Abstract
Investigations focused on the interplay between the human microbiome and cancer development, herein termed the ‘oncobiome’, have been growing at a rapid rate. However, these studies to date have primarily demonstrated associative relationships rather than causative ones. We pose the question of whether this emerging field of research is a ‘mirage’ without a clear picture, or truly represents a paradigm shift for cancer research. We propose the necessary steps needed to answer crucial questions and push the field forward to bring the mirage into a tangible reality.
The Oncobiome Mirage Appears
Of all human maladies, nothing strikes fear into our hearts, minds, and souls as cancer. A diagnosis of hypertension, diabetes, or any other litany of chronic diseases that can be controlled with medication will produce a very different response than that of cancer. Researchers therefore press forward, attempting to uncover the smoking gun to explain tumor susceptibility, initiation, and progression. This search has been tried countless times with similar, often discouraging, results. What then makes investigators think that work involving the host microbiota and cancer will be any different, or is it all only a mirage?
The microbiota encompass a wide variety of microorganisms (bacteria, viruses, protozoa, fungi, and archea) and this eclectic ecosystem shares the body space of every individual, creating a commensal, symbiotic, and pathobiont relationship that has garnered increasing attention regarding its role in carcinogenesis (see Glossary) [1–5]. Of all the body surface, the gastrointestinal tract harbors the greatest number and diversity of microbes in the human body, with bacteria representing the bulk of the microbiota (1012 bacteria/gm feces) [6]. Although the oncogenic role of viruses has been recognized [5], bacteria represent the chief member of the microbiota and will be the focus of this discussion. Perhaps the most recognized link between bacteria and cancer is the case of Helicobacter pylori and non-cardiagastric carcinoma [7,8]. This bacterium has been shown to secrete several virulence factors such as CagA (cytotoxin-associated gene A), VacA (vacuolating cytotoxin A), urease, and NapA2 (neutrophil-activating protein A) that result in oxidative stress, chronic inflammation, and host DNA damage that can lead to carcinoma [9–11]. Considering that H. pylori has been designated a type I carcinogen by the World Health Organization [12,13], several clinical trials have been performed to modulate the risk of gastric cancer by eradicating the bacteria in infected individuals [14,15]. A recent meta-analysis of six randomized controlled trials demonstrates a slight risk reduction of gastric cancer with H. pylori elimination [16]. Despite this link between a pathogenic organism and carcinogenesis, there still has been little direct evidence that the symbiotic microbiota modulates carcinogenesis in humans. The relationship between cancer and the host microbiota, to be termed the ‘oncobiome’, could be a mirage: we have an idea of an image in the distance but are uncertain of its true reality or significance. What is known is that more people are taking notice of this mirage – but is it real?
For our interpretation of the oncobiome to become clearer, we have much more to uncover. Much of oncobiome research is currently focused on colorectal cancer (CRC), which has been considered the ideal malignancy to study the effects of the host–microbe relationship on carcinogenesis and will serve as the model for most discussion points in this article. This focus is for obvious reasons as the large intestine harbors the greatest number and diversity of microbes in the human body (1012 bacteria/gm feces) [6]. Multiple studies using advanced genomic approaches have expanded the relationship between intestinal microbes and CRC development [17–26]. However, these investigations have mostly yielded circumstantial evidence implicating bacteria in CRC, and should give pause to those delving into the field because the mirage may be deceiving.
What Does the Mirage Look Like?
Our current interest and vision of the oncobiome developed over a century ago with the identification of bacteria in cancer specimens [27]. Since that time this association has been further explored [28] as well as the differences in fecal bacterial composition in populations at risk for CRC development [29]. Carcinogenesis is inherently a process of inflammation, with many proinflammatory and immunosuppressive pathways acting along the neoplastic process (Box 1) [30–32]. These immunological pathways have been functionally investigated in humans and mouse models of cancer, including CRC [33–40]. With a well-established impact of microbial products on the innate and adaptive immune system [41–45], one could speculate that bacteria could influence carcinogenesis through immune responses. The concept is clearly illustrated in a mouse model of impaired intestinal barrier function where the exposure of immune cells to the microbial product lipopolysaccharide (LPS) favors intestinal tumor growth through the action of interleukin (IL)-23/IL17 [46]. Although the link between microbial products, inflammation, and carcinogenesis is firmly established, the role of microbes acting as a consortium on neoplasia is less clear. Using genomic approaches, multiple studies have compared the intraluminal and mucosal surface microbiota between healthy patients and those with CRC [19,20,23,47,48]. Although no consensus ‘cancer-biota’ has emerged from these studies, it appears that the abundance of taxa associated with a protective function (e.g., Roseburia) decreased while taxa with potential deleterious effects (e.g., Escherichia/Shigella, Klebsiella, Fusobacterium) increased in either stool or mucosal location [49]. These studies therefore suggest that microbial dysbiosis is associated with CRC development. Whether these associations are causative and can therefore modulate initiation, progression, or metastasis remains unclear.
Nevertheless, the concept that dysbiosis could be linked to CRC pushed investigators to test whether microbial genes could serve as cancer biomarkers [50,51]. These studies showed that the detection of non-invasive, early-stage CRC could be feasible by using taxonomic microbial markers. Although microbial biomarkers do not need to be functionally linked to CRC to be clinically useful, the study of microbial genomics in relation to CRC pathogenesis must push beyond the associative phase to significantly contribute to disease knowledge. Adding complexity to this issue, microbiome data are not informative of the organizational level of microbial communities in a given niche. A recent study has reinforced the notion that genomic analysis of fecal samples alone may provide limited information on host–microbiota interaction in CRC [52]. Indeed, right-sided colon tumors are more likely associated with biofilm-producing bacteria because they were present in 89% of samples versus only 12% of left-sided tumors associated with biofilm-producing bacteria. In addition, subjects with biofilm-positive tumors possessed biofilm aggregates that were distant from their tumors and that were associated with normal mucosa, perhaps indicating susceptibility to such colonization. Clearly, microbes living in a planktonic state exhibit a different phenotypic and metabolomic profile than those organized in a biofilm community [53–56], and this must be accounted for in future investigations.
Even assuming a single causative organism, which is unlikely to be the case, difficulties culturing specific microbes to fulfill Koch's postulate have created barriers to establishing cancer causation. Nevertheless, evidence gathered from preclinical models hint at a functional role of microbes in CRC. For example, germ-free (GF) Fischer rats demonstrated decreased spontaneous tumor formation compared to conventionally housed rats, as well as decreased intestinal tumors in a 1,2-dimethylhydrazine-induced model of carcinogenesis [57,58]. In addition, utilizing the adenomatous polyposis coli (APC) multiple intestinal neoplasia (Min) murine model of colon carcinogenesis, which possesses a point mutation in the murine homolog of the human APC tumor-suppressor gene that results in multiple (>100) intestinal adenomas [59], a reduction in colon tumors was noted in GF ApcMin/+ mice compared to conventionally housed controls [60]. Finally, increased carcinogenesis was noted after the enteral introduction of Fusobacterium nucleatum or enterotoxigenic Bacteroides fragilis, in ApcMin/+ mice confirming the effect of bacteria on cancer formation in vivo [47,61]. However, as opposed to IBD where antibiotic usage has shown some clinical effectiveness [62–64], no such clinical studies are available for CRC. Despite these data in favor of the oncobiome, the microbiota may also prevent carcinogenesis through protective mechanisms, detoxification, or anticancer metabolites [65–67]. Precedence has nevertheless been established that supports a potential modulatory role for microbes in carcinogenesis [57,58,60,68]. However, these models do not replicate clinical reality, and CRC-causing bacteria in mouse models have not been confirmed by observational studies in humans.
The Ever-Changing Mirage
While the oncobiome mirage has not revealed the oasis of desired answers, the image is beginning to morph from bacterial association to causative pathways. Despite the fact that prospective/longitudinal studies have not been able to assess CRC risk in patients based on changes in their microbiota, animal studies have begun to interrogate the mechanistic details of bacteria-associated carcinogenesis. Current studies are focusing on the links between CRC and toxic bacterial metabolites, diet-induced changes, and bacteria-derived genotoxic substances, albeit unproven in human studies. For example, under eubiotic conditions the microflora ferments ethanol into acetaldehyde and carbohydrates into the three primary short-chain fatty acids, acetate, propionate, and butyrate [69,70]. It has been demonstrated that a correlation may exist between low-butyrate and high-acetate levels in patients with adenomatous polyp formation and colon cancer [71,72]. Although no difference in the overall bacterial community was demonstrated, titers of the butyrate-producing species Ruminococcus and Pseudobutyrivibrio ruminis were lower in CRC stool specimens, which correlated with lower butyrate levels [72]. While preclinical models have demonstrated a role of microbe-derived butyrate in dampening colitis-associated CRC development, similar studies have shown the opposite effect [73–77]. This may be secondary to host genetics or possible implications of dietary fiber [78], thus setting the stage for microbial activities being central to diet-induced carcinogenesis. The role of butyrate and other diet-induced metabolites in carcinogenesis likely requires further investigation.
In addition to the products of carbohydrate metabolism, toxins from bacterial metabolism have likewise been implicated in CRC. For example, a variety of ingested compounds and nutritional components are metabolized by host microbes into potentially pro- and anticarcinogenic metabolites [69,79,80], such as the metabolism of proteins into N-nitroso compounds, ammonia, polyamines, and hydrogen sulfide. Colonic epithelial exposure to these metabolites results in chronic inflammation [69,81–86]. The role of these compounds in CRC development is in many ways still hypothetical, and may be related to direct dietary ingestion rather than to byproducts of bacterial metabolism [87]. However, these studies are limited in that they rely on the local effects of microbiota-produced toxins, such as inflammation and epithelial cell damage in the case of CRC. Although potentially important, studies have not taken into consideration carcinogenic mediators that may act from distant sites [88–91]. It is currently unknown if systemic absorption of such metabolites infers the same potential cancer susceptibility as seen experimentally at the local (epithelial) level. While the production of pro- and anti-inflammatory metabolites by the commensal system has been implicated in CRC initiation and progression, the data regarding an actual causal relationship still do not exist [21,24,92]. Furthermore, fecal samples alone should not be relied upon for investigating the influence of microbial metabolites on carcinogenesis because metabolites from various sources can be detected in serum and urine samples, and may correlate with gastrointestinal dysbiosis or CRC risk. Metabolomics of serum and urine samples should therefore be undertaken to detect dysbiosis and cancer risk because fecal samples alone may not account for metabolites that have been absorbed by the host [93–97]. It is these metabolites, and their resultant influence on the host, that will potentially play a large role, if one exists, in cancer development and demands further investigation. The development of computational algorithms capable of integrating the vast amounts of heterogeneous biomedical data (metabolites, GWAS, etc.) may help to generate an interacting map between microbial metabolites and host cancer susceptibility, and foster design of hypothesis-driven experiments.
Moreover, bacteria-derived genotoxic substances have garnered attention for their direct ability to impart DNA damage, which is distinct from genotoxicity from byproducts of bacterial metabolism such as hydrogen sulfide and reactive oxygen species [69,82,85,86,92]. An example of such a genotoxin is colibactin, encoded by the polyketide synthase (pks) genotoxicity island, which is found primarily in the Enterobacteriaceae family of bacteria, of which E. coli of the B2 group represents the main carrier [98]. The genotoxic effect of pks-positive strains of E. coli is likely secondary to the induction of double-strand DNA breaks with subsequent cell cycle arrest and genomic instability [66,99,100]. Previous studies showed that colonic mucosal samples from patients with CRC had a higher prevalence of pks-positive E. coli compared to controls [99,101,102]. Although preclinical models showed that pks-positive E. coli strains promote CRC [99,101,103], the link between high E. coli prevalence, genotoxicity, and neoplasia in human CRC has not been demonstrated. Therefore, the microbiota-mediated mechanisms of cancer initiation and progression, at least as it currently stands for CRC, are potentially multifold. As the mirage changes, further details regarding a potential role for specific microbes, microbial metabolites, and/or genotoxic agents will be necessary to maintain a clear image.
How to Make the Mirage Clearer
To pass the correlative threshold of oncobiome research and move into causative territory, a variety of studies using human-derived, cancer-associated microbes are necessary (Figure 1). For example, it would be important to determine the oncogenic potential of both human biofilm-positive and luminal microbial communities in preclinical models and to define their carcinogenic activities. This is especially important given that studies using stools from either healthy subjects or CRC patients have provided surprising results on the relationship between luminal bacteria and CRC, such that CRC development was more prominent in GF mice transplanted with stools from healthy subjects than those from CRC patients [104]. Therefore, although CRC is communicable between mice [105], the transfer of carcinogenesis between human and mouse remains to be established. Preclinical models using transmission of these microbes would therefore help to define the natural history of both sporadic and hereditary forms of carcinogenesis, as has been used to study the impact of early E. coli pks+ colonization on intestinal mucosa [106].
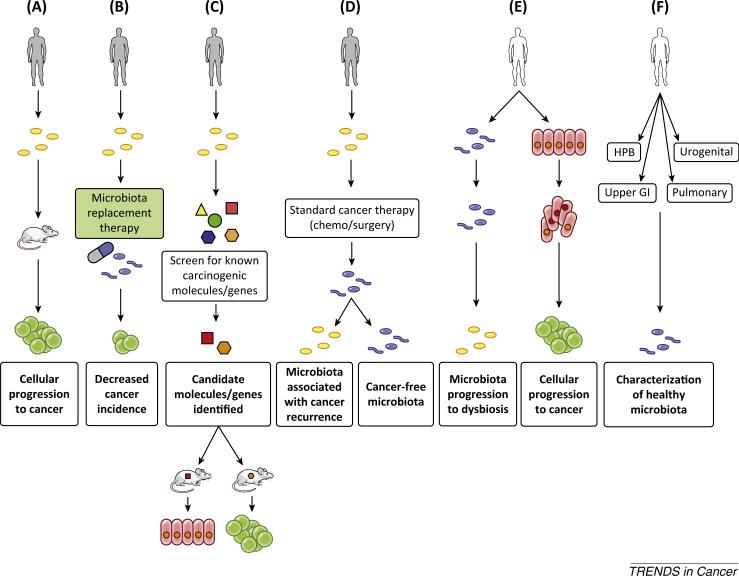
Figure 1. Proposed Oncobiome Studies Necessary to Progress the Field of Research.
Cancer subjects (gray) or healthy subjects (white) are presented for each area of needed investigation. (A) Cancer-associated microbiota (yellow) is transmitted to mice with humanized immune systems to investigate their interaction with the human immune system and their ability to cause cancer. (B) Patients at high risk or with a genetic predisposition to cancer are treated with microbiota replacement therapy to restore eubiosis (blue). Patients are compared to the general population and control subjects not treated with microbiota replacement therapy for differences in cancer incidence. (C) The microbiota of cancer patients are screened for known carcinogenic molecules and genes. Candidates are identified and tested in vivo for their ability to cause cancer formation. (D) The microbiota of cancer patients are determined before and after standard cancer treatment. Determination is made regarding restoration of eubiosis and if continued or recurrent dysbiosis is associated with cancer recurrence. (E) The microbiota of healthy subjects are determined prospectively and correlated with the development of precancerous and cancerous lesions. (F) The microbiota of healthy individuals are determined for various organ systems and body fluids. This will prove crucial for future investigations and to determine if body fluids/specimens from one site can act as a surrogate for a different disease site.
Moreover, it is important to understand if the acquisition of microbes with carcinogenic potential at birth influences CRC development later in life? It is known from kindred data and population studies that the age of onset for people with predispositions to cancers, such as hereditary breast or gastric, or those associated with chronic inflammatory states such as CRC with ulcerative colitis, is younger than for sporadic forms ([107–112]; National Cancer Institute Surveillance, Epidemiology, and End Results (SEER) Program Fact Sheets: Breast Cancer, http://seer.cancer.gov/statfacts/html/breast.html; Colon and Rectum Cancer, http://seer. cancer.gov/statfacts/html/colorect.html; Stomach Cancer, http://seer.cancer.gov/statfacts/html/stomach.html). This demonstrates a potential lead-time between genetically induced and microbe-induced cancers, and likely represents different timelines of progression. Regardless, the question remains: is microbial carcinogenic activity triggered early upon colonization or is it under the influence of external factors (diet, inflammation, or environment)? Evidence exists that our microbiota are established in utero and develop rapidly, but with stable diversity during the first year of life, and continue to increase in abundance throughout the first decade [113–115]. This may suggest that alteration of the microbiota during life via external factors (diet, environment) alters cancer susceptibility. The nding that the E. coli pks+ strain fails to promote CRC in a model of colitis-associated CRC highlights the complex interaction between microbes and host [116]. In this study, the authors showed, using II10−/− mice defective in mature T and B lymphocytes, that development of colitis-carcinogenesis led to transcriptional changes in E. coli gene repertoire, including genes present in the pks genotoxic island.
Furthermore, could microbe-derived carcinogenic molecules and genes be detected in CRC patients and do they correlate with malignancies? For example, expression of the F. nucleatum virulence factor FadA (Fusobacterium adhesin A), which is implicated in bacterial attachment and invasion, is increased in carcinoma tissues of CRC subjects and correlates with oncogenic and inflammatory host gene expression (Box 1) [117]. Similarly, the bft (Bacteroides fragilis toxin) gene responsible for the enterotoxic properties of enterotoxigenic Bacteroides fragilis and CRC development in ApcMin/+ mice [61] was found in a greater proportion of colon cancer specimens than healthy mucosal controls, suggesting a role for this bacterial toxin in carcinogenesis [118]. Finally, as the data on the interaction of the microbiota with the host immune system and tumor microenvironment mature [6,47,66,119], utilizing mice with humanized immune systems in conjunction with human microbes will prove crucial in dissecting the interaction between host microbes and the immune system in carcinogenesis [120,121].
As insight is gained regarding the oncobiome, focus will be placed on treatment of human diseases and mitigating cancer risk. Growing research has demonstrated the influence of the host microbiota on various chemotherapeutics [119,122,123]. Because not all chemotherapy trials result in drug efficacy against their targeted cancer, it can be hypothesized that this may be secondary to intestinal dysbiosis, which was not accounted for during trial design [124]. As such, it would be advantageous if these trials incorporated microbiota studies to correlate drug efficacy with microbial composition.
Finally, although the oncobiome in CRC is presently the most mature area, other malignancies demand attention. Gastrointestinal tumors (e.g., esophageal, gastric, hepatobiliary, and pancreatic) seem to be natural starting points, and the findings of CRC-microbiota research are potentially directly applicable to these malignancies as well. However, it is currently unknown what fluid/tissue sample(s) from these organs best reflect their unique microbiota. For example, is the microbiota of the bile, pancreatic fluid, or duodenum most reflective of hepato-pancreatobiliary (HPB) malignancies, or are these malignancies not influenced by their local microbiota but instead by microbiota from distant sites? The difficulty then becomes sample acquisition because obtaining such samples requires invasive procedures. However, one solution could be to recruit patients for trials who require upper endoscopy as part of their cancer workup. With these samples, future studies can initially focus on understanding the normal biota and later its possible connection between microorganisms and carcinogenesis.
Have We Reached the Mirage?
That the microbiota can cause cancer is a unique and potentially paradigm-shifting event. What then is to be done if particular bacterial species are confirmed to cause cancer? Hypothetical interventions would be based on the considerations of screening, treatment, and surveillance for each particular cancer (Figure 2). Using CRC as an example, screening begins at the age of 50 years for patients at average risk, and oncobiome tests could potentially augment or replace current screening modalities. For example, one National Comprehensive Cancer Network recommendation is that stool-based high-sensitivity guaiac or immunohistochemical testing be performed annually. This testing aims to detect occult blood and has been shown to reduce CRC mortality [125–127]. Because adenomatous (pre-malignant) polyps and early cancers may bleed only intermittently, if at all, this testing has the disadvantage of not being able to detect these lesions, prompting the recommendation to test three successive specimens. Emerging technologies rely on the detection of mutated APC or KRAS genes, or vimentin methylation in tumor cells sloughed in the stool. Overall, these tests have demonstrated poor sensitivity and specificity, and only one is currently available in the USA [128–130]. This method of screening is costly and, similarly to other screening methods, it relies on the detection of early signs or symptoms of cancer and thus is not necessarily preventative. However, oncobiome screening could potentially be designed to detect not only individual bacterial species associated with cancer but also dysbiosis long before adenomatous polyps or cancer have developed. Indeed a recent study characterized the microbiota from ‘healthy’ subjects and those with adenomas or CRC as confirmed by colonoscopy [51,131], and found that, when combined with known clinical risk factors for CRC (age, race, body mass index), combining six specific operational taxonomic units (OTU) of gut microbiota in stool samples significantly increased the ability to differentiate healthy subjects from those with adenomas or CRC. Likewise, taxonomic markers were identified through metagenomic sequencing of fecal samples to distinguish CRC patients from tumor-free patients [50]. While the sensitivity and specificity of the taxonomic markers was similar to the currently used screening method of fecal occult blood testing (FOBT: sensitivity 58% vs 49%, respectively; specificity 92%), their combined use with FOBT increased the sensitivity of CRC detection by more than 45% compared to FOBT alone (72% vs 49%, respectively), while maintaining the specificity (92%). While these data are limited, they demonstrate the possibility of utilizing microbiota composition to predict disease. It may therefore be envisaged that a proactive, as opposed to reactive, screening strategy could be implemented to prevent cancer formation, possibly through dietary modification as one example. However, while diet has previously been shown to alter the host microbiota, the direct effect of dietary modification on carcinogenesis is currently unresolved [132–134]. This screening strategy may also be useful for other malignancies as more evidence emerges on the oncobiome: saliva for oropharyngeal cancers, sputum for lung cancer, vaginal secretions for ovarian/uterine/cervical cancers, urine for renal and urinary bladder malignancies, and potentially feces for other gastrointestinal malignancies.
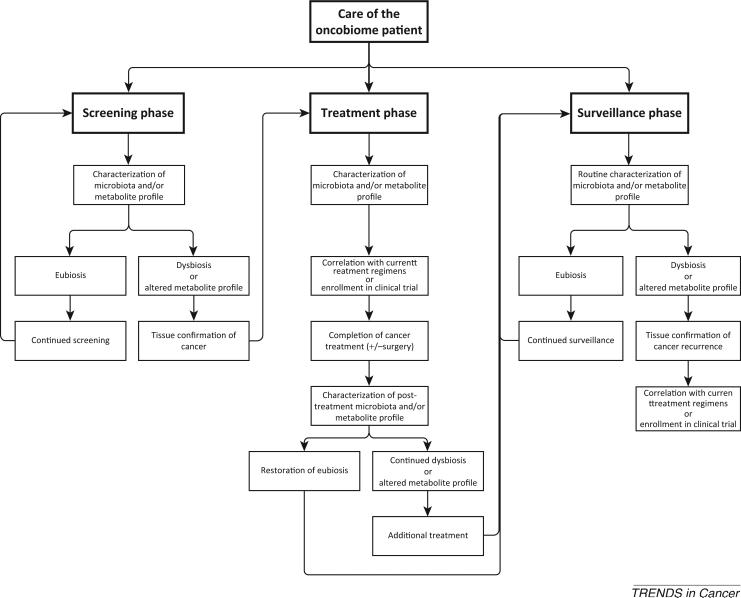
Figure 2. Hypothetical Integration of the Oncobiome into the Care of Cancer Patients.
Each area is divided into screening, treatment, and surveillance phases. Each phase characterizes the microbiota of the patient based on the cancer to be screened/treated (i.e., feces for CRC). Treatment is based upon tested regimens that have demonstrated efficacy with a particular microbiota or metabolite profile. It can be envisaged that patients who have restoration of eubiosis will have improved cancer survival compared to those who maintain or relapse to dysbiosis after treatment.
In the field of oncology, it is the hope that with the screening and diagnosis of cancer will come treatment options. Much effort has focused on individualized medicine, as evidenced by the use of gene arrays and patient-derived tumor xenografts to help guide patient discussions on cancer treatment and recurrence risk [135–140]. Assuming that the host microbiota plays a role in cancer, it too will provide an individualized approach to treatment. The potential influence of the microbiota on drug efficacy has been highlighted, and may have a great impact on future chemotherapy trials [119,123,141,142]. A situation can be envisaged whereby the microbiota of each patient is tested, before starting a chemotherapeutic treatment strategy, to choose the agents that will offer the greatest benefit. In this manner, the oncobiome will enter the arena of personalized medicine for cancer care with limitless possibilities.
Finally, as we gain a greater understanding of host eubiosis and the dysbiosis that occurs in various diseases, it is intriguing to think about restoration of eubiosis after cancer treatment. Typical cancer surveillance involves radiographic imaging which incurs a large financial burden to the patient and the medical community. However, if the curative treatment of cancer results in the restoration of eubiosis, this can be used to the advantage of the medical community for the purpose of cancer surveillance while limiting the use of current modalities. Much in the same way as the postoperative rise of serum carcinoembryonic antigen (CEA) levels may indicate CRC recurrence [143–145], a state of dysbiosis after postoperative restoration of eubiosis may indicate cancer recurrence or risk of recurrence. Tests for dysbiosis could potentially improve or augment the sensitivity and specificity of currently-available serum tumor markers, especially when one considers the fact that commonly used tumor markers have variable sensitivity and specificity for the diseases they aim to detect, are not produced in every clinical scenario by specific tumor types, and can have false positives even in the setting of curative surgery [146–152]. Cancer survival could be modified significantly by identifying patients who ‘relapse’ into a dysbiotic state. Lifestyle, dietary, or pharmacologic modifications could therefore be made to restore eubiosis and mitigate this risk. Alternatively, introduction of an entirely new microbiota could be envisaged, similarly to patients with recurrent Clostridium difficile infection [153].
Concluding Remarks
Despite the fact that there is, as yet, no direct evidence linking the human microbiota to cancer development and progression, the oncobiome field continues to grow rapidly with many unanswered questions (see Outstanding questions). The appeal of microbiota as an active component of diseases and health is too great to ignore, and intense investigation in this field of research, especially cancer, would likely shed light onto this novel paradigm. While some studies demonstrate the association of microbial populations with various cancers, others have begun to interrogate the intricate relationship between the host, its immune system, and its microbiota. Placing added focus on the oncobiome in the context of clinical chemotherapy trials will undoubtedly yield important information with regards to drug metabolism and efficacy. Finally, should elements of the host microbiota prove to have a direct role in cancer development, the implications for cancer screening, treatment, and surveillance are limitless. This dynamic field is only in its infancy, and advancing it will require a concerted effort between the medical and scientific communities to view the mirage as reality.
Trends.
The ‘oncobiome’ is the expanding field of research investigating the role of the microbiota and associated microbiome on human cancer development.
While particular bacteria, such as Escherichia coli, Bacteroides, and Fusobacterium, as well as associated toxins/genotoxins, have been associated with colon cancer development in mouse and human studies, there is no evidence that these microbes or metabolites directly cause cancer.
The oncobiome field is currently limited by studies of microbiota association with cancer, rather than with causation of cancer.
Should the influence of the oncobiome be confirmed, it can be envisaged that the screening, treatment, and surveillance of cancer patients will one day incorporate this research.
Box 1. The Interplay Between Microbes, Inflammation, and Cancer.
Although the majority of data for the interplay between host microbiota, inflammation, and carcinogenesis involve investigations on CRC, many of these same pathways may be applicable to other malignancies, particularly those that have a direct communication to the gastrointestinal tract. Bacteria may exert deleterious effects on their host in several ways, including metabolism of ingested material into toxic metabolites, direct secretion of toxic substances, and promotion of inflammatory pathways. For example, microorganism-associated molecular patterns (MAMPs) are components of the microbe such as lipopolysaccharide (LPS), flagellin, and nucleic acids that are recognized by the host immune system via pattern recognition receptors (PRRs) [154–156]. The best-characterized of these PRRs include Toll-like receptors (TLRs) and Nod-like receptors (NLRs) family [157,158]. Upon binding of MAMPs to these PRRs, various host responses occur that modify immune status. For example, LPS, present as part of the outer membrane on Gram-negative bacteria [154,159], binds to TLR4 [160,161] which upregulates IL-6 [162] and tumor necrosis factor (TNF) production, with subsequent recruitment of mononuclear cells, inhibition of T cell apoptosis, and inhibition of regulatory T cell (Treg) differentiation [32,163]. These events lead to persistent and unchecked inflammation. Furthermore, MAMPs serve to activate Th17 with subsequent upregulation of the proinflammatory cytokine, IL-23, and inhibition of IL-10, an anti-inflammatory cytokine [32]. These proinflammatory cytokines foster the neoplastic cascade by promoting cellular proliferation and inhibiting apoptosis [164,165]. Based on dietary carbohydrate consumption, byproducts of bacterial fermentation lead to the production of short-chain fatty acids (SCFA) which, through various G protein-coupled receptors located on colonic epithelium, activate CD4+ T cells with differentiation into regulatory T cells and the production of the anti-inflammatory cytokines IL-10 and transforming growth factor (TGF)-β [69]. These mediators also serve to inhibit the proinflammatory cytokines IL-6 and TNF. Therefore, in a state of dysbiosis whereby decreased fermentation of carbohydrates into SCFAs leads to a relative decrease in the anti-inflammatory signaling pathways, potentially leaving proinflammatory pathways unchecked. This may further lead to disruption of normal epithelial barriers, resulting in bacterial translocation and further aberrant signaling. Such dysregulation would potentially lead to increased host cellular proliferation, decreased apoptosis, and anchorage-independent growth – all hallmarks of malignant transformation [166].
Outstanding Questions.
What cancer-associated microbiota profiles result in cancer initiation in mice with humanized immune systems?
Does the acquisition of microbes with carcinogenic potential early in life influence cancer development later in life?
Is the carcinogenic activity of particular microbes triggered upon colonization, or is a second ‘hit’ from external factors (diet, environment, chronic inflammation) required?
What is the lead time between colonization with cancer-associated microbes and cancer development?
Can microbe-derived carcinogenic molecules and/or genes be detected in human specimens (stool, urine, saliva), and do they correlate with cancer presence, stage of disease, or response to chemotherapy?
What patient samples are most reflective of the microbiota of a particular organ or cancer? For example, is bile most reflective of hepato-pancreatobiliary malignancies or is stool an appropriate surrogate?
Is dysbiosis a hallmark of particular cancers compared to healthy controls, and is restoration of eubiosis after treatment for cancer an indicator of improved survival?
Can alteration of the microbiota of individuals at high-risk for cancer development mitigate their risk?
Glossary
- Commensal
describes the relationship between two organisms in which one benefits without affecting, or itself being beneficial, to the second organism.
- Dysbiosis
a state whereby the microbial composition of the host is unbalanced or skewed toward particular microorganisms as compared to the composition of a ‘healthy’ host.
- Eubiosis
a state whereby the microbial composition of the host is of a normal proportion that is typically found in ‘healthy’ individuals.
- Germ-free (GF)
refers to animals conceived, born, and raised in a sterile environment and thus lack any microorganisms (except endogenous viruses).
- Metabolomic
the study of the specific metabolites produced by microbes, either individually or collectively as part of the host microbiota.
- Oncobiome
the intricate interplay and study of the human microbiome and its influence on cancer development.
- Pathobiont
microorganisms that normally behave in a symbiotic manner with their host but exhibit pathogenic potential based on changes in their abundance or environmental conditions.
- Planktonic
microbes that exist as single cells in a free-floating environment that are typically fast-growing and susceptible to environmental influences/drugs, as opposed to microbes in a biofilm which are slower-growing communities of adherent bacteria that are more tolerant of environmental influences.
References
- 1.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajilic-Stojanovic M, et al. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 4.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sommer F, Backhed F. The gut microbiota – masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 7.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 8.Peek RM, Jr, Crabtree JE. Helicobacter infection and gastric neoplasia. J. Pathol. 2006;208:233–248. doi: 10.1002/path.1868. [DOI] [PubMed] [Google Scholar]
- 9.Hardbower DM, et al. Chronic inflammation and oxidative stress: the smoking gun for Helicobacter pylori-induced gastric cancer? Gut Microbes. 2013;4:475–481. doi: 10.4161/gmic.25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koeppel M, et al. Helicobacter pylori infection causes characteristic DNA damage patterns in human cells. Cell Rep. 2015;11:1703–1713. doi: 10.1016/j.celrep.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Wroblewski LE, Peek RM., Jr Helicobacter pylori in gastric carcinogenesis: mechanisms. Gastroenterol. Clin. North Am. 2013;42:285–298. doi: 10.1016/j.gtc.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.IARC Working Group . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans (Vol. 61) Schistosomes, Liver Flukes, and Helicobacter pylori. WHO International Agency for Research on Cancer; 1994. Infection with Helicobacter pylori. pp. 177–240. [Google Scholar]
- 13.de Martel C, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 14.Wong BC, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291:187–194. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 15.Pan KF, et al. A large randomised controlled intervention trial to prevent gastric cancer by eradication of Helicobacter pylori in Linqu County, China: baseline results and factors affecting the eradication. Gut Published online May. 2015;18:2015. doi: 10.1136/gutjnl-2015-309197. PMID: 25986943. [DOI] [PubMed] [Google Scholar]
- 16.Ford AC, et al. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: systematic review and meta-analysis of randomised controlled trials. BMJ. 2014;348:g3174. doi: 10.1136/bmj.g3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn J, et al. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchesi JR, et al. Towards the human colorectal cancer microbiome. PLoS ONE. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy AN, et al. Fusobacterium is associated with colorectal adenomas. PLoS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanapareddy N, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobhani I, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu N, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 27.Russell W. An address on a characteristic organism of cancer. Br. Med. J. 1890;2:1356–1360. doi: 10.1136/bmj.2.1563.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuerthele-Caspe V, et al. Cultural properties and pathogenicity of certain microorganisms obtained from various proliferative and neoplastic diseases. Am. J. Med. Sci. 1950;220:638–646. doi: 10.1097/00000441-195022060-00006. [DOI] [PubMed] [Google Scholar]
- 29.Aries V, et al. Bacteria and the aetiology of cancer of the large bowel. Gut. 1969;10:334–335. doi: 10.1136/gut.10.5.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 2015 doi: 10.1038/nrclinonc.2015.105. Published online June 30, 2015. http://dx.doi.org/10.1038/nrclinonc.2015.105. [DOI] [PubMed]
- 31.Mantovani A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 32.Elinav E, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 33.Punkenburg E, et al. Batf-dependent Th17 cells critically regulate IL-23 driven colitis-associated colon cancer. Gut. 2015 doi: 10.1136/gutjnl-2014-308227. Published online April 2, 2015. http://dx.doi.org/10.1136/gutjnl-2014-308227. [DOI] [PubMed]
- 34.Tiniakou I, et al. High-density lipoprotein attenuates Th1 and Th17 autoimmune responses by modulating dendritic cell maturation and function. J. Immunol. 2015;194:4676–4687. doi: 10.4049/jimmunol.1402870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipson EJ, Drake CG. Ipilimumab: an anti-CTLA-4 antibody for metastatic melanoma. Clin. Cancer Res. 2011;17:6958–6962. doi: 10.1158/1078-0432.CCR-11-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roshani R, et al. Inflammatory cytokines in human pancreatic cancer. Cancer Lett. 2014;345:157–163. doi: 10.1016/j.canlet.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 39.Ebrahimi B, et al. Cytokines in pancreatic carcinoma: correlation with phenotypic characteristics and prognosis. Cancer. 2004;101:2727–2736. doi: 10.1002/cncr.20672. [DOI] [PubMed] [Google Scholar]
- 40.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin. Immunopathol. 2013;35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooper LV, et al. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 45.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grivennikov SI, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen XJ, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jobin C. Colorectal cancer: looking for answers in the microbiota. Cancer Discov. 2013;3:384–387. doi: 10.1158/2159-8290.CD-13-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeller G, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zackular JP, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dejea CM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U.S. A. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson CH, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015;21:891–897. doi: 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnenburg JL, et al. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 55.Villanueva MT. Metabolism: bacterial biofilms may feed colon cancer. Nat. Rev. Cancer. 2015;15:320. [Google Scholar]
- 56.Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 57.Reddy BS, et al. Colon carcinogenesis with azoxymethane and dimethylhydrazine in germ-free rats. Cancer Res. 1975;35:287–290. [PubMed] [Google Scholar]
- 58.Sacksteder MR. Occurrence of spontaneous tumors in the germfree F344 rat. J. Natl. Cancer Inst. 1976;57:1371–1373. doi: 10.1093/jnci/57.6.1371. [DOI] [PubMed] [Google Scholar]
- 59.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, et al. Gut microbiota accelerate tumor growth via cjun and STAT3 phosphorylation in APC[r]Min/+[/r] mice. Carcinogenesis. 2012;33:1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 61.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh S, et al. Comparative efficacy of pharmacologic interventions in preventing relapse of Crohn's disease after surgery: a systematic review and network meta-analysis. Gastroenterology. 2015;148:64–76. doi: 10.1053/j.gastro.2014.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kostic AD, et al. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu XW, et al. Meta-analysis of ciprofloxacin in treatment of Crohn's disease. Biomed. Rep. 2015;3:70–74. doi: 10.3892/br.2014.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhan Y, et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 2013;73:7199–7210. doi: 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwabe RF, Jobin C. The microbiome and cancer. Nat. Rev. Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol. Rev. Camb. Philos. Soc. 2012;87:701–730. doi: 10.1111/j.1469-185X.2012.00218.x. [DOI] [PubMed] [Google Scholar]
- 68.Chen GY, et al. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Louis P, et al. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 70.Macfarlane G, Gibson G. In Gastrointestinal Microbiology. Chapman and Hall; 1997. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. pp. 269–318. [Google Scholar]
- 71.Weaver GA, et al. Short chain fatty acid distributions of enema samples from a sigmoidoscopy population: an association of high acetate and low butyrate ratios with adenomatous polyps and colon cancer. Gut. 1988;29:1539–1543. doi: 10.1136/gut.29.11.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weir TL, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh N, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donohoe DR, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4:1387–1397. doi: 10.1158/2159-8290.CD-14-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J. Nutr. 2004;134:479–482. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 76.Belcheva A, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 77.Ohland CL, Jobin C. Bugs and food: a recipe for cancer? Cell Metab. 2014;20:937–938. doi: 10.1016/j.cmet.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freeman HJ. Effects of differing concentrations of sodium butyrate on 1,2-dimethylhydrazine-induced rat intestinal neoplasia. Gastroenterology. 1986;91:596–602. doi: 10.1016/0016-5085(86)90628-1. [DOI] [PubMed] [Google Scholar]
- 79.Flint HJ, et al. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 80.Pomare EW, et al. Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J. Clin. Invest. 1985;75:1448–1454. doi: 10.1172/JCI111847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Windey K, et al. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012;56:184–196. doi: 10.1002/mnfr.201100542. [DOI] [PubMed] [Google Scholar]
- 82.Hughes R, Rowland IR. Metabolic activities of the gut microflora in relation to cancer. Microb. Ecol. Health Dis. 2000;2000(Suppl. 2):179–185. [Google Scholar]
- 83.Di Martino ML, et al. Polyamines: emerging players in bacteria–host interactions. Int. J. Med. Microbiol. 2013;303:484–491. doi: 10.1016/j.ijmm.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 84.Pegg AE. Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol. 2013;26:1782–1800. doi: 10.1021/tx400316s. [DOI] [PubMed] [Google Scholar]
- 85.Attene-Ramos MS, et al. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 86.Magee EA, et al. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 2000;72:1488–1494. doi: 10.1093/ajcn/72.6.1488. [DOI] [PubMed] [Google Scholar]
- 87.Loh YH, et al. N-Nitroso compounds and cancer incidence: the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am. J. Clin. Nutr. 2011;93:1053–1061. doi: 10.3945/ajcn.111.012377. [DOI] [PubMed] [Google Scholar]
- 88.Khor B, et al. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nestle FO, et al. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 2009;9:679–691. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pabst O. New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 91.Salzman NH, et al. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 92.Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp. Biol. Med. 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 93.Cross AJ, et al. A prospective study of serum metabolites and colorectal cancer risk. Cancer. 2014;120:3049–3057. doi: 10.1002/cncr.28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cross AJ, et al. Metabolites of tobacco smoking and colorectal cancer risk. Carcinogenesis. 2014;35:1516–1522. doi: 10.1093/carcin/bgu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guertin KA, et al. Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer. Am. J. Clin. Nutr. 2015;101:1000–1011. doi: 10.3945/ajcn.114.096099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marcobal A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, et al. Metagenomic and metabolomic analysis of the toxic effects of trichloroacetamide-induced gut microbiome and urine metabolome perturbations in mice. J. Proteome Res. 2015;14:1752–1761. doi: 10.1021/pr5011263. [DOI] [PubMed] [Google Scholar]
- 98.Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 99.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cuevas-Ramos G, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bonnet M, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 102.Buc E, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE. 2013;8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cougnoux A, et al. Small-molecule inhibitors prevent the genotoxic and protumoural effects induced by colibactin-producing bacteria. Gut. 2015 doi: 10.1136/gutjnl-2014-307241. Published online January 14, 2015. http://dx.doi.org/10.1136/gutjnl-2014-307241. [DOI] [PubMed]
- 104.Baxter NT, et al. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. PMID: 24967088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Payros D, et al. Maternally acquired genotoxic Escherichia coli alters offspring's intestinal homeostasis. Gut Microbes. 2014;5:313–325. doi: 10.4161/gmic.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fostira F, et al. Prevalence of BRCA1 mutations among 403 women with triple-negative breast cancer: implications for genetic screening selection criteria: a Hellenic Cooperative Oncology Group Study. Breast Cancer Res. Treat. 2012;134:353–362. doi: 10.1007/s10549-012-2021-9. [DOI] [PubMed] [Google Scholar]
- 108.Rebbeck TR, et al. Modification of BRCA1-associated breast and ovarian cancer risk by BRCA1-interacting genes. Cancer Res. 2011;71:5792–5805. doi: 10.1158/0008-5472.CAN-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van der Post RS, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J. Med. Genet. 2015;52:361–374. doi: 10.1136/jmedgenet-2015-103094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guilford P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 111.Chun N, Ford JM. Genetic testing by cancer site: stomach. Cancer J. 2012;18:355–363. doi: 10.1097/PPO.0b013e31826246dc. [DOI] [PubMed] [Google Scholar]
- 112.Choi PM, Zelig MP. Similarity of colorectal cancer in Crohn's disease and ulcerative colitis: implications for carcino-genesis and prevention. Gut. 1994;35:950–954. doi: 10.1136/gut.35.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Endo A, et al. Long-term monitoring of the human intestinal microbiota from the 2nd week to 13 years of age. Anaerobe. 2014;28:149–156. doi: 10.1016/j.anaerobe.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 114.Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rautava S, et al. Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 2012;9:565–576. doi: 10.1038/nrgastro.2012.144. [DOI] [PubMed] [Google Scholar]
- 116.Arthur JC, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boleij A, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ando K, et al. Humanizing bone marrow in immune-deficient mice. Curr. Top. Microbiol. Immunol. 2008;324:77–86. doi: 10.1007/978-3-540-75647-7_4. [DOI] [PubMed] [Google Scholar]
- 121.Werner-Klein M, et al. Immune humanization of immunodeficient mice using diagnostic bone marrow aspirates from carcinoma patients. PLoS ONE. 2014;9:e97860. doi: 10.1371/journal.pone.0097860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Justino PF, et al. Treatment with Saccharomyces boulardii reduces the inflammation and dysfunction of the gastrointestinal tract in 5-fluorouracil-induced intestinal mucositis in mice. Br. J. Nutr. 2014;111:1611–1621. doi: 10.1017/S0007114513004248. [DOI] [PubMed] [Google Scholar]
- 123.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Perez-Chanona E, Jobin C. From promotion to management: the wide impact of bacteria on cancer and its treatment. Bioessays. 2014;36:658–664. doi: 10.1002/bies.201400015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hardcastle JD, et al. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348:1472–1477. doi: 10.1016/S0140-6736(96)03386-7. [DOI] [PubMed] [Google Scholar]
- 126.Kronborg O, et al. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467–1471. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]
- 127.Mandel JS, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N. Engl. J. Med. 1993;328:1365–1371. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 128.Ned RM, et al. Fecal DNA testing for colorectal cancer screening: the ColoSure test. PLoS Curr. 2011;3:RRN1220. doi: 10.1371/currents.RRN1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ahlquist DA, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann. Intern. Med. 2008;149:441–450. W481. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Imperiale TF, et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 131.Narayanan V, et al. Human fecal microbiome-based biomarkers for colorectal cancer. Cancer Prev. Res. 2014;7:1108–1111. doi: 10.1158/1940-6207.CAPR-14-0273. [DOI] [PubMed] [Google Scholar]
- 132.Flint HJ. The impact of nutrition on the human micro-biome. Nutr. Rev. 2012;70(Suppl. 1):S10–S13. doi: 10.1111/j.1753-4887.2012.00499.x. [DOI] [PubMed] [Google Scholar]
- 133.Scott KP, et al. The influence of diet on the gut micro-biota. Pharmacol. Res. 2013;69:52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 134.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kraus S, et al. Recent advances in personalized colorectal cancer research. Cancer Lett. 2014;347:15–21. doi: 10.1016/j.canlet.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 136.Sivanand S, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci. Transl. Med. 2012;4:137ra175. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stebbing J, et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120:2006–2015. doi: 10.1002/cncr.28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Weroha SJ, et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin. Cancer Res. 2014;20:1288–1297. doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.You YN, et al. Oncotype DX colon cancer assay for prediction of recurrence risk in patients with stage II and III colon cancer: a review of the evidence. Surg. Oncol. 2015 doi: 10.1016/j.suronc.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 140.Thomas RM, et al. The canary in the coal mine: the growth of patient-derived tumorgrafts in mice predicts clinical recurrence after surgical resection of pancreatic ductal adeno-carcinoma. Ann. Surg. Oncol. 2015;22:1884–1892. doi: 10.1245/s10434-014-4241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee JR, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS ONE. 2015;10:e0122399. doi: 10.1371/journal.pone.0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Catry E, et al. Ezetimibe and simvastatin modulate gut microbiota and expression of genes related to cholesterol metabolism. Life Sci. 2015;132:77–84. doi: 10.1016/j.lfs.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 143.Desch CE, et al. Colorectal cancer surveillance: 2005 update of an American Society of Clinical Oncology practice guideline. J. Clin. Oncol. 2005;23:8512–8519. doi: 10.1200/JCO.2005.04.0063. [DOI] [PubMed] [Google Scholar]
- 144.Primrose JN, et al. Effect of 3 to 5 years of scheduled CEA and CT follow-up to detect recurrence of colorectal cancer: the FACS randomized clinical trial. JAMA. 2014;311:263–270. doi: 10.1001/jama.2013.285718. [DOI] [PubMed] [Google Scholar]
- 145.Locker GY, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 146.Palmqvist R, et al. Prediagnostic levels of carcinoembryonic antigen and CA 242 in colorectal cancer: a matched case-control study. Dis. Colon Rectum. 2003;46:1538–1544. doi: 10.1007/s10350-004-6810-z. [DOI] [PubMed] [Google Scholar]
- 147.Sorbye H, Dahl O. Carcinoembryonic antigen surge in metastatic colorectal cancer patients responding to oxaliplatin combination chemotherapy: implications for tumor marker monitoring and guidelines. J. Clin. Oncol. 2003;21:4466–4467. doi: 10.1200/JCO.2003.99.200. [DOI] [PubMed] [Google Scholar]
- 148.Ballesta AM, et al. Carcinoembryonic antigen in staging and follow-up of patients with solid tumors. Tumour Biol. 1995;16:32–41. doi: 10.1159/000217926. [DOI] [PubMed] [Google Scholar]
- 149.Litvak A, et al. False-positive elevations of carcinoembryonic antigen in patients with a history of resected colorectal cancer. J. Natl. Compr. Canc. Netw. 2014;12:907–913. doi: 10.6004/jnccn.2014.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tempero MA, et al. Relationship of carbohydrate antigen 19-9 and Lewis antigens in pancreatic cancer. Cancer Res. 1987;47:5501–5503. [PubMed] [Google Scholar]
- 151.Zhang B, Yang B. Combined alpha fetoprotein testing and ultrasonography as a screening test for primary liver cancer. J. Med. Screen. 1999;6:108–110. doi: 10.1136/jms.6.2.108. [DOI] [PubMed] [Google Scholar]
- 152.Farinati F, et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am. J. Gastroenterol. 2006;101:524–532. doi: 10.1111/j.1572-0241.2006.00443.x. [DOI] [PubMed] [Google Scholar]
- 153.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 154.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 156.Alexopoulou L, et al. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 157.Moresco EM, et al. Toll-like receptors. Curr. Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 158.Elinav E, et al. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 159.Galanos C, et al. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 1985;148:1–5. doi: 10.1111/j.1432-1033.1985.tb08798.x. [DOI] [PubMed] [Google Scholar]
- 160.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 161.Poltorak A, et al. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol. Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 162.Schromm AB, et al. The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 1998;161:5464–5471. [PubMed] [Google Scholar]
- 163.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 164.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- 165.Langowski JL, et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 166.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]