. (c) Schematic representation of wild type (Wt) and mutant (Mut) CD147 promoter regions. (d) LX-2 cells were co-tranfected with Wt CD147 or Mut CD147 promoters and pReceiver-M02-Smad4, CD147 promoter activity was detected by dual-luciferase reporter assay. (e) LX-2 cells were treated with TGF-β1 and subjected to ChIP assay. Positive control was anti-RNA polymerase. Two percent of the lysate was used as the input control. *P < 0.05.
Next, we developed a series of deletion constructs to identify the minimal promoter required for CD147 activation. We found that the most critical region for transcriptional activity of the CD147 promoter was located in positions between −338 and −644 which involved SBE sequence (Fig. 4b). To further identify the CD147 promoter core region, we then generated a Mut CD147 construct that contained mutations in the SBE domain by site-directed mutagenesis (Fig. 4c). We found that the promoter activity was completely abolished upon mutation of the Smad4-binding site in SBE, suggesting that SBE is required for activation of the CD147 promoter by Smad4 (Fig. 4d). To show direct binding of Smad4 to the putative binding site within the critical CD147 promoter region, chromatin immunoprecipitation assay (ChIP) was performed in LX-2 cells. The PCR-amplified fragment of the CD147 promoter region containing the SBE was retrieved from the immunoprecipitates using an anti-Smad4 antibody, whereas the control fragment could not be (Fig. 4e). These results showed that Smad4 activated CD147 transcription by direct interaction with the SBE domain that was located in the proximal promoter of the CD147 gene.
ERK1/2 and downstream Sp1 mediated CD147-induced TGF-β1, collagen I, and α-SMA expressions
The transient introduction of CD147 strongly induced profibrotic factors such as α-SMA, α1(I) collagen, and TGF-β1 expressions as shown in Fig. 5a. Meanwhile, the levels of phospho-ERK1/2 (p-ERK1/2) and Sp1 in LX-2-CD147 cells were increased compared with control (Fig. 5a). The CD147-upregulated profibrotic factors and Sp1 could be significantly inhibited by the selective ERK1/2 inhibitor PD098059 (Sigma) (Fig. 5b,c). All of these results indicated the crucial involvement of activated MAPK signaling in CD147-dependent TGF-β1 and collagen I production. To clarify the role of Sp1 in CD147-ERK1/2-dependent TGF-β1 and collagen I expression in HSCs, the Sp1 gene was transiently transfected into LX-2 cells followed by detection of fibrosis-related factors. As shown in Fig. 5d,e, over-expression of Sp1 transcription factor significantly induced the α-SMA, α1(I) collagen, and TGF-β1 mRNA and protein up-regulation.
Figure 5. ERK1/2 and Sp1 mediated CD147-induced TGF-β1 and collagen I expressions.
(a) Western blot analysis of CD147, ERK1/2, p-ERK1/2, Sp1, α-SMA, collagen I, and TGF-β1 in LX-2 cells transfected with pcDNA3.1-CD147, and (b) pre-incubated with 1 nM PD098059. (c) Western blot scanning densitometry for three independent experiments was analyzed with ImageJ software. Blots were probed for α-tubulin to ensure equal protein loading. (d) Real-time RT-PCR and (e) western blot analysis of molecules in LX-2 cells transfected with pcDNA3.1-Sp1. Cells were transfected with pcDNA3.1(+) as mock control. (f) Western blot analysis of molecules in LX-2-CD147 and LX-2 cells pre-incubated with mithramycin A or (g) treated with siRNA targeting Sp1 (si-Sp1). snc-RNA was used as a control. **P < 0.01, ***P < 0.001.
We next wondered whether the feedback effect of CD147 on TGF-β1 expression through Sp1. The treatment of mithramycin A (MITA, from Sigma), a drug known to modify GC-rich regions of the DNA and inhibit Sp1 binding down-regulated the expressions of CD147, α-SMA, α1(I) collagen, and TGF-β1 in both LX-2 and LX-2-CD147 cell lines (Fig. 5f). The RNA interference to knockdown Sp1 expression repressed both basal and CD147-induced α-SMA, α1(I) collagen, and TGF-β1 levels (Fig. 5g). These results suggested that ERK1/2 and downstream Sp1 mediated CD147-induced α-SMA, α1(I) collagen, and TGF-β1 expressions.
TGF-β1-CD147 positive feedback loop was activated in liver fibrosis
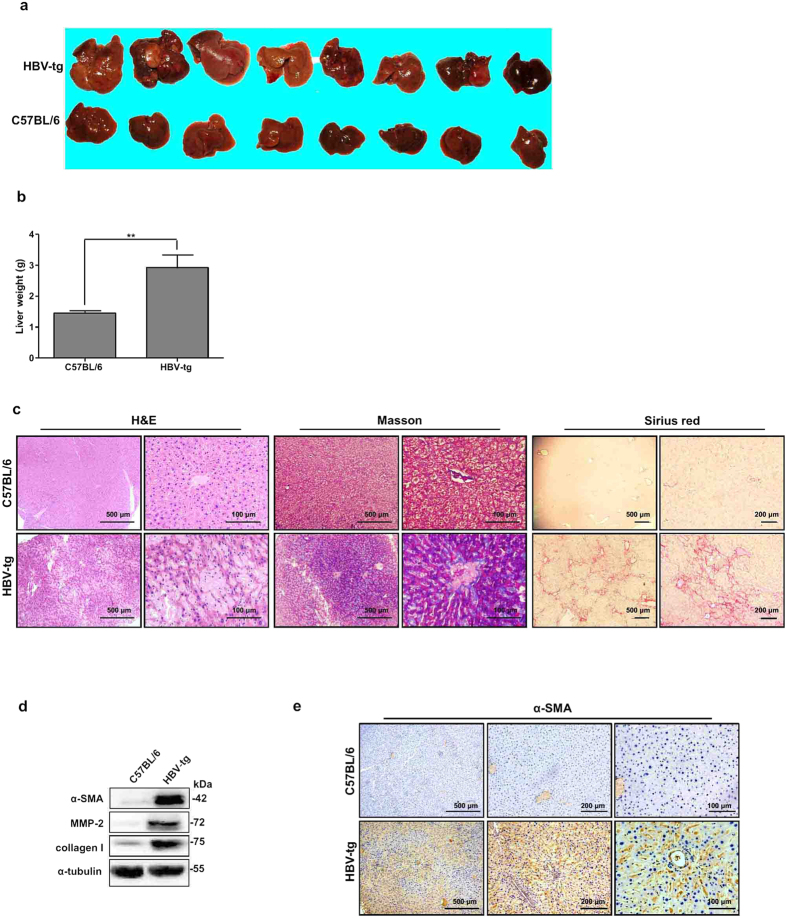
Spontaneous liver fibrosis is previously reported in HBV transgenic (HBV-tg) mice C57BL/6J-Tg(Alb1HBV)44Bri/J14. We used this strain of HBV-tg mice for assessing CD147-TGF-β1 positive feedback loop. It was found that the livers from 12-month-old HBV-tg mice appeared irregular shape and noticeable regenerative nodules with significantly higher weight compared to C57BL/6 control mice (Fig. 6a,b). H&E staining of liver tissues from 6-month-old C57BL/6 mice revealed normal cellular architecture, whereas liver tissues from HBV-tg mice demonstrated intense polymorphonuclear leukocyte and macrophage infiltration with severe centrilobular necrosis (Fig. 6c). Masson′s trichrome staining of the liver was performed to assess the collagen distribution and showed that extensive collagen deposition and pseudolobular formation was evident in liver tissues from HBV-tg mice compared with normal control, which was confirmed by Sirius red staining (Fig. 6c). Accordingly, western blotting and/or immunohistochemistry showed that α-SMA, MMP-2, and collagen I were remarkably increased in livers from HBV-tg mice (Fig. 6d,e). HBx was uniquely expressed (Supplemental Fig. 2a,b). These results verified that liver fibrosis was spontaneously developed in HBV-tg mice.
Figure 6. Spontaneous liver fibrosis in HBV-tg mice.
(a) Morphology of liver and (b) liver weight of 12-month-old C57BL/6 and HBV-tg mice (n = 8). **P < 0.01. (c) H&E, Masson′s trichrome, and Sirius red stainings of liver tissues from 6-month-old C57BL/6 and HBV-tg mice. (d) Western blot analysis of α-SMA, MMP-2, and collagen I expressions in liver tissues from 6-month-old C57BL/6 and HBV-tg mice. (e) Imm-unohistochemistry detected α-SMA expression in liver tissues from 6-month-old C57BL/6 and HBV-tg mice.
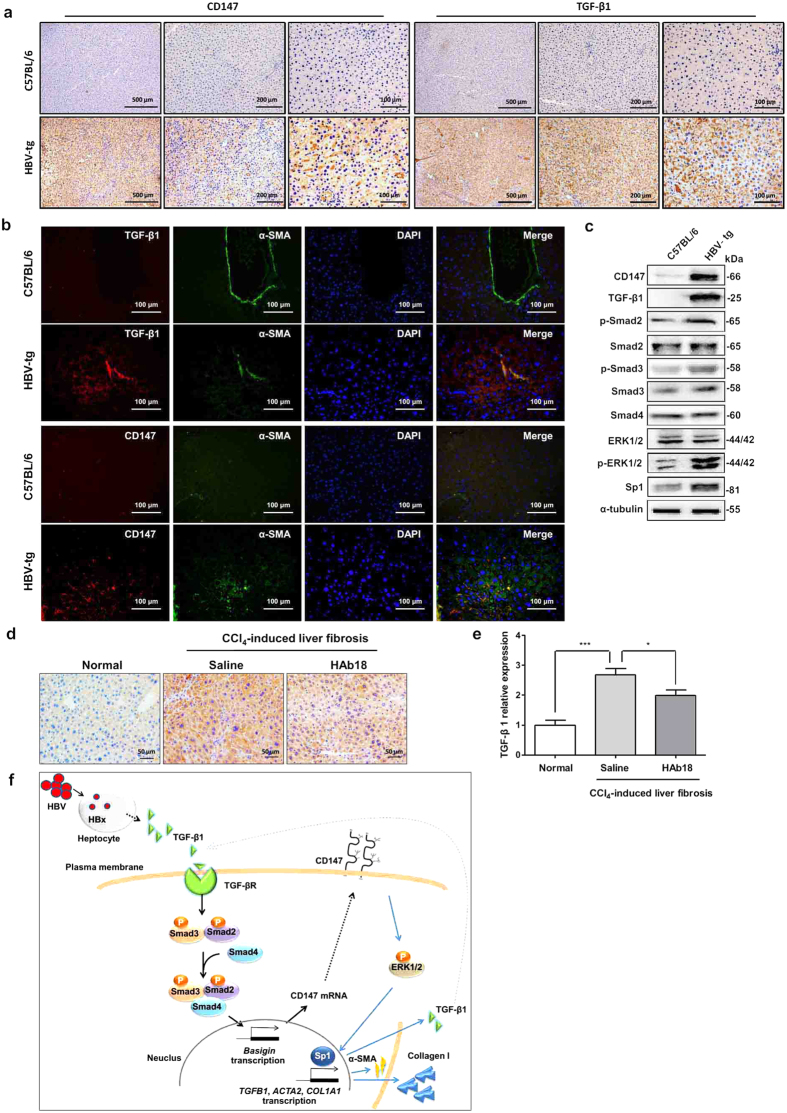
We then checked the expression of key molecules of TGF-β1-CD147 positive feedback loop in liver tissues. Immunohistochemistry analysis displayed a stronger staining of CD147 and TGF-β1 in liver tissues from 6-month-old HBV-tg mice, whereas rare staining of both molecules was observed in control mice (Fig. 7a). Double immunofluorescence detected that both TGF-β1 and CD147 co-localized with α-SMA-positive cells in fibrotic liver tissues, respectively, which suggested that TGF-β1-CD147 signaling in activated HSCs was involved in HBV-related liver fibrosis (Fig. 7b). The activation of all the key molecules (i.e. CD147, TGF-β1, p-Smad2, p-Smad3, p-ERK1/2, and Sp1) involved in the regulation of TGF-β1-CD147 positive feedback loop were elevated (Fig. 7c). In our previous work, we demonstrate that liver fibrosis induced by carbon tetrachloride (CCl4) is reversed by a monoclonal antibody (HAb18) against CD1479. A recent work by other group also shows the attenuation of liver fibrosis progression by blocking CD147 with a specific antibody15. To prove the positive feedback from CD147 to TGF-β1, TGF-β1 expression was then detected in CCl4-induced mouse liver tissues after treatment of HAb18 antibody. Immunohistochemistry showed that the increased TGF-β1 expression in CCl4-induced mouse liver tissue was suppressed by HAb18 antibody (Fig. 7d). The results were verified by real-time PCR analysis that TGF-β1 mRNA expression was significantly decreased in HAb18-treated group compared with saline group (Fig. 7e). Finally, a schematic model of the positive feedback loop was depicted in Fig. 7f.
Figure 7. Up-regulation of key molecules of TGF-β1-CD147 positive feedback loop in liver fibrosis mice.
(a) Immunohistochemistry analysis of CD147 and TGF-β1 expressions in liver tissues from 6-month-old C57BL/6 and HBV-tg mice. (b) Double immunofluorescence of TGF-β1 (red), CD147 (red), and α-SMA (green) in liver tissues from 6-month-old C57BL/6 and HBV-tg mice. Cell nuclei were stained with DAPI. (c) Western blot analysis of CD147, TGF-β1, p-Smad2, Smad2, p-Smad3, Smad3, Smad4, ERK1/2, p-ERK1/2, and Sp1 in liver tissues from 6-month-old C57BL/6 and HBV-tg mice. (d) Immunohistochemistry and (e) real-time RT-PCR analysis of TGF-β1 expression in liver tissues from CCl4-induced mice after treatment with CD147-specific antibody HAb18. n = 7. (f) Schematic model of TGF-β1-CD147 positive feedback loop in HSC activation.
Discussion
Chronic infection by HBV is a major cause of liver fibrosis and eventually leads to the development of cirrhosis and HCC3. Liver injury by HBV infection releases pro-inflammatory factors, which modulates HSC transdifferentiation contributing to the development of liver fibrosis16. HBV encoded-protein, HBx has been reported to induce TGF-β secretion in hepatocytes and subsequently to active HSCs by a paracrine way11. We previously find that over-expression of a transmembrane protein CD147 in HSC promotes up-regulation of fibrosis-related genes, including α-SMA, collagen I, and TGF-β1. CD147 is highly expressed in human HBV-related liver cirrhotic tissues and correlated with the Child-Pugh grade. TGF-β1 can also upregulate CD147 expression9. Based on the initial studies, it is rationale to assume HBx plays an important role by activation of HSCs through paracrine TGF-β1-CD147 signaling loop in HBV-related liver fibrosis.
TGF-β1/Smads signaling pathway has been reported to play a crucial role in liver fibrosis through induced deposition of ECM, especially type I collagen and secretion of fibrogenic cytokines in hepatocytes and HSCs5. However, the specific mechanisms for TGF-β1/Smads stimulating transcription of type I collagen gene in HSC are much unknown. Although the DNA-binding activity of Smad3 and Smad4 is weak and they usually associate with other DNA-binding transcription factors for target gene transcription, Smad4 directly participating the transcriptional regulation is disclosed in epithelial cells8,17. In the study, we found that the presence of Smad2, Smad3, and Smad4 was necessary for CD147 expression, and a Smad4-binding DNA element was located in the human basigin gene promoter for CD147 transcription in HSC cells.
Earlier studies show Sp1/3 independent or cooperative with Smad3/4 or Smad2/4 for α2(I) collagen (COL1A2) and α1(I) collagen (COL1A1) transcription18,19,20. Sp1 and AP-1 form a protein-DNA transcriptional regulatory complex for transcriptionally activating TGF-β1 (gene TGFB1) promoter in hepatitis C virus-infected hepatoma cells21. It is reported that the transcriptional activity of Sp1 in TGF-β1 regulation can be repressed with transcription factor Nrf2 by interacting with c-Jun22. Sp1/3 is also required for α-SMA (gene ACTA2) enhancer activation in response to TGF-β1 stimulation myofibroblast maturation23. We here demonstrated that the CD147 up-regulation by TGF-β1/Smad4 mediated Sp1 in induction of α1(I) collagen, TGF-β1, and α-SMA in human HSCs. As an upstream factor, ERK1/2 can modulate Sp1 activation and mediates TGF-β1 regulation for target genes24. As TGF-β1 can also up-regulate and modulate Sp1 expression and activation25 which was also proved in our study, we here showed CD147 as an upstream molecule to induce ERK1/2 phosphorylation, which resolved the signaling context between TGF-β1 and Sp1.
As integrin is important for HSC transdifferentiation and an interaction between CD147 and integrin is shown by our previous study26,27, CD147 mediated ERK1/2 activation probably by directly binding with integrin in HSCs. Taken together, the activation of TGF-β1-CD147 signaling loop was involved with Smads and ERK/Sp1 transduction to induce α-SMA and type I collagen over-production, which favored activated phenotypes of HSCs, such as enhanced wound closure, cell contraction, and cell proliferation9.
HBV transgenic mouse models are widely used for study of HCC development28,29. A strain of HBV transgenic mouse C57BL/6J-Tg(Alb1HBV)44Bri/J is also observed the development of liver fibrosis14,30, which was verified in our study. The HBV-tg mice used in this study contain sequence encoding the HBV large envelope polypeptide under the transcriptional control of the mouse albumin promoter. They also contain the HBV enhancer element and the HBV X open reading frame and its putative promoter sequences. Despite the donating investigators report a major decrease in the steady-state level of HBV envelope-specific transcripts and no HBV X-specific transcripts in tumors by Northern blot analysis31, we detected a high expression level of HBx by western blot and immunohistochemistry analysis in non-tumoral liver tissues from 6-month-old mice. Currently, a specific marker for the identification of activated HSCs is unavailable. However, α-SMA-positive HSCs can be distinguished from the other portal, interface, and septal myofibroblasts by their specific position in the liver perisinusoidal space32. In our previous work9, we identify the co-localization of CD147 molecule with α-SMA-positive cells in liver perisinusoidal space both in CCl4-induced fibrotic liver and human cirrhotic liver tissues. In our recently published work33, a significantly positive correlation is observed between CD147 and α-SMA (r = 0.8857, P = 0.0333) at N-diethylnitrosamine/phenobarbital initiation and early stage of liver tumor formation. As a mesenchymal phenotype marker, α-SMA is also expressed in transformed hepatocytes during the process of epithelial-mesenchymal transition34. Our previous study finds that α-SMA is detected in the cytoplasm of hyperplastic hepatocytes in CCl4-induced mouse fibrotic liver sections35. In the spontaneous liver fibrosis HBV-tg model, we also observed the positive staining of α-SMA in hepatocytes as well as the cells in perisinusoidal space in liver tissues (Fig. 7b, green fluorescence). The merge of CD147-positive red fluorescence with α-SMA-positive green fluorescence in perisinusoidal space was obviously presented, which indicated that CD147 was over-expressed in activated HSCs in HBV-tg mouse liver.
In summary, we here demonstrate that TGF-β1 released from HBx-expressing hepatocytes promotes the activation of HSCs through up-regulating CD147 by TGF-β1/Smads signaling in the development of liver fibrosis. As a bridge molecule, CD147 links TGF-β1 and Sp1 signaling transduction for transcription of collagen I, α-SMA, and TGF-β1 itself, therefore a positive feedback between TGF-β1 and CD147 is proposed in HSC transdifferentiation. Our findings provide new insights into HBV-mediated fibrogenesis. A new strategy using combination of TGF-β receptor inhibitor and anti-CD147 antibody may be promised to reverse the liver fibrosis in future.
Materials and Methods
Cell culture
Human HSC cell line, LX-2 and human normal liver cell line, L02 were cultured at 37 °C in a humidified atmosphere containing 5% CO2 with Dulbecco′s Modified Eagle Medium (DMEM) (Hycolon, Logan, USA) containing 10% fetal bovine serum, 100 U/ml of penicillin, and 100 mg/ml of streptomycin. The L02-HBx cell line was established by stable transfection of eukaryotic expression plasmid pcDNA3.1-HBx with hygromycin B selection. LX-2-CD147 cell line was established by infecting lentivirus-mediated human CD147 cDNA into LX-2 cells with puromycin selection for stable transfection.
Western blot
Western blot was performed as previously described36. The primary antibodies used were mouse-anti human CD147 antibody (1:1000) prepared by our laboratory37, rat anti-mouse CD147 (1:500), anti-HBx (1:1000), anti-α-SMA (1:1000), anti-α1(I) collagen (1:1000) (Abcam, Cambridge, UK), anti-Smad2 (1:500), anti-phospho-Smad2 (1:500), anti-ERK1/2 (1:500), anti-phospho-ERK1/2 (1:500) (Cell Signaling Technology, Danvers, USA), anti-MMP-2 (1:500), anti-Sp1 (1:500), anti-Smad3 (1:500), anti-phospho-Smad3 (1:500) (Epitomic, California, USA), anti-Smad4 (1:1000) (Santa Cruz Biotechnology, Santa Cruz, USA), anti-TGF-β1 (1:200), and anti-α-tubulin (1:200) (Santa Cruz Biotechnology). Secondary antibodies included horseradish peroxidase-conjugated goat anti-mouse IgG(H+L) (1:3000), goat anti-rat IgG(H+L) (1:2000) and sheep anti-rabbit IgG(H+L) (1:2000) (Pierce Biotechnology, Rockford, USA). The Western-Light chemiluminescent detection system (Image Station 4000 MM Pro, XLS180, Kodak, USA) was used to visualize the signals. The gray values were analyzed with ImageJ software.
Enzyme-linked immunosorbent assay (ELISA)
L02 and L02-HBx cells were plated in 6-well cell culture plates at a density of 1 × 106 cells per well for 24 h. Cells were washed and starved for 24 h with serum-free DMEM. Then conditioned medium was harvested and centrifuged at 3000 g for 5 min to remove cell debris and stored at −20 °C until use. Soluble TGF-β1 was performed using a TGF-β1 Emax® Immune Assay System ELISA kit (Peprotech, Madison, WI) according to the manufacturer′s instructions. The concentration in each sample well was determined by interpolation from a standard curve. Each sample was tested in duplicate.
Real-time RT-PCR
RNA was extracted with the Total RNA Kit II (Omega, Riverside, USA) and reverse transcribed into cDNA using a PrimeScript™ RT reagent kit (TaKaRaBio, Otsu, Japan). Single-stranded cDNA was amplified by quantitative RT-PCR using a SYBR Premix ExTaq™ kit (TaKaRaBio) on the Stratagene Mx3005P™ Real-Time PCR System (Agilent Technologies, Germany). Glyceraldehyde-3-phosphate dehydrogenase mRNA was used to normalize RNA inputs. RT reactions were performed at 42 °C for 60 min followed by 70 °C for 10 min. The qRT-PCR was conducted at 95 °C for 20 sec, followed by 40 cycles of 95 °C for 10 sec, 60 °C for 20 sec, and then 70 °C for 10 sec. The occurrence of primer dimers and secondary products was inspected using melting curve analysis. Our data indicated that the amplification was specific. There was only one PCR product amplified for individual set of primers. All primers were synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. and listed in Supplemental Table 1.
Wound healing assay
LX-2 cells were grown to 100% confluence on 18-mm-diameter dishes. The growth medium was removed, cells were washed and a wound was produced (0.5 mm). Cells were rinsed to remove non-adherent cells, and fresh medium with 1% fetal bovine serum was added. Digital images were taken at time points with an inverted microscope (CKX41; Olympus) equipped with digital camera.
Transwell migration assay
LX-2 and LX-2-CD147 cells were harvested and re-suspended to give a final concentration of 1.0 × 106/ml in serum-free DMEM. Cell suspensions (100 μl) were added to the upper compartment, and the filter chamber was inserted in a 24-well plate with DMEM containing 10% fetal bovine serum. The plate was incubated for 12 h at 37 °C in a 5% CO2 atmosphere. Then the filters were fixed with methanol for 10 min and stained with 0.5% methylrosaniline chloride for 20 min. The cells on the upper surface of the filters were removed with a cotton swab. The cells on the reverse side were counted under a microscope in five random fields at a magnification of ×100. Each assay was performed in triplicate.
Cell contraction assay
A standard kit assay was used (Cell Biolabs, San Diego, CA, USA). LX-2 cells were harvested and re-suspended in DMEM, two parts of cells were mixed with eight parts of collagen gel lattice mixture and plated for 1 h at 37 °C. After the gel was polymerized, 1 ml of medium was added and incubated for 2 days. Next the gels were released from the sides of wells, and 24 h later the images were taken. The changes of collagen gel size were analyzed using ImageJ software and normalized to areas of the well.
RNA Interference
Transfection of small interfering RNAs (siRNA) was performed using Lipofectamine 2000 (Invitrogen). All siRNA sequences were synthesized by Shanghai GenePharma Co, Ltd. and were listed in Supplemental Table 2. The snc-RNA was used as a negative control under similar conditions.
Immunofluorescence
Immunofluorescence was carried out as described previously36. Primary antibodies included rabbit anti-Smad4 (Santa Cruz Biotechnology), mouse anti-α-SMA (Abcam), rabbit anti-TGF-β1 (Biorbyt, Cambridge, UK), and rat anti-mouse CD147 antibodies (Abcam). Alexa Fluor 488-conjugated donkey anti-rabbit IgG(H+L) (Molecular probes), Alexa Fluor 488-conjugated goat anti-mouse IgG(H+L) Alexa Fluor 594-conjugated donkey anti-rabbit IgG(H+L), and Alexa Fluor 594-conjugate donkey anti-rat IgG(H+L) (Pierce, Rockford, USA) were used as secondary antibodies. The 4′,6-diamidino-2-phenylindole (DAPI) was used as a nuclear counterstain (Molecular probes, Eugene, USA). The co-localization expression was visualized under a confocal fluorescence microscopy (Nikon, Japan).
Immunohistochemistry
Liver tissues were fixed with 4% formalin and embedded in paraffin. Sections were deparaffinized and incubated with primary antibodies including anti-HBx, anti-α-SMA, anti-CD147 (Abcam), and anti-TGF-β1 (Biorbyt), followed by visualization with the Histostain®-Plus Kit (Invitrogen).
Collagen staining
Collagen accumulation was detected with Sirius red and Masson’s trichrome, which were performed as previously described38. Liver tissues were fixed with 4% formalin and embedded in paraffin. Sections were cut at a 5 μm thickness. Following deparaffinization and hydration, the sections were stained with Sirius red and Masson′s trichrome (Sigma, USA).
Dual-luciferase reporter assay
The 1659-base pair cDNA containing the coding region of human Smad4 (pReceiver-M02-Smad4) was purchased from GeneCopoeia TM (Rockville, MD, USA). A reporter construct plasmid P(−1761/+37), which contained a 1798-bp genomic DNA fragment spanning the 5′ upstream region of CD147, and a series of deletion constructs were generated by our laboratory39. To generate site-directed mutants of CD147 binding elements at −449 to −444, the QuikChange® Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was used. The incorporation of the mutations was verified by sequencing (Shanghai Sangon). The CD147 promoter plasmids containing the firefly luciferase reporter were co-transfected with pReceiver-M02-Smad4 and internal control, pRL-TK (Promega, Madison, WI, USA), using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, cells were detected for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega).
Chromatin immunoprecipitation (ChIP)
This was done essentially as described previously36. LX-2 cells were treated with 4 ng/ml of TGF-β1 for 24 h. ChIP was performed using the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore Bedford, MA, USA). Cell lysates were incubated with anti-Smad4 (R&D Systems, Minneapolis, USA) or IgG antibodies. The immunoprecipitated DNA was amplified by the promoter-specific primers: forward 5′-AGGTCACTTCCTCCCACC-3′; reverse 5′-TCTGAGTTAAACACGGGC-3′. The PCR products were analyzed on 1% agarose gel.
Mice
HBV transgenic mice C57BL/6J-Tg(Alb1HBV)44Bri/J, which contains HBV genome S, pre-S, and X domains under the mouse albumin promoter, were provided by The Jackson Laboratory (Bar Harbor, ME). C57BL/6 mice were purchased from VITALRIVER experiment animal company (Beijing, China). The establishment and evaluation of liver fibrosis mouse model and treatment with antibody against CD147 were reported in our previous study9. All experimental protocols were approved by Laboratory Animal Ethics Committee of Fourth Military Medical University and performed in strict accordance with the People′s Republic of China Legislation Regarding the Use and Care of Laboratory Animals.
Statistical analysis
Each experiment was repeated at least three times. Student′s t-test was used to compare the two mean values. A one-way analysis of variance was performed to compare the multiple mean values. Data were presented as the mean ± SD from three independent experiments unless otherwise indicated. The Graphpad Prism software and SPSS 17.0 software were used for statistical analysis.
Additional Information
How to cite this article: Li, H.-Y. et al. Activation of TGF-β1-CD147 positive feedback loop in hepatic stellate cells promotes liver fibrosis. Sci. Rep. 5, 16552; doi: 10.1038/srep16552 (2015).
Supplementary Material
Acknowledgments
We thank Prof. Scott L. Friedman (Mount Sinai School of Medicine, USA) for providing LX-2 cell line. This work was supported by grants from the National Natural Science Foundation of China (No. 81172144, No. 31571434) and the National Science and Technology Major Project (No. 2012ZX10002-015, No. 2012AA020806, No. 2015CB553701).
Footnotes
Author Contributions H.L. and D.J. designed the experiments, acquired and analyzed data, wrote the manuscript and prepared the figures; D.Z. and H.L. acquired parts of data; L.K., Y.G. and X.W. provided technical support; Z.C. and H.B. overall designed and supervised the study; H.B. critically revised the manuscript. All authors reviewed the manuscript.
References
- Schuppan D. & Kim Y. O. Evolving therapies for liver fibrosis. J Clin Invest 123, 1887–1901 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puche J. E., Saiman Y. & Friedman S. L. Hepatic stellate cells and liver fibrosis. Compr Physiol 3, 1473–1492 (2013). [DOI] [PubMed] [Google Scholar]
- Papatheodoridis G. V., Chan H. L., Hansen B. E., Janssen H. L. & Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: Assessment and modification with current antiviral therapy. J Hepatol 62, 956–967 (2015). [DOI] [PubMed] [Google Scholar]
- Weng H. L. et al. The etiology of liver damage imparts cytokines transforming growth factor beta1 or interleukin-13 as driving forces in fibrogenesis. Hepatology 50, 230–243 (2009). [DOI] [PubMed] [Google Scholar]
- Inagaki Y. & Okazaki I. Emerging insights into transforming growth factor beta Smad signal in hepatic fibrogenesis. Gut 56, 284–292 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y. D. et al. Regulation of transforming growth factor-beta 1 expression by the hepatitis B virus (HBV) X transactivator. Role in HBV pathogenesis. J Clin Invest 97, 388–395 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Smad phosphoisoform signals in acute and chronic liver injury: similarities and differences between epithelial and mesenchymal cells. Cell Tissue Res 347, 225–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D. et al. Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci 100, 2133–2142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. W. et al. HAb18G/CD147 promotes activation of hepatic stellate cells and is a target for antibody therapy of liver fibrosis. J Hepatol 57, 1283–1291 (2012). [DOI] [PubMed] [Google Scholar]
- Kornek M., Popov Y., Libermann T. A., Afdhal N. H. & Schuppan D. Human T cell microparticles circulate in blood of hepatitis patients and induce fibrolytic activation of hepatic stellate cells. Hepatology 53, 230–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Vílchez S. et al. The hepatitis B virus X protein induces paracrine activation of human hepatic stellate cells. Hepatology 47, 1872–1883 (2008). [DOI] [PubMed] [Google Scholar]
- Guo G. H., Tan D. M., Zhu P. A. & Liu F. Hepatitis B virus X protein promotes proliferation and upregulates TGF-beta1 and CTGF in human hepatic stellate cell line, LX-2. Hepatobiliary Pancreat Dis Int 8, 59–64 (2009). [PubMed] [Google Scholar]
- Zawel L. et al. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1, 611–617 (1998). [DOI] [PubMed] [Google Scholar]
- Jin Z. et al. Accelerated liver fibrosis in hepatitis B virus transgenic mice: involvement of natural killer T cells. Hepatology 53, 219–229 (2011). [DOI] [PubMed] [Google Scholar]
- Yan Z. et al. CD147 promotes liver fibrosis progression via VEGF-A/VEGFR2 signalling-mediated cross-talk between hepatocytes and sinusoidal endothelial cells. Clin Sci (Lond). 129, 699–710 (2015). [DOI] [PubMed] [Google Scholar]
- Lin J., Wu J. F., Zhang Q., Zhang H. W. & Cao G. W. Virus-related liver cirrhosis: molecular basis and therapeutic options. World J Gastroenterol 20, 6457–6469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. et al. SMAD4 regulates cell motility through transcription of N-cadherin in human pancreatic ductal epithelium. PloS one 9, e107948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ou J., Inagaki Y., Greenwel P. & Ramirez F. Synergistic cooperation between Sp1 and Smad3/Smad4 mediates transforming growth factor beta1 stimulation of alpha 2(I)-collagen (COL1A2) transcription. J Biol Chem 275, 39237–39245 (2000). [DOI] [PubMed] [Google Scholar]
- Sysa P., Potter J. J., Liu X. & Mezey E. Transforming growth factor-beta1 up-regulation of human alpha(1)(I) collagen is mediated by Sp1 and Smad2 transacting factors. DNA Cell Biol 28, 425–434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Artlett C. M., Jimenez S. A., Hall D. J. & Varga J. Positive regulation of human alpha 1 (I) collagen promoter activity by transcription factor Sp1. Gene 164, 229–234 (1995). [DOI] [PubMed] [Google Scholar]
- Presser L. D., McRae S. & Waris G. Activation of TGF-β1 promoter by hepatitis C virus-induced AP-1 and Sp1: role of TGF-β1 in hepatic stellate cell activation and invasion. PLoS One 8, e56367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gao P. et al. Nrf2 ameliorates diabetic nephropathy progression by transcriptional repression of TGFβ1 through interactions with c-Jun and SP1. Biochim Biophys Acta 1839, 1110–1120 (2014). [DOI] [PubMed] [Google Scholar]
- Cogan J. G., Subramanian S. V., Polikandriotis J. A., Kelm R. J. Jr. & Strauch A. R. Vascular smooth muscle alpha-actin gene transcription during myofibroblast differentiation requires Sp1/3 protein binding proximal to the MCAT enhancer. J Biol Chem 277, 36433–36442 (2002). [DOI] [PubMed] [Google Scholar]
- Qureshi H. Y., Sylvester J., El M. M. & Zafarullah M. TGF-beta-induced expression of tissue inhibitor of metalloproteinases-3 gene in chondrocytes is mediated by extracellular signal-regulated kinase pathway and Sp1 transcription factor. J Cell Physiol 203, 345–352 (2005). [DOI] [PubMed] [Google Scholar]
- Greenwel P., Inagaki Y., Hu W., Walsh M. & Ramirez F. Sp1 is required for the early response of alpha2(I) collagen to transforming growth factor-beta1. J Biol Chem 272, 19738–19745 (1997). [DOI] [PubMed] [Google Scholar]
- Henderson N. C. et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med 19, 1617–1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Extracellular membrane-proximal domain of HAb18G/CD147 binds to metal ion-dependent adhesion site (MIDAS) motif of integrin β1 to modulate malignant properties of hepatoma cells. J Biol Chem 287, 4759–4772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M. et al. HBx induces hypomethylation of distal intragenic CpG islands required for active expression of developmental regulators. Proc Natl Acad Sci USA 111, 9555–9560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarocchi M. et al. Telomerase activated thymidine analogue pro-drug is a new molecule targeting hepatocellular carcinoma. J Hepatol 61, 1064–1072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. Pathological functions of interleukin-22 in chronic liver inflammation and fibrosis with hepatitis B virus infection by promoting T helper 17 cell recruitment. Hepatology 59, 1331–1342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari F. V. et al. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell 59, 1145–1156 (1989). [DOI] [PubMed] [Google Scholar]
- Carpino G. et al. Alpha-SMA expression in hepatic stellate cells and quantitative analysis of hepatic fibrosis in cirrhosis and in recurrent chronic hepatitis after liver transplantation. Dig Liver Dis 37, 349–356 (2005). [DOI] [PubMed] [Google Scholar]
- Lu M. et al. Cell expression patterns of CD147 in N-diethylnitrosamine/phenobarbital-induced mouse hepatocellular carcinoma. J Mol Histol 46, 79–91 (2015). [DOI] [PubMed] [Google Scholar]
- Zeisberg M. et al. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem 282, 23337–23347 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang D. W. & Bian H. J. No contribution to liver fibrosis, but possible carcinogenesis? Hepatology 51, 1468–1469 (2010). [DOI] [PubMed] [Google Scholar]
- Wu J. et al. HAb18G/CD147 promotes epithelial-mesenchymal transition through TGF-beta signaling and is transcriptionally regulated by Slug. Oncogene 30, 4410–4427 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. HAb18G (CD147), a cancer-associated biomarker and its role in cancer detection. Histopathology 54, 677–687 (2009). [DOI] [PubMed] [Google Scholar]
- Li Y. L. et al. Newcastle disease virus represses the activation of human hepatic stellate cells and reverses the development of hepatic fibrosis in mice. Liver Int 29, 593–602 (2009). [DOI] [PubMed] [Google Scholar]
- Kong L. M. et al. Promoter hypomethylation up-regulates CD147 expression through increasing Sp1 binding and associates with poor prognosis in human hepatocellular carcinoma. J Cell Mol Med 15, 1415–1428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.