Abstract
Potential for interspecies hydrogen transfer within paddy soil enrichments obtained via addition of magnetite nanoparticles and ethanol (named as PEM) was investigated. To do this, PEM derived from rice field of Hangzhou (named as PEM-HZ) was employed, because it offered the best methane production performance. Methane production and Fe (III) reduction proceeded in parallel in the presence of magnetite. Inhibition experiments with 2-bromoethane sulfonate (BES) or phosphate showed that interspecies hydrogen transfer and Fe (III) reduction also occurred in methane production from ethanol. 16S rRNA-based Illumina sequencing results showed that Dechloromonas, Thauera, Desulfovibrio and Clostridium were the dominant putative Fe (III) -reducers, and that hydrogenotrophic Methanobacterium accounted for about 88% of the total archaeal community. These results indicated that magnetite nanoparticles that acted as electron acceptor could facilitate rapid oxidation of ethanol by members of the Fe (III) -reducers in PEM-HZ and establishment of the syntrophic relationship of Fe (III) -reducers with Methanobacterium via interspecies hydrogen transfer. Our results could offer a model to understand the microbial interaction with magnetite from a novel angle during methanogenesis.
Ethanol is one of the intermediates from anaerobic degradation of organic substrates. The conversion of ethanol to methane depends on effective interspecies electron transfer (IET) between ethanol oxidizers and methanogens1. Two mechanisms for IET have been reported. The former was IET via hydrogen, in which ethanol oxidizers metabolize ethanol and reduce protons to generate H2 and the methanogens utilize hydrogen to reduce CO21. The latter was direct IET, in which the methanogens can directly use electrons releasing from ethanol oxidizers for reducing of CO2 to methane through biological electrical connections2.
Magnetite nanoparticles, one of the most common minerals in the soil, are able to facilitate microbial extracellular electron transfer3,4. This biochemical reaction has strong impact on the conversion of ethanol to methane. Data have revealed that the addition of magnetite to paddy soil had a stimulatory effect on ethanol degradation and methanogenesis, concomitant with the enrichment Fe (III)-reducer Geobacter and methanogen Methanosarcina5. In this case, the researchers speculated that magnetite could act as a conduit of electrons to facilitate direct IET between Geobacter and Methanosarcina, providing a rapid methane production.
However, besides Geobacter, iron oxides added to paddy soil resulted in the enrichment of other Fe (III)-reducing bacteria, such as Clostridium6,7,8. Existing evidence has shown that Clostridium can utilize acetate to reduce Fe (III) in the supplementary akaganeite slurry, and produce significant amounts of H2 which was subsequently consumed9. Although a significant abundance of Clostridia were also detected in the magnetite-added paddy soil, researchers focused on only direct IET, without much consideration for interspecies hydrogen transfer5. H2 might be produced and subsequently converted to methane by Methanosarcina, which was able to perform hydrogenotrophic methanogenesis10. In our preliminary experiment significant H2 production and Clostridia were detected in paddy soil amended with magnetite and ethanol. Additionally, in syntrophic methanogenesis, ethanol oxidation is thermodynamically unfavorable (ΔG0′=+9.6 kJ/mol) and can occur only when a very low H2 pressure is kept by hydrogen-scavenging methanogens11.
Thus, in the light of this context, we hypothesized that magnetite could facilitate methane production from ethanol in other manners in addition to acting as a conduit of electrons. To test this hypothesis, paddy soil enrichments obtained in the presence of magnetite nanoparticles and ethanol (named as PEM) were firstly established in this work. Using the resulted enrichments, we subsequently determined metabolites, Fe (III) reduction, and microbial community structure. Through inhibitor experiments, we then probed the methanogenic mechanisms.
Results
Paddy soil enrichments
To investigate the effects of magnetite on methane production from ethanol, PEM was firstly established. The methanogenic characteristics of paddy soil enrichments from different rice filed (GZ, HZ, and TZ) were evaluated. As shown in Supplementary Figure S1 and Table 1, magnetite added to paddy soil led to a significant increase in the maximum methane production rate and a significant reduction in the lag-phase time compared with paddy soil enrichments obtained in the absence of magnetite nanoparticles (named as PEC). PEM-HZ was selected as inoculum for subsequent semi-continuous enrichment cultivations because PEM-HZ showed the shortest lag-phase time with comparable maximum methane production rate.
Table 1. Kinetic parameters for methane production.
| Group | Ps (mol CH4/mol EtOH) | Rm (mL/d) | λ(d) | R2 |
|---|---|---|---|---|
| PEC-GZ | 1.45 | 1.3 | 19.9 | 0.99 |
| PEM-GZ | 1.65 | 2.0 | 9.3 | 0.99 |
| PEC-HZ | 1.52 | 1.9 | 26.3 | 0.96 |
| PEM-HZ | 1.64 | 4.5 | 8.1 | 0.99 |
| PEC-TZ | 1.39 | 2.1 | 29.2 | 0.99 |
| PEM-TZ | 1.59 | 4.5 | 9.2 | 0.99 |
Methane production during semi-continuous enrichment cultivation was shown in Supplementary Figure S2 and Table 2. As for each generation of enrichments, the maximum methane production rate of PEM-HZ was higher than that of PEC-HZ. The lag-phase time of methane production in PEM-HZ showed a reduction that was always shorter than that in the corresponding PEC-HZ. However, the methane yield and maximum production rate of PEC-HZ showed a declined tendency with the increase of transfers. Metabolite analysis showed that ethanol were not efficiently removed in PEC-HZ at each generation (Table 3). Interestingly, Table 3 also illustrates that considerable amounts of volatile fatty acids (VFAs) (e.g. butyrate and caproate) were accumulated in PEC-HZ that were higher than those in the corresponding PEM-HZ at each generation. The accumulation of VFAs in PEC-HZ indicated that parts of carbon flux of ethanol might shift towards carboxylic acid production rather than methane production due to the decreased methanogen populations (Fig. 1b). In PEC-HZ,0.07 mmol and 0.01 mmol of H2 were produced and subsequently consumed during generation 1 and 2, respectively. It has been reported that anaerobic biotransformation of ethanol can generate acetate, propionate or butyrate12, and that consumption of H2 and acetate can generate caproate13. Thus, it was likely that parts of ethanol was converted to the carboxylic acid in PEC-HZ.
Table 2. Methane yield, methane production rate and lag-phase time in the semi-continuous subculture.
| Ps (mol CH4/mol ethanol) | Rm (mmol/d) | λ(d) | R2 | |
|---|---|---|---|---|
| PEC-HZ | ||||
| Start-up | 1.52 | 0.08 | 26.3 | 0.96 |
| Generation l | 0.29 | 0.004 | 4.9 | 0.95 |
| Generation 2 | 0.38 | 0.01 | 8.9 | 0.99 |
| PEM-HZ | ||||
| Start-up | 1.64 | 0.20 | 8.1 | 0.99 |
| Generation l | 1.52 | 0.11 | 3.0 | 0.99 |
| Generation 2 | 1.59 | 0.07 | 0.9 | 0.99 |
Table 3. Metabolite analysis in the semi-continuous subculture.
| Ethanol (mM) | Acetate (mM) | Propionate (mM) | Butyrate (mM) | Caproate (mM) | |
|---|---|---|---|---|---|
| PEC-HZ | |||||
| Start-up | 0.64 ± 0.10 | 1.26 ± 0.08 | 0.26 ± 0.02 | 0.11 ± 0.08 | 0.43 ± 0.18 |
| Generation l | 8.91 ± 4.64 | 5.28 ± 0.44 | 2.07 ± 0.23 | 1.48 ± 0.09 | 1.10 ± 0.08 |
| Generation 2 | 7.55 ± 0.07 | 1.31 ± 0.10 | 0.02 ± 0.00 | 8.30 ± 0.30 | 4.72 ± 0.00 |
| PEM-HZ | |||||
| Start-up | 0.59 ± 0.34 | 0.72 ± 0.29 | 0.37 ± 0.16 | 0.15 ± 0.10 | 0.37 ± 0.27 |
| Generation l | 0 | 5.84 ± 2.55 | 2.46 ± 1.76 | 0.54 ± 0.51 | 0.80 ± 0.33 |
| Generation 2 | 0 | 5.06 ± 0.36 | 0.65 ± 0.54 | 0.21 ± 0.05 | 0 |
Figure 1. Taxonomic classification of bacteria and communities.
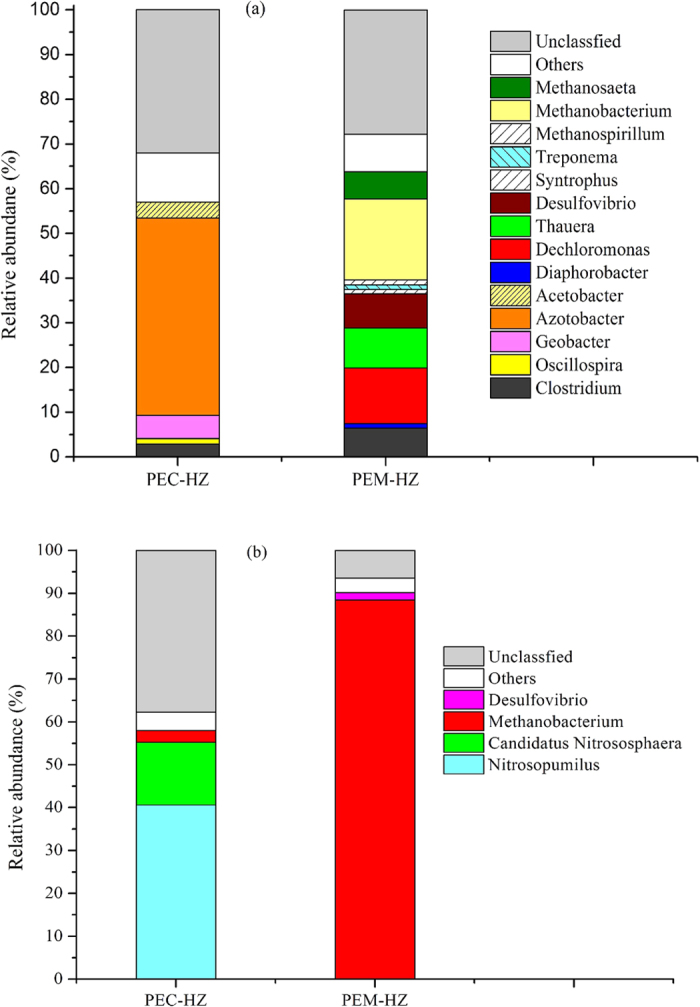
The taxonomy of OTU representative sequences was phylogenetically assigned to taxonomic classifications using RDP Classifier and Greengenes with a confidence threshold of 80%. Genus accounting for less than 1% of total composition were classified as “others”. (a) Bacteria; and (b) Archaea.
Structure of microbial communities
The microbial diversities of the paddy soil enrichments from the second generation were analyzed. A small portion of archaeal and bacterial sequences were recovered, respectively (Fig. 1), due to non-specificity of primer pairs targeting V4 region of bacterial and archaeal 16S rRNA genes. Results from Supplementary Table S2 and Figure S3 showed that magnetite added to the paddy soil altered the structure of microbial communities.
High-throughput Illumina sequencing analysis showed clear differences in communities between PEM-HZ and PEC-HZ. Figure 1a shows the phylogenetic classification of bacterial OTUs at the genus level. 32% (PEC-HZ) and 27.8% (PEM-HZ) of the total reads were not classified at the genus level, indicating that these bacteria are unknown. Dechloromonas, Thauera, Desulfovibrio and Clostridium in PEM-HZ accounted for 12.4%, 9%, 7.6% and 6.4% of total bacterial sequences, respectively, while those of PEC-HZ were 0.2%, 0.1%, 0.1% and 2.93%. However, a higher percentage of Azotobacter, Geobacter and Acetobacter was observed in PEC-HZ compared with those in PEM-HZ. Azotobacter remained as the most abundant (44.2%) in PEC-HZ but decreased to 0.6% in PEM-HZ. Members of Geobacter and Acetobacter accounted for 5.3% and 3.5% of the bacterial reads sequenced in PEC-HZ, respectively, but the percentage of those in PEM-HZ decreased to 0.7% and 0.03%. Acetobacter can transform ethanol to acetate14, and Azotobacter can utilize acetate to support growth metabolism15,16. Members of Azotobacter genus in PEC-HZ may utilize acetate produced by Acetobacter to maintain their growth. Minor genera (relative abundance was less than or equal to 1%) in both two groups were Oscillospira, Treponema, Syntrophus and Diaphorobacter, but their role in ethanol degradation remained unclear in this work.
Archaeal community analysis showed that Methanobacterium accounted for 88.4% in PEM-HZ, while the dominant species for PEC-HZ was Nitrosopumilus (40.6%) (Fig. 1b). Only 2.8% of Methanobacterium was detected in PEC-HZ. Nitrosopumilus has been described as the ammonia-oxidizing archaea17, indicating that the ammonia-oxidizing process may occur in PEC-HZ. In addition, 6.1% of acetoclastic methanogen Methanosaeta in PEM-HZ was recovered with the bacterial primer set (Fig. 1a).
It should be noted that the maximum methane production rate and methane yield of PEC-HZ significantly declined compared to start-up in the semi-continuous subculture (Table 2). This may be due to the declined Methanobacterium populations. It was possible that methanogens were washed out after two generations of subculture. The decreased Methanobacterium populations cannot outcompete the dominant species (e.g. Azotobacter, Geobacter, Acetobacter and Clostridium) in terms of competition of substrate (e.g. ethanol, acetate and H2), causing the suppression of methanogenesis due to competitive exclusion.
Intermediate metabolites
If methane is produced via intermediates, intermediates might be detected and increase with time due to product accumulation after methanogens are inhibited by inhibitors. Thus, both 2-bromoethane sulfonate (BES) and phosphate were used to determine the most probable methanogenic pathways. Figure 2 shows that 0.035 mmol H2 was produced and then consumed in PEM-HZ control group (Group I). In group III, BES completely inhibited methane production by PEM-HZ, accompanied with significant amounts of H2. Besides small amounts of methane, considerable amounts of hydrogen were produced in group II, indicating that hydrogenotrophic methanogenesis might be partially inhibited by 100 mM phosphate. This was not in agreement with Conrad′s observation18, which showed that phosphate can specifically inhibit acetoclastic methanogenesis rather than hydrogenotrophic methanogenesis. Additionally, although Fe (II) concentrations throughout the incubations were below 0.2 mM in three groups (Supplementary Figure S4), X-ray diffraction (XRD) analysis showed that magnetite was transformed into Fe (II)3(PO4)2 at the end of incubation (Fig. 3), indicating that the best parts of Fe (III) in magnetite were reduced to Fe (II).
Figure 2. Methane and hydrogen production in the presence of inhibitor.
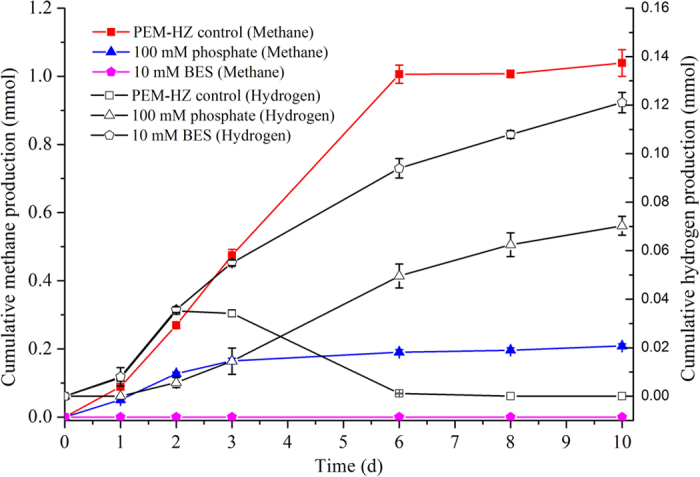
The second generation of PEM-HZ was used as inocula. The inhibitor was added at the beginning of incubation. The addition of BES (10 mM) or phosphate (100 mM) resulted in a significant decrease in methane production and an increase in hydrogen production.
Figure 3. The X ray diffraction spectra of the formed Fe (II)3(PO4)2 at the end of incubation.
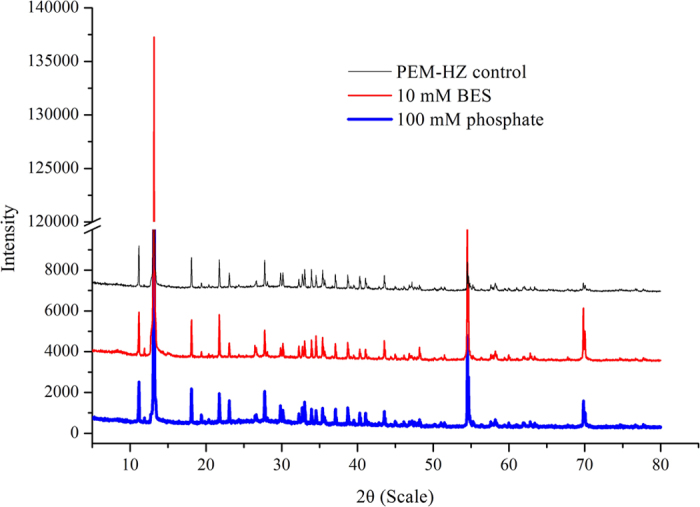
Magnetite was transformed into Fe (II)3(PO4)2, the pattern of which was consistent with the standard value of Fe (II)3(PO4)2 • 8H2O (00-030-0662).
Besides H2, volatile fatty acids (VFAs) (e.g. acetate, propionate and butyrate) were detected (Supplementary Figure S5), while no formate was detected in three groups. On one hand, acetate was accumulated to maximum concentrations of 3.2 mM, and subsequently consumed in group I. In group II and III, acetate was accumulated to maximum concentrations of 2.89 to 6.92 mM at the end of incubation (Supplementary Figure S3). On the other hand, propionate (1–1.52 mM) and n-butyrate (0.5–0.76 mM) accumulated at early stage of incubation in three groups, indicating that methanogenesis and net accumulation of propionate and n-butyrate may proceed in parallel. The produced propionate and n-butyrate were completely depleted at the end of the incubation in three groups. Comparative analysis showed that these acids were further metabolized to acetate in group II and III.
Discussion
The results showed that magnetite addition to paddy soil altered the microbial ecosystem, accelerated methane production, and led to the dominance of hydrogenotrophic methanogenesis. In this work, magnetite that acted as electron acceptor could facilitate methane production from ethanol by PEM-HZ. This presented an apparent discrepancy with literature reports5, which have shown that magnetite that could act as a conduit of electrons facilitated methanogenesis in paddy soil amended with ethanol. This discrepancy might be attributed to the difference in communities between this work and the previous reports. Although Geobacter can establish a syntrophic relationship with Methanosaeta for converting ethanol to methane via direct IET2, only 0.65% of Geobacter and 6.1% of Methanosaeta were recovered, suggesting that DIET in Geobacter might not be the predominant mechanism for methanogenesis in PEM-HZ. Members of Methanosaeta genus detected in PEM-HZ might be involved in conversion of acetate to methane as revealed by inhibition experiment with phosphate (Fig. 2 and Supplementary Figure S5).
In the presence of magnetite nanoparticles, Fe (II) formation and methane production were detected (Figs 2 and 3). The Fe (III) reduction was not inhibited by inhibitors (Fig. 3 and Supplementary Figure S4), suggesting that methanogens did not participate in Fe (III) reduction. 16S rRNA-based Illumina sequencing results from PEM-HZ revealed that Dechloromonas, Thauera, Desulfovibrio and Clostridium were the most abundant bacterial species. These dominant species have been demonstrated to reduce Fe (III) of iron oxides via respiratory6,7,8,19,20,21. Thus, these species detected in PEM-HZ indicated a strong possibility that they were closely related to reducing Fe (III) of magnetite. It is noteworthy that these Fe (III)-reducers may act as competitors of electron donors (e.g. ethanol and acetate) with Geobacter. Geobacter cannot outcompete the dominant Fe (III)-reducers in terms of competition of electron donors. Thus, only a small amount of Geobacter (0.65%) was enriched due to competitive exclusion in PEM-HZ. Additionally, archaeal community analysis for PEM-HZ revealed that Methanobacterium was dominant species, which use H2 to reduce CO2 and produce methane22. These findings showed the possibility that magnetite facilitated the establishment of IET between Fe (III)-reducing bacteria and methanogens based on interspecies hydrogen transfer.
Inhibition experiments showed that the degradation of ethanol occurred, accompanied with the production of H2 in the presence of BES or phosphate. This further indicated that methane was likely produced via interspecies hydrogen transfer. It is known that syntrophic oxidation of ethanol becomes thermodynamically favorable reaction only when H2 concentration must be maintained very low by hydrogenotrophic methanogens1. However, in the presence of magnetite, significant amounts of H2 and Fe (II) were formed in group II and III. This suggested that magnetite acted as electron acceptor may facilitated syntrophic ethanol oxidation in a manner similar to Jiang’s report9, which observed that considerable amounts of hydrogen gas were produced by acetate oxidizing and Fe (III)-reducing bacteria in the presence of akaganeite and hypothesized that when Fe (III) acted as the electron acceptor, anaerobic acetate-oxidation was an easy oxidation access to acetate with production of H2.
Therefore, these observations, combined with the observation that methane production and Fe (III) reduction proceeded in parallel in group I, suggested that, magnetite nanoparticles that acted as electron acceptor could facilitate rapid oxidation of ethanol by members of the Fe (III)-reducing bacteria in PEM-HZ and establishment of the syntrophic relationship of Fe (III)-reducers with Methanobacterium via interspecies H2 transfer. Similar mechanism might also occur in conversion of the intermediates (e.g. propionate, n-butyrate and caproate) of ethanol degradation to methane. This may result in a rapid methanogenic reaction as observed in this work.
Our results could offer a model to understand the microbial interaction with magnetite from a novel angle during methanogenesis.
Methods
Paddy soil sample and preparation of magnetite nanoparticles
Three paddy soil samples were collected from Guangzhou (GZ), Hangzhou (HZ) and Taizhou (TZ), China, respectively. Their characteristics were listed in Table 4. Magnetite nanoparticles (8–10 nm) were synthesized according to Kang′s methods23, and identified via X-ray diffraction (XRD) analysis.
Table 4. The characteristic of paddy soil sample.
| Inoculum source | Total solid (TS) (w/w) | Volatile solid (VS) (w/w) |
|---|---|---|
| Guangzhou (GZ) | 61.0 ± 0.13% | 8.12 ± 0.21% |
| Hangzhou (HZ) | 62.9 ± 0.67% | 6.07 ± 0.07% |
| Taizhou (TZ) | 62.32 ± 0.98% | 4.34 ± 0.21% |
Methane production by batch culture.
A series of batch tests were performed at 30 oC in 60 mL of anaerobic bottles with a working volume of 20 mL, except for test 3 where 40 mL of working volume in 120 mL bottles was employed. Each test was carried out in triplicate without shaking. The mixture containing inocula and medium was loaded into the bottles. The bottles were subsequently purged with high purity N2 for 2 min and sealed with butyl rubber stoppers. The medium (pH 7.0) was composed of 20 mM ethanol, and other components were described previously24.
In test 1, three paddy soil samples (GZ, HZ and TZ) were assessed to screen the optimum enrichments in terms of methane production performance. Enrichment cultivation was initiated by inoculated 1.5 g-VS of soil samples into an anaerobic fresh medium. Cultivations were performed in the absence and presence of magnetite (20 mM) respectively. In test 2, the resulted enrichments of HZ (PEC-HZ and PEM-HZ) from test 1 were selected to further conduct semi-continuous subcultures. Specifically, as for the first generation of subculture, 5 mL of PEM-HZ from test 1 (start-up) were added into fresh medium amended with 20 mM magnetite. When methane production reached to the highest yield, the second generation of subculture was initiated by transferring the first generation enrichments (5 mL) to fresh medium supplemented with 20 mM magnetite. The same procedures for two generations of subculture of PEC-HZ were carried out in the absence of magnetite.
In test 3, Methanogenic pathways of PEM-HZ were determined by inhibitor addition (Supplementary Table S1). In inhibition experiments, 0.14 g-VS/L of PEM-HZ from the second generation was grown in the fresh medium containing 20 mM magnetite. The inhibitor was added into the medium at the beginning of incubation. In group II, acetoclastic methanogenesis was detected by inhibition with phosphate (100 mM), an inhibitor of acetoclastic methanogenesis18. In group III, hydrogenotrophic methanogenesis was determined by inhibition with BES (10 mM), a specific inhibitor for methanogens25.
Analytical methods.
The volume of biogas produced was measured using water displacement method. CH4 and H2 was periodically analyzed using a gas chromatograph26. Ethanol, individual VFAs, TS and VS were determined as described previously24. Concentrations of Fe (II) extracted with 0.5 N HCl were determined using a ferrozine method27. The modified Gompertz equation was used to calculate the maximum methane production rate (Rm) and the lag-phase time (λ)26. The methane yield Ps was calculated by dividing cumulative methane production with the total ethanol used. An XRD spectrum was analyzed by a conventional 2θ/θ method using wide angle X-ray diffraction (Bruker D8 Advance).
DNA extraction, PCR amplification and 16S rDNA sequencing.
Samples collected from the second generation of paddy soil enrichments were used for total genomic DNA extraction according to a CTAB/SDS method. DNA was purified, quantified and stored at -20°C before PCR amplification. Amplicon libraries were constructed for Illumina sequencing using primer pairs 515f/806r for bacteria28 and 519f/ 825r for archaea29. PCR, library construction, Illumina sequencing and sequence analysis were performed as described previously24. The sequences were deposited to the GeneBank Sequence Read Archive with accession No. SRR1784993.
Additional Information
How to cite this article: Yang, Z. et al. Magnetite nanoparticles facilitate methane production from ethanol via acting as electron acceptors. Sci. Rep. 5, 16118; doi: 10.1038/srep16118 (2015).
Supplementary Material
Acknowledgments
The authors appreciate Associated Prof. Yajie Zhang (Ningbo Institute of Industrial Technology, Chinese Academy of Sciences), Associated Prof. Bucai Zhang (Jiangsu Agri-animal Husbandry Vocational College), and Associated Prof. Li Zhuang (Guangdong Institute of Eco-Environmental and Soil Sciences) for providing the paddy soil. This work was funded by the National Natural Science Foundation of China (No. 21307143, No. 41276143), 863 Project (2011AA060905), Key Deployment Project of the Chinese Academy of Sciences (KGCX2-EW-317), Key Research Program of the Chinese Academy of Sciences (No. KGZD-EW-304).
Footnotes
Author Contributions R.G. conceived of the study. Z.Y. and R.G. designed experiment. Z.Y. performed experiment. X.S. and C.W. conducted microbial community analyzes. L.W. and R.G. carried out data interpretation. Z.Y. and R.G. drafted the manuscript. All the authors have read and approved the final manuscript.
References
- Sieber J. R., McInerney M. J. & Gunsalus R. P. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66, 429–452 (2012). [DOI] [PubMed] [Google Scholar]
- Rotaru A.E. et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415 (2014). [Google Scholar]
- Liu F. et al. Magnetite compensates for the lack of a pilin-associated c-type cytochrome in extracellular electron exchange. Environ. Microbiol. 17, 648–655 (2015). [DOI] [PubMed] [Google Scholar]
- Braunschweig J., Bosch J. & Meckenstock R. U. Iron oxide nanoparticles in geomicrobiology: from biogeochemistry to bioremediation. New Biotechnol. 30, 793–802 (2013). [DOI] [PubMed] [Google Scholar]
- Kato S., Hashimoto K. & Watanabe K. Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals. Environ. Microbiol. 14, 1646–1654 (2012). [DOI] [PubMed] [Google Scholar]
- Li H., Peng J., Weber K. & Zhu Y. Phylogenetic diversity of Fe (III)-reducing microorganisms in rice paddy soil: enrichment cultures with different short-chain fatty acids as electron donors. J. Soils Sediments 11, 1234–1242 (2011). [Google Scholar]
- Ding L. J., Su J. Q., Xu H. J., Jia Z. J. & Zhu Y. G. Long-term nitrogen fertilization of paddy soil shifts iron-reducing microbial community revealed by RNA-13C-acetate probing coupled with pyrosequencing. ISME J 9, 721–734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.J., Yang J., Chen X. P., Sun G. X. & Zhu Y. G. Phylogenetic diversity of dissimilatory ferric iron reducers in paddy soil of Hunan, South China. J Soils Sediments 9, 568–577 (2009). [Google Scholar]
- Jiang S. et al. Methanogenesis facilitated by geobiochemical iron cycle in a novel syntrophic methanogenic microbial community. Environ. Sci. Technol. 47, 10078–10084 (2013). [DOI] [PubMed] [Google Scholar]
- Janssen P. H. & Frenzel P. Inhibition of methanogenesis by methyl fluoride: studies of pure and defined mixed cultures of anaerobic bacteria and archaea. Appl. Environ. Microbiol. 63, 4552–7 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. J., Schink B., Pfennig N. & Conrad R. Energetics of syntrophic ethanol oxidation in defined chemostat cocultures. Arch. Microbiol. 155, 82–88 (1990). [Google Scholar]
- Schink B., Phelps T. J., Eichler B. & Zeikus J. G. Comparison of ethanol degradation pathways in anoxic freshwater environments. J. Gen. Microbiol. 131, 651–660 (1985). [Google Scholar]
- Steinbusch K. J. J., Hamelers H. V. M., Plugge C. M. & Buisman C. J. N. Biological formation of caproate and caprylate from acetate: fuel and chemical production from low grade biomass. Energy Environ. Sci. 4, 216–224 (2011). [Google Scholar]
- Zhang W. X. et al. Analysis of the bacterial community in Zaopei during production of Chinese Luzhou-flavor liquor. J. Inst. Brewing 111, 215–222 (2005). [Google Scholar]
- Tauchert K., Jahn A. & Oelze J. Control of diauxic growth of Azotobacter vinelandii on acetate and glucose. J. Bacteriol. 172, 6447–6451 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S. E., Costenbader C. J. & Melton T. Diauxic growth in Azotobacter vinelandii. J. Bacteriol. 164, 866–871 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratscher J., Dumont M. G. & Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. PNAS 108, 4170–4175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R., Klose M. & Claus P. Phosphate inhibits acetotrophic methanogenesis on rice roots. Appl. Environ. Microbiol. 66, 828–831 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Yu Z., Lu Q., Zhuang L. & Zhou S. G. Anaerobic humus and Fe (III) reduction and electron transport pathway by a novel humus-reducing bacterium, Thauera humireducens SgZ-1. Appl. Microbiol. Biotechnol. 99, 3619–3628 (2015). [DOI] [PubMed] [Google Scholar]
- Hori T., Muller A., Igarashi Y., Conrad R. & Friedrich M. W. Identification of iron-reducing microorganisms in anoxic rice paddy soil by 13C-acetate probing. ISME J 4, 267–278 (2009). [DOI] [PubMed] [Google Scholar]
- Vandieken V., Knoblauch C. & Jørgensen B. B. Desulfovibrio frigidus sp. nov. and Desulfovibrio ferrireducens sp. nov., psychrotolerant bacteria isolated from Arctic fjord sediments (Svalbard) with the ability to reduce Fe (III). Int. J. Syst. Evol. Microbiol. 56, 681–685 (2006). [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W. & Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 (2008). [DOI] [PubMed] [Google Scholar]
- Kang Y. S., Risbud S., Rabolt J. F. & Stroeve P. Synthesis and characterization of nanometer-size Fe3O4 and γ-Fe2O3 particles. Chem. Materials 8, 2209–2211 (1996). [Google Scholar]
- Yang Z., Xu X., Guo R., Fan X. & Zhao X. Accelerated methanogenesis from effluents of hydrogen-producing stage in anaerobic digestion by mixed cultures enriched with acetate and nano-sized magnetite particles. Bioresour. Technol. 190, 132–139 (2015). [DOI] [PubMed] [Google Scholar]
- Chidthaisong A. & Conrad R. Specificity of chloroform, 2-bromoethanesulfonate and fluoroacetate to inhibit methanogenesis and other anaerobic processes in anoxic rice field soil. Soil Biol. Biochem. 32, 977–988 (2000). [Google Scholar]
- Yang Z., Guo R., Xu X., Fan X. & Luo S. Hydrogen and methane production from lipid-extracted microalgal biomass residues. Int. J. Hydrogen Energy 36, 3465–3470 (2011). [Google Scholar]
- Lovley D. R. & Phillips E. J. P. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51, 683–689 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason A. P et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. PNAS 110, 6548–6553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehab N., Li D., Amy G., Logan B. & Saikaly P. Characterization of bacterial and archaeal communities in air-cathode microbial fuel cells, open circuit and sealed-off reactors. Appl. Microbiol. Biotechnol. 97, 9885–9895 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


