Abstract
Accumulation of mutant polyglutamine proteins in intraneuronal inclusions is a hallmark of polyglutamine diseases. Impairment of protein clearance systems and sequestration of clearance-related proteins into inclusions occur in many protein folding diseases, including the polyglutamine diseases. The ubiquitin-binding and proteasome adaptor protein UBQLN2 participates in protein homeostasis and localizes to inclusions in various neurodegenerative diseases. Employing mouse models and human brain tissue of Huntington's disease (HD) and spinocerebellar ataxia type 3 (SCA3), we show that UBQLN2 is selectively recruited to inclusions in HD but not SCA3. Consistent with this result, in a cell-based system mutant HTT interacts with UBQLN2 through the UBA domain while the SCA3 disease protein ATXN3, a deubiquitinating enzyme, does not interact with UBQLN2. Differential recruitment of UBQLN2 to aggregates in HD and SCA3 underscores the heterogeneity of inclusions in polyglutamine diseases and suggests that components of neuronal protein quality control may be differentially perturbed in distinct polyQ diseases.
Keywords: UBQLN2, SCA3, Huntington's disease, polyglutamine, UBA, UBL
Introduction
Polyglutamine (polyQ) neurodegenerative diseases are caused by abnormally long polyQ tracts that promote protein misfolding and aggregation of the mutant disease proteins. A shared neuropathological hallmark of polyglutamine diseases is accumulation of ubiquitin-positive inclusions in susceptible brain regions, typically within neurons (Orr and Zoghbi, 2000; Paulson et al., 1997). Two cellular systems are principally responsible for the clearance of misfolded proteins, the ubiquitin-proteasome pathway (UPP) and the autophagy-lysosome system (Nijholt et al., 2011). Impairment of both systems and sequestration of clearance-related proteins into aggregates formed by disease proteins may contribute to polyglutamine disease pathogenesis (Rubinsztein, 2006; Zhang and Saunders, 2009; Zhang et al., 2014).
Here we investigate the involvement of the clearance-related protein ubiquilin-2 (UBQLN2) in polyglutamine disease. UBQLNs are a family of multi-functional proteins that contain a N-terminal ubiquitin-like (UBL) domain and a C-terminal ubiquitin-associated (UBA) domain (Lee and Brown, 2012). The human genome encodes four UBQLNs, numbered 1 through 4;(Lee and Brown, 2012) of which UBQLNs 1,2 and 4 are expressed in brain (Safren et al., 2015). The presence of UBA and UBL domains, which respectively bind ubiquitinated proteins and the proteasome (Ko et al., 2004; Lee and Brown, 2012; Walters et al., 2002), supports the view that UBQLNs act as shuttle proteins targeting proteins for proteasomal degradation (Lim et al., 2009). UBQLNs may also regulate autophagy-lysosome function (Rothenberg et al., 2010) among other potential functions (Gilpin et al., 2015). The best studied family member, UBQLN1, also known as protein linking integrin-associated protein and cytoskeleton-1 (PLIC-1), is reported to interact with the polyQ disease proteins huntingtin (HTT) and ataxin-3 (ATXN3) (Doi et al., 2004; Heir et al., 2006). Overexpression of UBQLN1 in numerous model systems of Huntington's disease (HD) delays polyQ aggregation and increases lifespan (Rothenberg et al., 2010; Safren et al., 2014; Wang et al., 2006; Wang and Monteiro, 2007), while knockdown of UBQLN1 exacerbates polyQ toxicity in nematode models (Wang et al., 2006). UBQLN1 also interacts with the Alzheimer's disease (AD) associated proteins presenilins-1, -2, and Amyloid Precursor Protein, modulates γ-secretase cleavage, and accumulates in Hirano bodies in AD hippocampus (Mah et al., 2000)((Satoh et al., 2013; Stieren et al., 2011; Viswanathan et al., 2011). These results suggest that UBQLN1 plays a role in the pathogenesis of or cellular response to mutant proteins in several neurodegenerative diseases.
UBQLN2 is closely related to UBQLN1 except for a PXX repeat region unique to UBQLN2. Thus, the various functions ascribed to UBQLN1 may apply to UBQLN2 as well (Mah et al., 2000; Zhang and Saunders, 2009). UBQLN2 is prominently expressed in brain and smooth muscle (Wu et al., 1999). It is primarily localized in the cytoplasm (Deng et al., 2011; Osaka et al., 2014). Mutations in this protein directly cause hereditary neurodegenerative disease along the amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD) spectrum (Deng et al., 2011; Zhang and Saunders, 2009). Furthermore, studies of human brain tissue indicate that UBQLN1 and/or UBQLN2 localize to many different types of inclusions including neurofibrillary tangles in AD, Lewy bodies in Parkinson disease, and neuronal nuclear inclusions in polyQ diseases (Mori et al., 2012). There are, however, inconsistencies among published reports regarding the aggregate pathologies sequestering UBQLNs. These discrepancies may reflect use of different antibodies to define co-localization (Mori et al., 2012; Rutherford et al., 2013). Pathological screening of several mouse models of neurodegenerative disease and human disease brains indicates that UBQLN2 is likely associated with polyQ disease protein aggregates but does not associate with Lewy bodies in PD, tau pathology in AD, progressive supranuclear palsy or FTD (Nolle et al., 2013; Rutherford et al., 2013).
Divergency in the literature prompted us to revisit whether UBQLN2 accumulates in polyQ inclusions in two polyQ diseases, HD and SCA3. Using a specific anti-UBQLN2 antibody on mouse disease model and human disease brain tissue, we demonstrate that UBQLN2 localizes to nuclear inclusions in HD but not in SCA3. Further studies in cell models establish that UBQLN2 selectively interacts with mutant HTT but not with ATXN3.
Materials and Methods
Mouse models
To model Huntington's disease, we utilized the HD-KI (Q150) (Lin et al., 2001) and HDKI (Q200) (Heng et al., 2010; Zeng et al., 2013) mouse models expressing murine huntingtin with ~150 and ~200 CAG repeats respectively.
To model SCA3 disease, we utilized SCA3 YAC transgenic mice (Cemal et al., 2002; Costa Mdo et al., 2013) and SCA3 KI mice (Ramani et al., 2014). The SCA3 YAC transgenic mouse lines express the full-length human ATXN3 gene with either 15 CAG repeats (normal length) or 84 CAG repeats (disease length). The SCA3 KI mouse line expresses murine full length ATXN3 containing 82 glutamine repeats and develops robust nuclear inclusion pathology of ATXN3.
All animals were housed and handled in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Animals were housed in cages with a maximum number of five animals and maintained in a specific pathogenic-free condition with a 12h light/dark cycle maintained at 23°C.
Genotyping
DNA was extracted from mouse tail with DNeasy Bood & Tissue Kit (Qiagen, Valencia, CA). HD-KI (Q200) mice were genotyped as described previously (Zeng et al., 2013), using the following primer sequences: forward primer CCC ATT CAT TGC CTT GCT G; the reverse primer GCG GCT GAG GGG GTT GA. LA Taq with GC buffer II (Takara Bio Inc) was used for PCR amplification. PCR conditions were as follows: 98°C for 5 min, followed by 30 cycles at 98°C for 30 s, 53°C for 30 s, 72°C for 1.5 min, and a final extension at 72°C for 5 min. For SCA3 KI mouse line, the ATXN3 gene was amplified using the forward primer (TTC ACG TTT GAA TGT TTC AGG) and reverse primer (ATA TGA AAG GGG TCC AGG TCG) with the following cycling conditions: 95°C for 2 min, then 30 cycles at 95°C for 30 s, 51°C for 30 s, 72°C for 1 min, followed by 72°C for 10 min, and terminating with 4°C. The HD-KI (150) mouse tails were sized for CAG repeat length by Laragen Inc. (Los Angeles, CA). YACMJD genotyping was performed as described previously (Cemal et al., 2002; Costa Mdo et al., 2013).
Mouse brain tissue harvesting
Animals were anesthetized with ketamine/xylazine mixture and perfused transcardially with 0.1 M phosphate buffer. Brains were dissected and divided sagittally. One half was immediately placed on dry ice and stored at −80°C for biochemical studies while the other half was fixed in 4% paraformaldehyde at 4°C for 24 hours, and cryoprotected in 30% sucrose in 0.1 M phosphate buffer for an additional 24 hours at 4°C. Fixed hemispheres were sectioned at 30 μm sagittally through the entire hemisphere. Free floating sections were stored at −20°C for immunostaining.
Immunohistochemistry
Immunohistochemistry was performed as described previously (Costa Mdo et al., 2013; Zeng et al., 2013). Briefly, free floating sections were labeled with goat anti-N-terminal HTT (1:250; Santa Cruz Biotechnology, Santa Cruz, CA) or mouse anti-ATXN3 (1H9) (1:1000, MAB5360; Millipore). Immunostaining was performed using the Vectastain Elite Kit (Vector Laboratories, Burlingame, CA) or Vector MOM immunodetection kit (Vector Laboratories) when a mouse primary antibody was used. Sections were developed in ImmPACT DAB (Vector Laboratories), mounted on Superfrost slides (Fisher Scientific, Pittsburgh, PA) and air dried after dehydration with graded ethanol and xylene. Coverslips were affixed with DPX (Electron Microscopy Sciences, Hatfield PA, USA). All sections were imaged with an Olympus BX51 microscope (Olympus, Center Valley, PA).
Human disease brain tissue
Disease brain tissue was obtained through the University of Michigan Brain Bank. Three HD brains were evaluated, all of which showed typical neuropathological features of HD upon autopsy. All three individuals had adult onset HD with the triad of progressive movement, psychiatric and cognitive symptoms, were followed through the course of their disease by UM Neurology, and were from families with molecularly confirmed HD. Approximate age of onset and age of death were: late 30s, age 56; mid 50s, age 67; and mid 40s, age 73. CAG repeat length was not determined in these individuals. The evaluated SCA3 disease brain was from an individual with so called Type II SCA3, an adult-onset progressive form of ataxia that began in his early 40s, resulting in death at age 59. CAG repeat length was 72 repeats. Autopsy revealed characteristic features of Type II SCA3 with prominent cerebellar and brainstem atrophy.
Immunofluorescence staining of mouse and human tissue
For human tissue, brain sections were deparaffinized in xylene and rehydrated by immersion in a descending series of ethanol. Sections underwent antigen retrieval in a 80°C citrate buffer for 15-20 min and then were incubated with goat anti-N-terminal HTT (1:200; Santa Cruz), mouse anti-HTT EM48 (1:200; Millipore), mouse anti-ATXN3 1H9 (1:200, MAB5360; Millipore), rabbit anti-polyubiquitin (1:200; Dako, Carpinteria, CA), mouse anti-polyubiquitin (1:200; Millipore), rabbit anti-UBQLN2 (1:200; Novus), rabbit anti-UBQLN1/2 (1:250; clone 5F5, Abnova, Walnut, CA) or guinea pig anti-p62 C-terminal (1:200; Progen). Sections were then incubated with corresponding secondary antibodies: Donkey anti-goat FITC (1:500; Jackson ImmunoResearch), Alex Fluor 568, 488 and/or 647 antibodies (1:500; Invitrogen). All sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI), mounted with Prolong Gold Antifade Reagent (Invitrogen), and imaged using a Nicon A1 confocal microscope.
Plasmids
pcDNA3.1-GFP-Huntingtin exon I with 25 or 103 CAG repeats was kindly provided by Dr Shengyun Fang and have been reported previously (Yang et al., 2007). Plasmid p4455 FLAG-hPLIC-2 was from Addgene plasmid 8661 (Kleijnen et al., 2000). FLAG-hPLIC-2-ΔUBL and FLAG-hPLIC-2-ΔUBA were created by PCR using FLAG-hPLIC-2 as a template and cloned into the same vector. Plasmids of pEGFP-C1-Ataxin3Q28 and pEGFP-C1-Ataxin3Q84 have been described previously (Chai et al., 2002).
Transfection and immunoprecipitation
Human embryonic kidney 293 (HEK293) cells were cultured in DMEM, supplemented with 10% FBS, 100U/ml penicillin/streptomycin. Transfections were carried out with lipofectamine 2000 (invitrogen) as described previously (Zeng et al., 2012). Transfected cells were lysed with 1% Triton X-100 lysis buffer containing 150 mM NaCl, 20 mM Tris/HCl, pH 8.0, 5 mM EDTA, 2 mM N-ethylmaleimide (Life technologies) and complete Mini Protease Inhibitor tablets (Roche). Nuclei and insoluble material were removed by centrifugation. Antigen-antibody complexes were immunoprecipitated with anti-GFP (Invitrogen) or anti-ATXN3 1H9 and protein G beads (Sigma) at 4°C overnight with rotation. After washing three times in lysis buffer, the immunoprecipitates were dissolved in SDS loading buffer and loaded on 10% SDS/PAGE and transferred to polyvinylidene difluoride membranes. The membranes were incubated with the following antibodies: rabbit anti-GFP (1:1000; Santa Cruz biotechnology), rabbit anti-tubulin-α (1:10,000; cell signaling), rabbit anti-flag (1: 5000; Sigma), mouse anti-ATXN3 1H9 (1:1000; Millipore). After incubation with secondary antibodies, blots were developed with blue basic autorad films (GeneMate).
Results
UBQLN2 localizes to aggregates in mouse models of HD but not SCA3
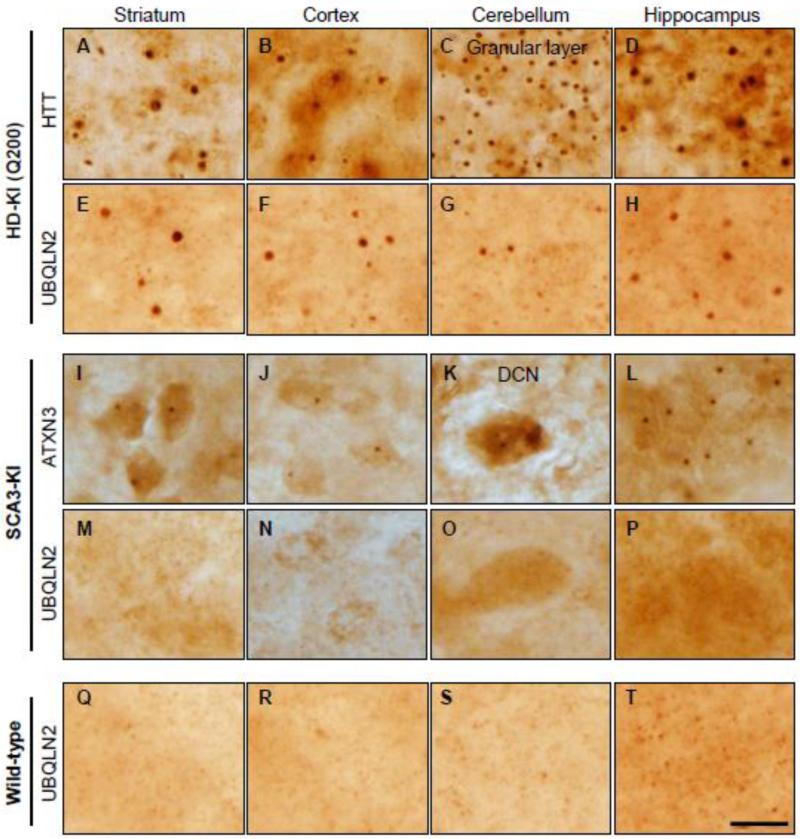
To investigate whether UBQLN2 is implicated in protein aggregation in the two most common polyQ diseases, HD and SCA3, we investigated knock-in (KI) mouse models of each disease: the HD-KI (Q200) (Heng et al., 2010; Zeng et al., 2013) and the SCA3-KI (Ramani et al., 2014) models. The HD-KI (Q200) mice exhibit robust nuclear inclusions and motor deficits by 50 weeks of age (Zeng et al., 2013), and SCA3-KI mice show nuclear inclusions as early as 12-weeks (Ramani et al., 2014). Several brain regions from HD and SCA3 KI mice were analyzed at an age when intraneuronal inclusions formed by the disease proteins are robustly present: 75 weeks in HD-KI (Q200) mice and 52 weeks in SCA3-KI mice (Fig 1.A-D, I-L). UBQLN2 robustly localized to HTT inclusions in HD-KI (Q200) mice (Fig 1. E-H), but did not localize to inclusions in any SCA3-KI brain regions. Instead, UBQLN2 immunoreactivity remained diffusely distributed throughout neurons in SCA3-KI and WT mice (Fig 1. M-T).
Fig.1.
UBQLN2 localizes to polyglutamine inclusions in HD-KI (Q200) mice but not SCA3-KI mice. Immunohistochemistry with anti-HTT (A-D) or anti-UBQLN2 (E-H, Q-T) antibody was performed on brain sections from 75-week-old HD-KI (Q200) mice (A-H) or WT littermates (Q-T). HD-KI (Q200) mice show robust inclusions in the indicated brain regions that often co-immunostain positively for UBQLN2. Immunohistochemistry with anti-ATXN3 1H9 (I-L) or anti-UBQLN2 (M-P) antibody was performed on brain sections from 52-week-old SCA3 KI mice. ATXN3 inclusions in SCA3-KI mice do not coimmunostain for UBQLN2. Scale bars = 20 μm.
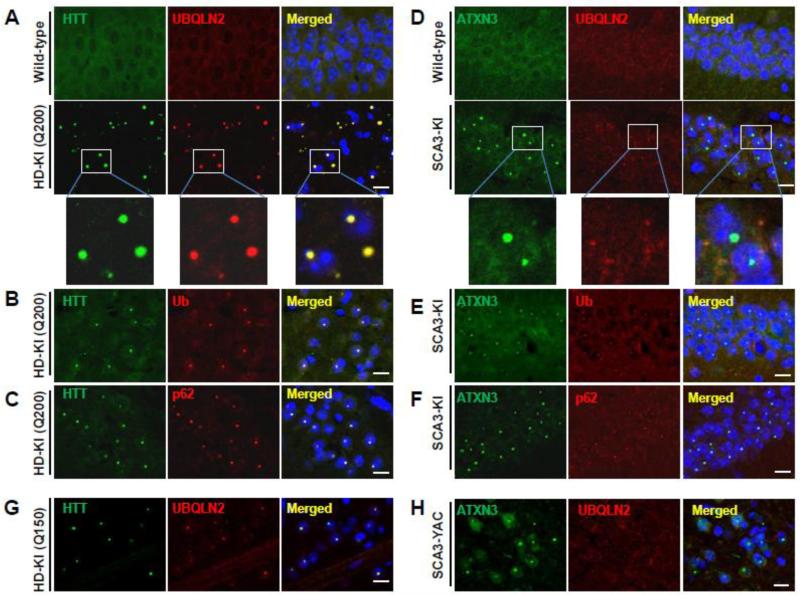
To further characterize this difference in inclusion composition, we performed double immunofluorescence for UBQLN2 with HTT, ATXN3, ubiquitin or p62. In HD-KI mice, UBQLN2 immunostaining exhibited complete overlap with HTT-immunoreactive nuclear inclusions (Fig. 2A), but in SCA3-KI mice did not overlap with intranuclear ATXN3- immunoreactive inclusions, which are particularly abundant in the hippocampus (Fig. 2D). Similar findings were observed in other brain regions (data not shown). In contrast, ubiquitin and p62, two critical components for cellular handling of misfolded proteins, localized to disease protein inclusions in both HD- and SCA3-KI mice (Fig. 2B, 2C, 2E, 2F).
Fig.2.
UBQLN2 localizes to inclusions in mouse models of HD but not SCA3. (A-C) Brain sections from 75-week-old heterozygous HD-KI (Q200) mice were double immunolabeled for (A) HTT and UBQLN2, (B) HTT and ubiquitin or (C) HTT and p62. UBQLN2, ubiquitin and p62 all localize to HTT intranuclear inclusions in the hippocampus. Scale bars = 10 μm. (D-F) Brain sections from 52-week-old homozygous SCA3-KI mice were double immunolabeled for (D) ATXN3 and UBQLN2, (E) ATXN3 and ubiquitin, or (F) ATXN3 and p62. UBQLN2 does not localize to SCA3 nuclear inclusions in the hippocampus whereas ubiquitin and p62 do. (G) Double labeling of HTT (green) and UBQLN2 (red) shows colocalization in nuclear inclusions in the striatum of a second HD model, the HD-KI (Q150) mouse, at 70 weeks of age. (H) Double labeling of ATXN3 (green) and UBQLN2 (red) indicates UBQLN2 is not recruited into SCA3 nuclear inclusions in the cortex of a second SCA3 model, homozygous SCA3 YAC mice, at 45 weeks of age. Scale bars = 10 μm.
We confirmed differential accumulation of UBQLN2 into inclusions in two additional HD and SCA3 disease mouse models: the HD-KI (Q150) and SCA3 YACMJD84 transgenic mice. HD-KI (Q150) mice develop behavioral abnormalities and neuropathologic features later than HD-KI (Q200) mice, reflecting their shorter repeat length (Lin et al., 2001). SCA3 YACMJD84 transgenic mice express the full-length human ATXN3 gene with an expanded repeat (Q84). By 8 weeks of age, both heterozygous and homozygous YAC Q84 mice develop progressive intranuclear accumulation of ATXN3 in various brain regions (Costa Mdo et al., 2013). We performed double immunofluorescence analysis on 70-week-old heterozygous HD-KI (Q150) and 45-week-old homozygous SCA3 YAC Q84 brains. As in HD-KI (Q200) mice and SCA3-KI mice, UBQLN2 co-localized to HTT-positive intranuclear inclusions in striatal neurons of HD-KI (Q150) mice (Fig.2G), but did not co-localize with ATXN3-positive inclusions in various brain regions of SCA3 YAC Q84 mice (Fig. 2H).
Early recruitment of UBQLN2 to inclusions of HD-KI mice but not SCA3-KI mice
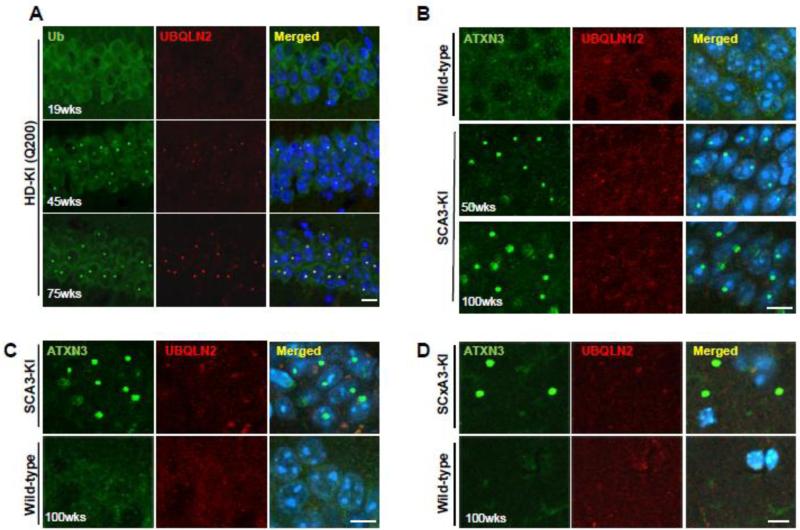
Given the strong association of UBQLN2 with inclusions in HD-KI mice, we spatially and temporally investigated the recruitment of UBQLN2 to HTT-immunoreactive nuclear inclusions. HD-KI (Q200) mice have few HTT inclusions at 20 weeks but exhibit robust nuclear inclusions by 40 weeks which continue to accumulate to 80 weeks, at which time the mice manifest significant motor deficits and striatal dopamine receptor loss (Heng et al., 2010). Co-immunostaining for UBQLN2 and ubiquitin in 19, 45 and 75 week old HD-KI (Q200) mice revealed that recruitment of UBQLN2 and ubiquitin to nuclear inclusions of HD-KI (Q200) mice paralleled the timing and spatial distribution of HTT-immunoreactive inclusion formation in several brain regions (Fig.3A, Fig.S2,Table 1). UBQLN2 localized with ubiquitin-positive inclusions 100% of the time except in the cerebellum where, in 45-week-old HD-KI (Q200) mice, inclusions in granule cells are much smaller than in other brain regions and UBQLN2 staining remains diffuse (Table 1). In contrast, even in 100-week-old SCA3 KI mice, when intranuclear and extranuclear inclusion pathology in the brain is maximally robust, UBQLN2 did not colocalize to nuclear inclusions in the CA1 region (Fig.3C) or to extranuclear inclusions in the striatum radiatum (Fig.3D). UBQLN1, closely related to UBQLN2, has been reported to localize to SCA3 nuclear inclusions (Mori et al., 2012). However, immunostaining with an anti-UBQLN antibody that recognizes both UBQLN1 and UBQLN2 (Fig.S1) in 50 and 100-week-old SCA3-KI mice showed neither UBQLN1 nor UBQLN2 colocalization to SCA3 nuclear inclusions (Fig. 3B). These findings reveal unexpected, differential accumulation of UBQLN2 within inclusions formed by two distinct polyQ disease proteins: early co-localization of UBQLN2 to HTT-immunoreactive inclusions in HD mice but little if any UBQLN2 co-localization to ATXN3-immunoreactive inclusions in SCA3 mice.
Fig.3.
Early recruitment of UBQLN2 to inclusions of HD-KI (Q200) mice but not SCA3-KI mice. (A) Double immunostaining for ubiquitin (green) and UBQLN2 (red) was performed in hippocampus of 19, 45 and 75 week old heterozygous HD-KI (Q200) mice. The presence of ubiquitin and UBQLN2 immunoreactivity within HTT nuclear inclusions paralleled the timing of HTT inclusion formation in the hippocampus. Scale bars = 10 μm. (B) Double immunostaining for ATXN3 (green) and UBQLN1/2 (red) was performed in hippocampus of 50 and 100-week-old homozygous SCA3-KI mice; the anti-UBQLN antibody used in B recognizes both UBQLN1 and UBQLN2. Even in 2-year old SCA3-KI mice, UBQLN1 and UBQLN2 do not localize to SCA3 intranuclear inclusions. (C) Double immunofluorescence for ATXN3 (green) and UBQLN2 (red) in the CA1 of hippocampus of 100-week-old SCA3-KI mice and age-matched wild type mice showing that UBQLN2 is not recruited into SCA3 nuclear inclusions even at an advanced age. Scale bars = 10 μm. (D) Double immunofluorescence for ATXN3 (green) and UBQLN2 (red) in the striatum radiatum (SR) of the hippocampus from 100-week-old SCA3-KI mice and age-matched wild type mice showing that UBQLN2 is not recruited into large extranuclear ATXN3 inclusions.
Table 1.
Time course of UBQLN2 and ubiquitin recruitment to HTT neuronal nuclear inclusions in HD-KI (Q200) mice
| Hippocampus/cortex/striatum | Cerebellum | |||||
|---|---|---|---|---|---|---|
| Staining | 20wks | 45wks | 75wks | 20wks | 45wks | 75wks |
| Anti-HTT | - | ++ | +++ | - | ++ | +++ |
| Anti-UBQLN2 | - | ++ | +++ | - | - | ++ |
| Anti-Ubiquitin | - | ++ | +++ | - | ++ | +++ |
‘-’ diffuse nuclear staining only; ‘++’ obvious immunoreactive nuclear inclusions; ‘+++’ increasing numbers of immunoreactive nuclear inclusions.
UBQLN2-positive aggregates in human HD brain but not in SCA3 brain
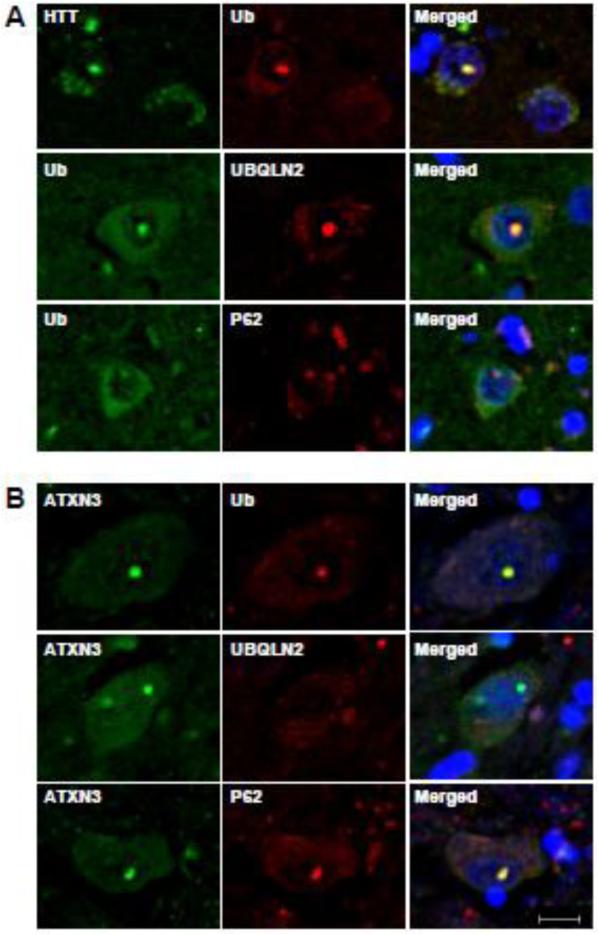
Studies of human disease brain tissue have suggested that UBQLN2 localizes to many types of inclusions in neurodegenerative diseases including HD and SCA3 (Mori et al., 2012). These published results differ from our findings in mouse models of HD and SCA3, in which SCA3 nuclear inclusions do not immunostain for UBQLN2. To assess UBQLN2 pathology in human disease brain, we performed immunofluorescence analysis for UBQLN2 in human HD and SCA3 disease brains. Consistent with our results in the four mouse models, HTT-immunoreactive intranuclear inclusions in brain sections from all three HD brains also immunolabeled with antibodies to UBQLN2, ubiquitin and p62 (Fig.4A). In contrast, UBQLN2 colocalization to nuclear inclusions was not observed in SCA3 disease brain even though the inclusions did immunostain with antibodies to ubiquitin and p62 (Fig.4B).
Fig.4.
Accumulation of UBQLN2 in inclusions of human HD brain but not SCA3 brain. (A) Sections from HD basal ganglia were double immunolabeled for HTT and ubiquitin, ubiquitin and UBQLN2 or ubiquitin and p62. UBQLN2, ubiquitin and p62 colocalize with HTT nuclear inclusions (N=3). B) Brain sections from SCA3 pons were double immunolabeled for ATXN3 and ubiquitin, ATXN3 and UBQLN2, or ATXN3 and p62. Ubiquitin and p62, but not UBQLN2, colocalize to SCA3 nuclear inclusions (N=1). Scale bars = 10 μm.
UBQLN2 selectively interacts with mutant HTT through the UBA domain
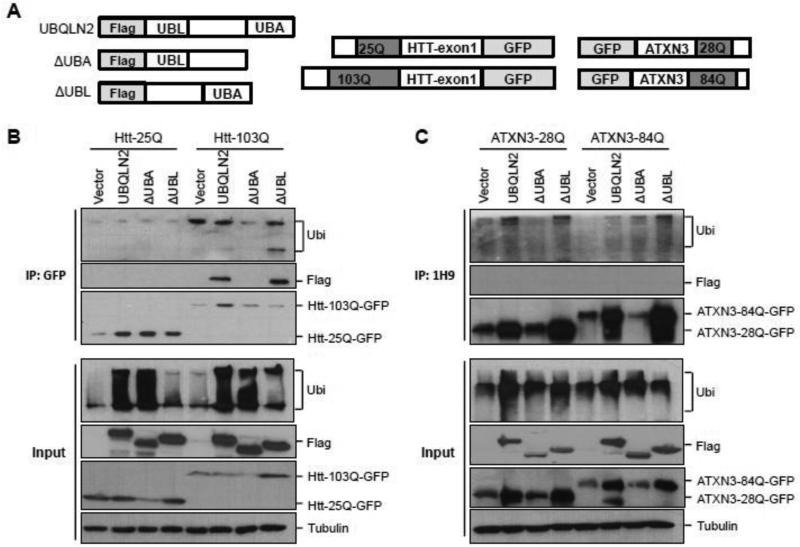
Given the difference in recruitment of UBQLN2 to disease protein inclusions in HD and SCA3, we investigated whether UBQLN2 interacts differentially with HTT and ATXN3. For these studies, we generated flag-tagged full length UBQLN2 and two truncated flag- tagged forms of UBQLN2 lacking the UBA or UBL domains (ΔUBA and ΔUBL respectively) (Fig.5A). In HEK293 cells co-expressing flag-UBQLN2 with normal or expanded HTT-GFP fusions, UBQLN2 co-precipitated with expanded mutant, but not normal HTT (Fig.5B). UBQLN2 lacking the UBL continued to interact with mutant HTT, but deletion of the UBA domain prevented co-precipitation of UBQLN2 with HTT-103Q. These results suggest that the UBA domain of UBQLN2 mediates the recruitment of UBQLN2 to disease HTT inclusions. Because UBA domains bind ubiquitin, the interaction between UBQLN2 and HTT could be mediated by ubiquitin itself. However, our assessment of coprecipitated ubiquitin in these studies (Fig 5B) did not reveal ubiquitinated HTT species and neither ruled in or out a ubiquitin-dependent interaction.
Fig. 5.
UBQLN2 selectively interacts with mutant HTT through the UBA domain. (A) Diagram of UBQLN2, HTT and ATXN3 expression constructs. (B-C) In HEK293 cells, flag-tagged UBQLN2, ΔUBA or ΔUBL was co-expressed with GFP-HTT25Q or GFPHTT103Q (B) or with GFP-ATXN3-28Q or GFP-ATXN3-84Q (C). HTT or ATXN3 in lysates from transfected cells was immunoprecipitated (IP) with anti-GFP antibody (B) or anti-1H9 antibody (C). Co-precipitated UBQLN2 was detected by IB with anti-FLAG antibody (B-C), shown on top panels. Input levels of the expressed proteins are shown on the bottom. UBQLN2 selectively interacts with an N-terminal HTT fragment with a 103Q expansion but not with normal or expanded ATXN3; deletion of the UBA domain of UBQLN2 attenuates interaction with HTT-103Q. Lysates and IP samples were also probed with anti-ubiquitin antibody, which did not reveal specific ubiquitinated forms of HTT or ATXN3. In B-C, vector represents co-transfected empty vector plasmid.
To examine whether ATXN3 similarly interacts with UBQLN2, we coexpressed flag-UBQLN2 or truncated UBQLN2 mutants with normal (28Q) or expanded (84Q) GFPATXN3 in HEK293 cells. Co-IP results with ATXN3 differed markedly from those with HTT. As shown in Fig.5C, UBQLN2 failed to interact with normal or expanded ATXN3. In summary, UBQLN2 selectively binds to mutant HTT protein but not ATXN3, and the UBA domain is necessary for the interaction with mutant HTT.
Discussion
PolyQ diseases share the common neuropathological feature of intraneuronal inclusions containing the disease proteins as well as ubiquitin, p62, proteasome subunits and other protein quality control components. Our results demonstrating differential recruitment of the ubiquitin pathway adaptor protein UBQLN2 to inclusions in HD and SCA3 underscore the fact that, despite similar appearance across diseases, polyQ inclusions are heterogenous structures whose composition may shed light on disease-related pathways in specific diseases. The fact that HTT aggregates robustly recruit UBQLN2 while ATXN3 aggregates do not, both in mouse models and in human disease tissue, suggests a functional connection between UBQLN2 and HTT that does not exist for ATXN3. At minimum, UBQLN2 represents a neuropathological disease marker that distinguishes HD and SCA3 nuclear inclusions.
The closely related proteins UBQLN1 and UBQLN2 have both been reported to interact with HTT (Doi et al., 2004; Wang and Monteiro, 2007) and localize to HTT inclusions (Rutherford et al., 2013; Safren et al., 2015). Prior studies showed that overexpression of UBQLN1 in C. elegans (Wang et al., 2006) or R6/2 mouse models of HD disease (Safren et al., 2014) reduces polyQ aggregation and increases lifespan. Consistent with these findings, we found that UBQLN2 colocalizes with ubiquitin and p62 in HTT intranuclear inclusions in two HD mouse models and human HD disease brain. Co-precipitation studies in cells demonstrated that mutant HTT interacts with UBQLN2 through its UBA domain. These findings are consistent with the results of Ko and colleagues, who reported that the UBA domain of UBQLN2 interacts with polyubiquitinated proteins and targets misfolded proteins to the proteasome for degradation (Ko et al., 2004) and the recent work of Safren and colleagues (Safren et al 2105) showing that all three brain expressed UBQLNs localize to inclusions in the R6/2 mouse model of HD. Our data suggest that HTT is a likely substrate for UBQLN2-mediated clearance and that accumulation of UBQLN2 in HTT inclusions may reflect an attempt to degrade misfolded HTT protein by the proteasome or the autophagylysosome system. To further assess the potential neuroprotective function of UBQLN2 in HD disease, efforts to knock out or overexpress UBQLN2 in HD-KI (Q200) mice would be informative.
UBQLNs were previously reported to localize to protein aggregates in SCA3 human tissue (Mori et al., 2012). Using a UBQLN2 selective antibody (Fig.S1), however, we did not detect UBQLN2 immunoreactivity in ATXN3 inclusions in mouse models of SCA3 or in human disease tissue. Mori et al used an anti-UBQLN antibody that non-selectively recognizes both UBQLN1 and UBQLN2 (Fig.S1) and thus prior results may not reflect actual colocalization of UBQLN2 to ATXN3 inclusions (Brettschneider et al., 2012; Mori et al., 2012). In addition, UBQLN1 and UBQLN2 both can form cytoplasmic puncta when overexpressed in cells, even as wild type protein (Heir et al., 2006; Osaka et al., 2014; Rothenberg et al., 2010). In the absence of co-immunostaining with ATXN3, it is difficult to know whether UBQLN2-positive inclusions detected by immunohistochemistry in cell systems or human tissue reflect ATXN3 aggregates. Consistent with the results from SCA3 human disease tissue and mouse models, co- precipitation studies in cultured cells demonstrated that UBQLN2 fails to interact with normal ATXN3 or expanded ATXN3 protein. These findings suggest that ATXN3 may not be a substrate for UBQLN2-mediated clearance.
In HD, SCA3, and other polyQ disorders, impairment of protein homeostasis and sequestration of clearance-related proteins may represent common pathomechanisms (Rubinsztein, 2006; Zhang and Saunders, 2009; Zhang et al., 2014). As ubiquitin pathway adaptor proteins, UBQLN2 and other UBQLNs have been suggested to function in a general clearance-related pathway in polyglutamine diseases (Mori et al., 2012; Zhang et al., 2014). But our data showing differential recruitment of UBQLN2 to nuclear inclusions in HD and SCA3 indicate that these components of neuronal protein quality control may be differentially perturbed in different polyQ diseases. The fact that UBQLN2-positive inclusions are absent in SCA3 disease does not support a role for UBQLN2 as a unifying contributor to disease pathways in polyQ disease. Differential sequestration of UBQLN2, however, could contribute to differing regional patterns of pathology in different polyQ diseases.
Supplementary Material
Highlights.
UBQLN2 localizes to aggregates in mouse models of HD but not SCA3.
Early recruitment of UBQLN2 to inclusions in HD-KI mice.
UBQLN2-positive aggregates in human HD brain but not in SCA3 brain.
HTT, the HD disease protein, rather than ATXN3, the SCA3 disease protein interacts with UBQLN2.
Acknowledgments
This work was supported by NIH grants 1R01 AG034228 and R01 NS038712 (HLP), and the UM Protein Folding Diseases Initiative. We thank Dr. Shengyun Fang for pcDNA3.1-GFP-huntingtin exon I with 25 and 103 CAG repeat plasmids. We thank Dr. Peter Howley for p4455 FLAG-hPLIC-2 plasmid. We thank the University of Michigan Brain Bank and the Michigan Alzheimer Disease Center for patient brain tissue.
Abbreviations
- polyQ
polyglutamine
- HD
Huntington's disease
- HTT
Huntingtin protein
- SCA3
Spinocerebellar ataxia type 3
- UPP
Ubiquitin-proteasome pathway
- UBL
Ubiquitin-like domain
- UBA
Ubiquitin-associated domain
- PLIC-1
Integrin-associated protein and cytoskeleton-1
- SCA3-KI
SCA3 knock-in mouse
- HD-KI (Q200)
Huntingtin knock-in mouse with 200 CAG repeats
- HD-KI (Q150)
Huntingtin knock-in mouse with 150 CAG repeats
- SCA3 YAC
SCA3 YACMJD84 transgenic mouse
- AD
Alzheimer's disease
- APP
Amyloid Precursor Protein
- ALS
Amyloid lateral sclerosis
- VCP
Valosin-containing protein
- SR
striatum radiatum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- Brettschneider J, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 2012;123:825–39. doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemal CK, et al. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet. 2002;11:1075–94. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- Chai Y, et al. Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc Natl Acad Sci U S A. 2002;99:9310–5. doi: 10.1073/pnas.152101299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Mdo C, et al. Toward RNAi therapy for the polyglutamine disease Machado-Joseph disease. Mol Ther. 2013;21:1898–908. doi: 10.1038/mt.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, et al. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 2004;571:171–6. doi: 10.1016/j.febslet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- Gilpin KM, et al. ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum Mol Genet. 2015;24:2565–77. doi: 10.1093/hmg/ddv020. [DOI] [PubMed] [Google Scholar]
- Heir R, et al. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–8. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng MY, et al. Early autophagic response in a novel knock-in model of Huntington disease. Hum Mol Genet. 2010;19:3702–20. doi: 10.1093/hmg/ddq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen MF, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–19. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Ko HS, et al. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–4. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Lee DY, Brown EJ. Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem. 2012;393:441–7. doi: 10.1515/hsz-2012-0120. [DOI] [PubMed] [Google Scholar]
- Lim PJ, et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187:201–17. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum Mol Genet. 2001;10:137–44. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- Mah AL, et al. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–62. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori F, et al. Ubiquilin immunoreactivity in cytoplasmic and nuclear inclusions in synucleinopathies, polyglutamine diseases and intranuclear inclusion body disease. Acta Neuropathol. 2012;124:149–51. doi: 10.1007/s00401-012-0999-z. [DOI] [PubMed] [Google Scholar]
- Nijholt DA, et al. Removing protein aggregates: the role of proteolysis in neurodegeneration. Curr Med Chem. 2011;18:2459–76. doi: 10.2174/092986711795843236. [DOI] [PubMed] [Google Scholar]
- Nolle A, et al. Ubiquilin 2 is not associated with tau pathology. PLoS One. 2013;8:e76598. doi: 10.1371/journal.pone.0076598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Reversing neurodegeneration: a promise unfolds. Cell. 2000;101:1–4. doi: 10.1016/S0092-8674(00)80617-0. [DOI] [PubMed] [Google Scholar]
- Osaka M, et al. Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu575. [DOI] [PubMed] [Google Scholar]
- Paulson HL, et al. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–44. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- Ramani B, et al. A knock-in mouse model of Spinocerebellar ataxia type 3 exhibits prominent aggregate pathology and aberrant splicing of the disease gene transcript. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddx176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–32. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, et al. Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res. 2013;1524:62–73. doi: 10.1016/j.brainres.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safren N, et al. Signature changes in ubiquilin expression in the R6/2 mouse model of Huntington's disease. Brain Res. 2015;1597:37–46. doi: 10.1016/j.brainres.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safren N, et al. Ubiquilin-1 overexpression increases the lifespan and delays accumulation of Huntingtin aggregates in the R6/2 mouse model of Huntington's disease. PLoS One. 2014;9:e87513. doi: 10.1371/journal.pone.0087513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J, et al. Ubiquilin-1 immunoreactivity is concentrated on Hirano bodies and dystrophic neurites in Alzheimer's disease brains. Neuropathol Appl Neurobiol. 2013;39:817–30. doi: 10.1111/nan.12036. [DOI] [PubMed] [Google Scholar]
- Stieren ES, et al. Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J Biol Chem. 2011;286:35689–98. doi: 10.1074/jbc.M111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan J, et al. Alzheimer's disease-associated ubiquilin-1 regulates presenilin-1 accumulation and aggresome formation. Traffic. 2011;12:330–48. doi: 10.1111/j.1600-0854.2010.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, et al. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–77. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet. 2006;15:1025–41. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–7. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AL, et al. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol Cell. 1999;4:619–25. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- Yang H, et al. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp Cell Res. 2007;313:538–50. doi: 10.1016/j.yexcr.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Zeng L, et al. The de-ubiquitinating enzyme ataxin-3 does not modulate disease progression in a knock-in mouse model of Huntington disease. J Huntingtons Dis. 2013;2:201–15. doi: 10.3233/JHD-130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, et al. Laforin is required for the functional activation of malin in endoplasmic reticulum stress resistance in neuronal cells. FEBS J. 2012;279:2467–78. doi: 10.1111/j.1742-4658.2012.08627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Saunders AJ. An emerging role for Ubiquilin 1 in regulating protein quality control system and in disease pathogenesis. Discov Med. 2009;8:18–22. [PMC free article] [PubMed] [Google Scholar]
- Zhang KY, et al. Ubiquilin 2: a component of the ubiquitin-proteasome system with an emerging role in neurodegeneration. Int J Biochem Cell Biol. 2014;50:123–6. doi: 10.1016/j.biocel.2014.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.