Abstract
Objective
Evaluating HIV-1 specific T-cell response in African populations is sometimes compromised by extensive virus diversity and paucity of non-clade B reagents. We evaluated whether consensus group M (ConM) peptides could serve as comparable substitutes for detecting immune responses in clade A and clade D HIV-1 infection.
Methods
Frequencies, breadths and polyfunctionality (≥3 functions: IFN-γ, IL-2, TNF-α and Perforin) of HIV-specific responses utilizing ConM, ConA and ConD Gag and Nef peptides was compared.
Results
Median genetic distances of infecting gag sequences from consensus group M were (8.9%, IQR 8.2–9.7 and 9%, IQR 3.3–10) for consensus A and D, respectively. Of 24 subjects infected with A and D clade virus, Gag responses were detected in comparable proportions of subjects when using ConM peptides 22/24, ConA peptides 17/24, and ConD peptides 21/24; p = 0.12. Nef responses were also detected at similar proportions of subjects when using ConM peptides 15/23, ConA peptides 19/23, and ConD peptides 16/23, p = 0.39. Virus-specific CD4+ and CD8+ T-cell polyfunctionality were also detected in similar proportions of infected individuals when using different peptide sets.
Conclusions
These data support the use of consensus group M overlapping peptide sets as reagents for detecting HIV-specific responses in a clade A and D infected population, but underscore the limitations of utilizing these reagents when evaluating the breadth of virus-specific responses.
Keywords: HIV-1 diversity, T-cell responses, Group M peptides
1. Introduction
HIV-1 infection naturally occurs through a single infecting viral variant [1]. This initial viral homogeneity is progressively followed by rapid generation of multitudes of distinct viral sequences at population level [2–4]. Even virus isolates from the same infected individual can considerably differ [5]. Consequently, HIV-1 group M is characterized by the existence of 9 genetically distinct subtypes (A–D, F–H, J, and K), and over 58 circulating recombinant forms (CRFs, http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html, last accessed 06.05.13). This enormous antigenic diversity is a key obstacle to formulating reagents for monitoring virus specific immune responses in HIV-1 infected populations. Peptides that more closely match the autologous HIV-1 sequences are the ideal; and undoubtedly improve detection of breadths and magnitudes of virus-specific T-cell responses [6,7]. Even reagents based on locally circulating strain sequences are better at detecting virus-specific responses than the equivalent clade or group consensus [8]. Nevertheless, there are real limitations to the use of autologous peptides in general population monitoring. Virus sequences can differ by up to 20% and 35% within clade and between clades, respectively [9]. Despite known limitations of consensus peptides, they have been used to comprehensively screen various populations for breadths and magnitudes of virus-specific T-cell responses to the entire HIV proteome [10–13]. Practical efforts to address HIV-1 antigenic diversity have partly focused on the more conserved regions of HIV-1 to reduce genetic distance. Consensus reagents overcome genetic variability of a given viral protein by adopting the most common amino acid at each position [14]. This can result in reduction in genetic distance from circulating strains by up to a half [15]. Several studies have evaluated group M and clade-specific consensus reagents for their ability to detect virus specific T-cell responses in infected populations. Given the high degree of the Gag protein conservation [14], comparable detection by the different consensus Gag reagents is not unexpected. Consequently, Bansal et al. [16] reported comparable virus-specific T-cell detection by group and clade-specific Gag reagents in clade B-infected American and clade C-infected Zambian subjects. Surprisingly, this equal performance has not been consistently observed in all populations. While group M and clade-based reagents detected similar frequencies of consensus Gag and Nef responses in both clade B- and F1-infected subjects; in clade C infected subjects, these group-based reagents detected lower levels of consensus response than clade C peptides [17]. These data underscore the necessity to assess virus-specific T cell detection abilities of consensus reagents in populations with diverse co-circulating HIV-1 clades.
Although clade A and D contribute only 7.4% and 3.4% of the global HIV-1 epidemic, respectively [18], they dominate the co-circulating strains in Uganda [19]. Characterization of immune responses to clade A and D viruses has been partly limited by the scarceness of clade-based reagents. Use of the readily available group-based consensus peptides might address the challenges of clade A and D reagent scarcity. Previous detection of high levels of consensus response using group M peptides support their use in our mixed clade A and D infected population [20]. Previous studies suggested that relative performance of consensus group M reagents depends on the clade involved [17]; underscoring need to assess performance of these reagents in populations with other different HIV-1 diversities. It is not known how group M, clade A and clade D consensus peptides compare in detection of virus-specific responses in mixed clade A1- and D-infected populations.
The objective of this study was to therefore compare how group- and clade-based consensus peptides detect virus-specific T-cell responses in a mixed clade A1- and D-infected population. We selected Gag and Nef because they are the most frequently targeted HIV-1 proteins [10,11,21–23]. Gag was also shown to contain highly cross-reactive regions across clades A, B, C and D [24]. Furthermore, T-cell recognition of Gag and Nef by clade A-infected subjects was shown to be similar to that seen in subtypes B and C infection [25]. Besides, consensus Gag and Nef peptides are readily available in reagent repositories. We sought to determine whether consensus group M peptides could serve as useful alternative reagents for monitoring virus specific responses in this population.
2. Materials and methods
2.1. Study population
We previously assessed 50 HIV-1 infected, treatment-naïve subjects for recognition of group M consensus peptides. Their CD4+ T-cell counts, plasma viral loads infecting clades and genetic distances were previously determined as described before [20]. Here, we evaluated cryopreserved peripheral blood mononuclear cells (PBMC) from 24 of the above subjects [20]. They were selected based on cell availability and previous ConM responsiveness. Detection of virus-specific FN-γ responses by group and clade-based consensus Gag and Nef peptides was compared (Table 1). We additionally selected 29 ConM-, 17 ConA- and 14 ConD-responsive subjects to evaluate detection of virus-specific polyfunctional responses using intracellular cytokine staining assay (Table 2). All study subjects provided written informed consent for participation. Uganda Virus Research Institute Ethics Review Board and Uganda National Council of Science and Technology approved this study.
Table 1.
Demographics of subjects evaluated to compare detection of IFN-γ responses by group M and clade-based consensus peptides.
| Parameter; median (IQR) | HIV clades (number of subjects) |
|||
|---|---|---|---|---|
| Clade A1 (14) | Clade D (10) | Overall (24) | P value | |
| Gender ratio (male/female) | 3/11 | 2/8 | 5/19 | 1.0 |
| Age (years) | 39 (36–44) | 39 (31–58) | 39 (35–45) | 0.97 |
| Plasma viral load (RNA copies/ml) | 21950 (1900–117800) | 12150 (2923–30950) | 12700 (2748–41700) | 0.785 |
| CD4+ T-cell counts (cells/μl) | 571 (499–787) | 645 (569–822) | 616 (514–794) | 0.4 |
| WHO clinical stage [%]: (I, II and III) | 14, 64 and 21 | 10, 40 and 50 | 33, 54 and 13 | 0.34 |
| Genetic distance from ConM (%) | 8.9 (8.2–9.7) | 9.0 (8.3–1.0) | 9.0 (8.3–1.0) | 0.55 |
| Genetic distance from ConA (%) | 5.3 (4.4–6.5) | 13.6 (13.2–14.3) | 6.6 (5.2–13.3) | 0.0001a |
| Genetic distance from ConD (%) | 13 (12–13) | 5.6 (5.2–5.8) | 11.0 (5.8–13.0) | 0.0006a |
Mann–Whitney test.
Table 2.
Demographics of subjects evaluated to compare ability of group M and clade-based consensus peptides to detect of HIV-specific functional T-cell subsets.
| Parameter; median (IQR) | Consensus peptide set (number of subjects evaluated) |
P value | ||
|---|---|---|---|---|
| ConM (29) | ConA (12) | ConD (17) | ||
| Gender ratio (male/female) | 8/21 | 7/5 | 4/13 | 0.1 |
| Age (years) | 36 (29–43) | 38 (33–44) | 38 (34–44) | 0.83 |
| Infecting clade (A1/D) | 13/16 | 6/6 | 8/9 | 0.95 |
| Plasma viral load (RNA copies/ml) | 12,700 (4340–97,300) | 14,200 (5673–290,300) | 11,600 (2285–27,100) | 0.57 |
| CD4+ T-cell counts (cells/μl) | 551 (496–714) | 566 (514–655) | 650 (530–943) | 0.19 |
| WHO clinical stage [%]: (I, II and III) | 17, 69 and 14 | 25, 58 and 17 | 35, 53 and 12 | 0.73 |
| Genetic distance from ConM (%) | 9.3 (8.5–10.0) | 9.3 (8.4–12.0) | 8.9 (8.3–10.0) | 0.84 |
2.2. Synthetic peptides
Consensus group M (ConM) and consensus clade-based Gag peptides were obtained through the NIH AIDS Reference Reagent Repository (https://www.aidsreagent.org/Index.cfm). The ConM reagent set contained 129 overlapping peptides covering the entire HIV-1 Gag region. Most peptides were 15 amino acids long, with 11-amino acid overlaps between sequential peptides. Clade-based consensus reagents comprised 49 ConA and 49 ConD peptides; most peptides were 20 amino acids in length, and overlapping by 10-amino acids. Consensus Nef peptides were 15 amino acids long overlapping by 11, and comprised 53 ConM (https://www.aidsreagent.org/Index.cfm), 27 ConA and 26 ConD peptides (http://www.nmitt.de/peptide-en.html). Sequences of the evaluated peptides are illustrated in supplementary Table 1.
2.3. Sequence comparisons across peptide sets
Sequence conservation across corresponding peptide regions was estimated by conventional assessments of similarities and dissimilarities of aligned peptide sequences, as described elsewhere [26]. Briefly, analogous peptide sequence pairs were aligned and assessed for corresponding amino acid similarity using an identity matrix. Identical amino acids were scored as 1, while mismatched amino acids were either scored as 0. Sequence identity was derived from the proportion of identical corresponding amino acids, and expressed as the percentage match. Sequences with identity scores of ≥90% were considered conserved. Sequences with identities below 90% were considered unique.
2.4. Designing of peptide pools and matrices
Individual peptides were mapped by ELISpot assay using a pool-matrix design, as previously described [20]. Briefly, all the 227 Gag peptides were assembled into three matrices comprising pools of group M, clade A and clade D consensus peptides, respectively. Up to 10 peptides were pooled per matrix so that each peptide occurred in two distinct pools only. Accordingly, 23 Gag pools were prepared comprising 13 ConM, 5 ConA and 5 ConD peptide pools. Likewise, all the 106 Nef peptides were grouped to yield three matrices containing 6 ConM, 3 ConA and 3 ConD peptide pools. Deconvolution of the matrix enabled mapping of the responding peptide based on detection of responses in the two corresponding pools. Deconvoluted peptides were subsequently retested in duplicate to confirm the response. Mapped Gag peptides were consequently pooled per subject, and evaluated for the detection of virus-specific polyfunctional responses using intracellular cytokine assay. Individual peptides within pools occurred at a final concentration of 2 μg/ml.
2.5. Enzyme-linked Immunospot (ELISpot) assay
Virus-specific responses were primarily identified using IFN-γELISpot assay, as previously described [20]. Positive responses were defined as those with ≥100 spot forming units (SFU)/106 PBMC above three times the media and cells only background. Response magnitude was defined as the total SFU/106 PBMC. Breadth was defined as the number of detected peptides. Targeting two adjacent peptides was considered as detection of one epitope. Targeting three adjacent peptides was considered as detection of two epitopes; the greater of the two responses was used when evaluating magnitude. Magnitudes were divided by the number of evaluated peptides to correct for differences in the number of peptides contained in each peptide set. Adjusted magnitudes are used in all analyses comparing medians.
2.6. Fluorochrome antibodies
Aqua (L34957, Invitrogen), CD19 APC Alexafluor750 (1072337A, Invitrogen) and CD14 APC Alexafluor750 antibodies (773927B, Invitrogen) were used to exclude dead cells, B cells, and monocytes, respectively. The T-cells were identified by their expression of CD3 (brilliant violet 570, B152103, Biolegend), CD8 (pacific blue, 22416, BD Bioscience) and CD4 (PE-Cy5.5, 1049514A, ebiosciences) surface markers. Virus-specific response was quantified by intracellular detection of IFN-γ (Alexafluor 700, 21128, BD Biosciences), IL-2 (APC, 341116BD Biosciences), Perforin (FITC B-D48 clone, F111124, Diaclone) and TNF-α antibodies (PE-Cy7, E07677-1632, ebiosciences).
2.7. Preparation of PBMCs and staining for flow cytometry
Cryopreserved PBMCs that were previously determined to be IFN-γ responsive, were thawed, rested and incubated with the pooled peptides for 6 h at 37 °C, in the presence of 5% CO2 [20]. Negative controls were PBMCs incubated without any peptides. Positive controls were PBMCs incubated with Staphyloccoccus enterotoxin B (SEB). After incubation, the PBMCs were washed once in PBS (2% FBS), and stained with Aqua viability dye to discriminate dead cells, before subsequent staining for surface and intracellular markers, as previously described [20].
2.8. Flow cytometry data analysis
At least 500,000 events were acquired on an LSRII flow cytometer (Becton Dickinson). Data was analyzed using FlowJo (version 9.5.3, TreeStar), Pestle (version 1.6.2) and Spice softwares (Version 5.3101) [27]. The gating strategy used for defining the virus-specific T-cell response is summarized in Supplementary Fig. 1. Briefly, we gated on lymphocytes, singlets, dump− cells, viable cells (Aqua Blue+), CD3+, CD4+ and CD8+ cells to determine single positive CD3+CD8−CD4+ and CD3+CD4−CD8+ T-cells, before defining the antigen-specific cytokine secreting T cells.
2.9. Statistical analysis
Statistical analyses were performed using Epi Info™ and Graph Pad 5.0 (GraphPad Software, Inc., San Diego, California, USA). Continuous data is summarized as medians with interquartile ranges (IQR). Medians were compared using Mann–Whitney test (if two groups) and Kruskal–Wallis Rank Sum test (if three groups). Proportions were compared using Fisher’s Exact and Chi Square tests. Means were compared using the Student’s t-test. Correlations between continuous variables were evaluated using Spearman Rank Correlation test. P values ≥0.05 were considered significant.
3. Results
3.1. Study population for comparing detection of IFN-γ response
Demographics of the 24 subjects assessed for HIV-specific IFN-γ detection by the three consensus peptide sets are summarized in Table 1. Their median plasma viral loads and CD4+ counts were 12,700 (2748–41,700) RNA copies/ml and 616 (514–794) cells/μl, respectively. The subjects’ age, gender distribution, plasma viral loads, CD4+ T-cell counts and WHO clinical staging did not significantly differ by clade. Over 99% infecting gag sequences were highly divergent (genetic distance >3%) from the corresponding clade based consensus sequences. Infecting gag sequences were more divergent from Group M consensus sequences (9.0% [IQR 8.3–1.0]) than from clade A (5.3%; [IQR 4.4–6.5]) or clade D consensus sequences (5.6%; [IQR 5.2–5.8]); p < 0.001, Mann–Whitney test. Divergence from consensus group M was similar between clade A (8.9%; [IQR 8.2–9.7]) and D infecting gag sequences (9.0%; [IQR 8.3–1.0]), Table 1.
3.2. Virus-specific IFN-γ was detected in similar proportions of subjects across Gag peptide sets
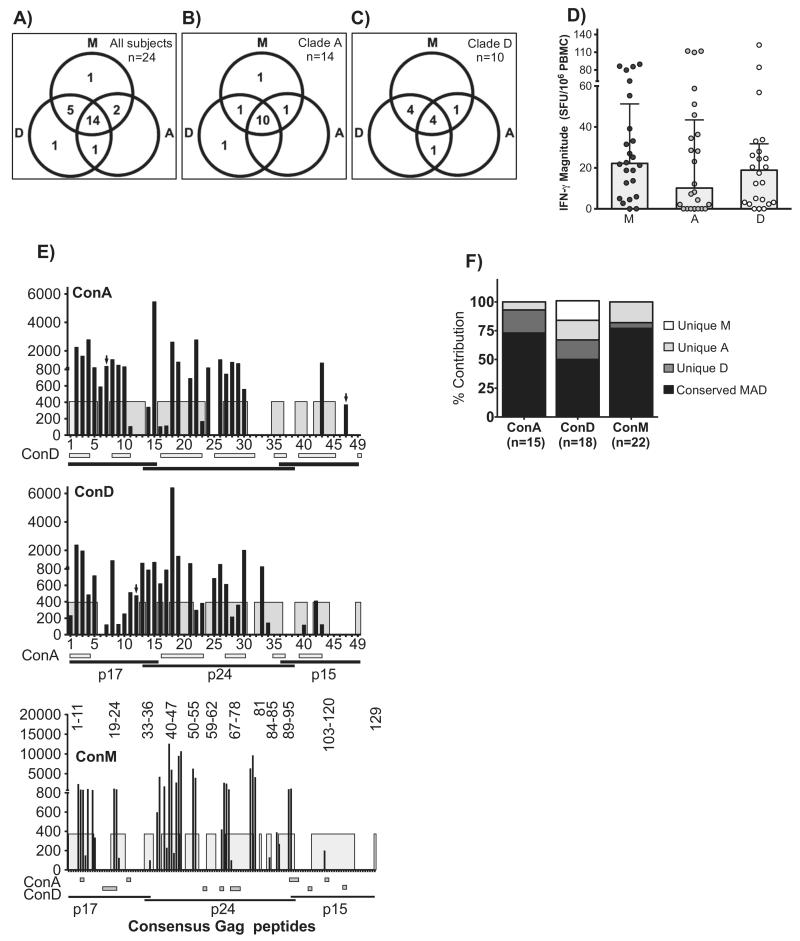
We first evaluated the consensus Gag peptide sets for rates of detection of virus-specific IFN-γ in 24 subjects. All comparisons are given as ConM, ConA and ConD, respectively. Of 24 subjects, 22(92%), 17(71%) and 21(88%) had measurable Gag-specific IFN-γ responses, respectively, p = 0.12, Chi square test. Fourteen cross-recognized all three sets; 5 cross-recognized only ConM and ConD; 2 cross-recognized only ConM and ConA; 1 cross-recognized only ConA and ConD; 1 uniquely recognized only ConM and 1 uniquely recognized ConD only. Combined use of three sets improved overall detection by 29% (17/24–24/24) subjects, p = 0.009, Fisher’s Exact test. When stratified by infecting Gag clade, the proportion of detected subjects did not significantly differ among clade A1- (13/14, 11/14, 12/14, (Fig. 1B) and D-infected subjects (9/10, 6/10, 9/10, (Fig. 1C). Overall, the adjusted median IFN-γ magnitudes did not significantly vary across peptide sets [(22.2: IQR 5.6–51.2), (10.2; IQR 0–43) and (18.9; IQR 3.1–31.8)], Fig. 1D.
Fig. 1.
Gag-specific IFN-γ was detected in similar proportions of subjects across peptide sets, but ConM underestimated the breadths of response. The IFN-γ ELISpot assay was used to evaluate detection of Gag-specific IFN-γ response to ConM (n = 129), ConA (n = 49) and ConD peptides (n = 49) in 24 HIV clade A1 and D chronically infected subjects. Recognition of ConM, ConA and ConD peptides is illustrated in (A); and is subsequently stratified by clade A1- (B; n = 14) and D-infected subjects (C; n = 10). For each peptide set, total magnitudes were divided by the number of peptides evaluated to adjust for the varying numbers of peptides contained in each set. For example, magnitudes for the group M peptide set were divided by 129; and 49 divided magnitudes of the clade-based consensus peptides. (D) illustrates evaluations of adjusted IFN-γ magnitudes across peptide sets. Targeting individual ConA, ConD and ConM peptides by the 24 study subjects is chronologically illustrated in (E). Gray sections within the plot areas define regions with ≥90% sequence identity across sequences. The Y-axis indicates SFU/106 PBMCs while the X-axis displays the individual peptides. The ConM set has 129 peptides and cannot display on the X-axis; ConM peptide within the highly region across ConM/ConA/ConM are hence displayed as peptide numbers in the plot area. Horizontal bars below the X-axis define regions with ≥90% sequence identity between a given consensus clade peptide sequence and the plotted consensus peptide. Black arrows show peptides that uniquely detected virus-specific IFN-γ responses. Relative distribution of the detected IFN-γ breadth throughout unique and highly conserved regions across ConM, ConA and ConD is illustrated in (F).
3.3. Consensus group M Gag peptides detected significantly lower IFN-γ breadths
We then assessed for the proportions of individual peptide recognition by each reagent set. Virus-specific IFN-γ responses were detected in all the three major Gag sub-regions; Gag p24 was the most targeted while p15 was targeted least. Overall, 26% more ConA peptides (55%: 27/49), and 34% more ConD (65%: 31/49) were targeted than ConM (29%: 37/129); p < 0.0001, Chi Square test, Fig. 1E. To estimate breadths of response, targeting two adjacent peptides was considered as detection of a single epitope, while targeting three adjacent peptides was considered as detection of two epitopes. Gag breadth was significantly lower to ConM (22/129) than to ConA (15/49) and ConD (18/49), p < 0.012, Chi Square test. These breadths were primarily contributed by peptides within highly conserved regions across ConM (77%; 17/22), ConA (73%; 11/15) and ConD (50%; 9/18)] reagent sets, Fig. 1F.
3.4. ConD better detected HIV-specific IFN-γ in mixed clade A and D infection
We then compared performance of the three reagent sets across clade A1- and D-infected subjects. Overall proportions of targeted peptides improved when the evaluated consensus reagent was based on the infecting clade. Thus, clade A1-infected subjects targeted 30% more ConA peptides (51%: 25/49, p = 0.00024) than ConM (21%: 27/129). Consensus clade D peptides were better able to detect virus-specific responses across mixed clade A/D infection. Although not statistically significant, ConD detected more breadth (17/49) in clade A1 subjects than ConA (14/49); the targeted peptides spread across all three major Gag sub-regions, supplementary Fig. 2A. Likewise, clade D-infected subjects targeted 17% more ConD peptides (37%: 18/49) than ConM (20%: 26/129), p = 0.02, Fisher’s exact test. However, clade D-infected subjects targeted fewer ConA peptides than ConD; and this targeting occurred in the highly conserved p24 Gag sub-region only, supplementary Fig. 2B. Detected breadths to ConM were similar across clades. In both clade A1- and D-infected subjects, ConD breadth was attributed to broader targeting of peptides located in highly conserved regions across ConM, ConA and ConD; and substantial contributions from targeting sequences unique to ConA, ConD and ConM peptides. On the contrary, ConA breadths mainly focused on conserved regions across ConM, ConA and ConD, supplementary Fig. 2C.
3.5. Mismatched sequences contributed to the detected virus-specific IFN-γ
We used the subject Gag sequences to conservatively estimate proportions of IFN-γ responses that were attributed to mismatched sequences, supplementary Table 2.
Of the targeted peptides, 55 were attributed to peptides that were fully matched to the viral sequence, 62 were attributed to peptides with 1 mismatch, 23 were attributed to peptides with 2 mismatches, 11 to peptide with 3 mismatches, and 39 to peptides with more than 3 mismatches, as illustrated in supplementary in Table 1. Peptides that were mismatched at more than 3 positions in 15-mer and 20-mer peptides were tolerated to still yield functional virus-specific responses.
3.6. Consensus Nef peptide sets detected similar frequencies of IFN-γ, but ConM underestimated the breadths of response
We next evaluated 23 subjects for detection of HIV-specific IFN-γ responses by three consensus Nef peptide sets. All comparisons are given as ConM, ConA and ConD, respectively. Nef-specific IFN-γ responses were detected in 96% (22/23) subjects. Proportions of subjects with detectable IFN-γ were similar across reagent sets: 65%(15/23), 83%(19/23) and 70%(16/23); p = 0.39, Chi Square test, respectively. Of the 23 subjects, 9 cross-recognized all three peptides sets; 5 cross-recognized only ConM and ConA; 4 cross-recognized only ConD and ConA; 1 cross-recognized only ConM and ConD; two were uniquely recognized by ConD and 1 was uniquely recognized by ConA. In one subject, none of the sets detected any virus-specific responses. Use of 3 sets significantly improved overall detection by up to 31%(15/23 to 22/23), p = 0.04, Fisher’s exact test, supplementary Fig. 3A. Response magnitudes were comparable across peptide sets (data not shown).
Overall, 70% more ConA peptides (96%: 26/27%, p < 0.0001), and 43% more ConD (69%: 18/26%, p = 0.00034) were targeted compared to ConM (26%: 14/53). Thus, ConM detected significantly lower Nef-specific IFN-γ breadths (7/53) than ConA (13/27; p = 0.002) and ConD (11/26; p = 0.01), Fisher’s exact test, supplementary Fig. 3B. Consistent with this, ConM breadth was attributed to more focused targeting of regions conserved across the three peptide sets, while ConA and ConD more broadly targeted both the highly conserved as well as the unique consensus clade sequences, Fig. 3C.
3.7. IFN-γ, IL-2, TNF-α and Perforin magnitudes were similar across peptide sets
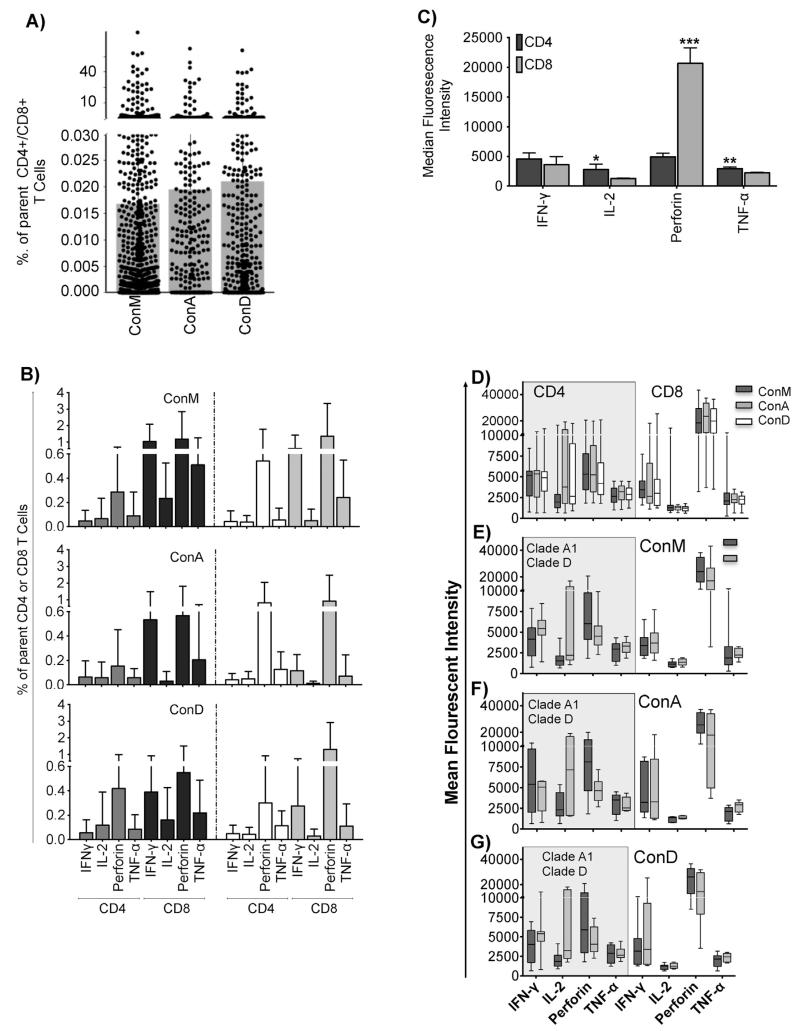
We next used the ConM, ConA and ConD peptide sets to evaluate detection of HIV-specific IFN-γ, IL-2, TNF-α and Perforin in 29,12 and 17 subjects, respectively. Subject characteristics were similar across the three groups; Table 2. Subject selection was based on predetermined IFN-γ responsiveness and cell availability. Comparisons are given as ConM, ConA and ConD, respectively. All the subjects responded to SEB in the CD3+CD8−CD4+ (3.83; IQR 0.68–8.99%) and CD3+CD4−CD8+ (4.60; IQR 1.14–13.00%) T-cell phenotypically defined subsets. Magnitudes of the detected responses were similar across the reagent sets (Fig. 2A); and did not significantly differ between clade A1 and D (Fig. 2B). Expression of Perforin predominated in the CD8+ T-cells; TNF-α was higher in the CD4+ T-cells, IL-2 tended to be higher in CD4+ T-cells, while IFN-γ was comparable across CD4+ and CD8+ T-cells, Fig. 2C. Expression of IFN-γ, IL-2, TNF-α and Perforin did not significantly differ across peptide sets (Fig. 2D) or by infecting clades for ConM (Fig. 2E), ConA (Fig. 2F) and ConD (Fig. 2G).
Fig. 2.
Gag-specific T cells were detected at similar frequencies across peptide sets. Magnitudes of all detected HIV-specific CD4+ and CD8+ T-cell functions (IFN-γ, IL-2, Perforin and TNF-α) are indicated in (A). The detected magnitudes were subsequently stratified according to the infecting clade, and according to the CD4+ and CD8+ parent T-cells. All bars indicate medians and interquartile ranges, (B). The overall average expression of T-cell functions (IFN-γ, IL-2, Perforin and TNF-α) in CD3+ and CD4+ T cell compartments is compared as Median Fluorescence Intensity (MFI), (C). Bars show means and standard deviations. Significant differences are highlighted as: *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001; Unpaired t test. Analyses were corrected for multiple comparisons using the Sidak-Bonferroni method; hence, only p values ≤ 0.01 are considered statistically significant. Expression (MFI) of HIV-1 specific IFN-γ, IL-2, Perforin and TNF-α ni CD4+ and CD8+ T cells following stimulation with ConM, ConA and ConD is compared in (D); and is and then stratified by clade for ConM (E), ConA (F) and ConD (G).
3.8. Functional T-cell subsets were detected at similar frequencies across peptide sets
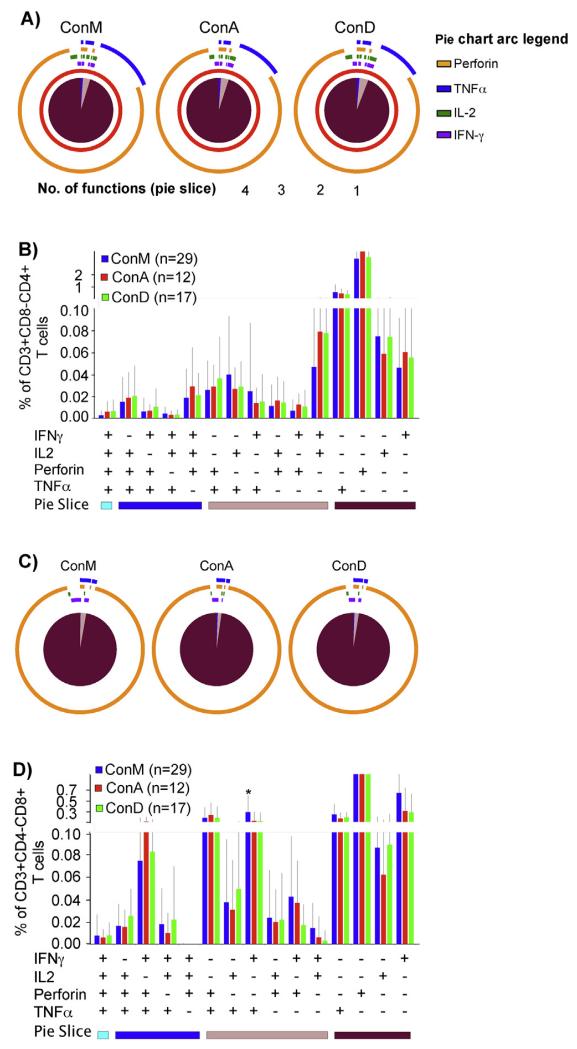
We then assessed ConM, ConA and ConD reagents for detection of co-expressed virus-specific T-cell functions. Frequencies of the detected monofunctional (95%, 95% and 94%), polyfunctional (1.1%, 1.1% and 1.3%) and bi-functional (3.8%, 4.2% and 4.4%) CD3+CD8−CD4+ responses were comparable (Fig. 3A); and uniformly distributed throughout the fifteen CD4+ T cell functional subsets, Fig. 3B. Similarly, frequencies of the detected monofunctional (97.2%, 97.8% and 96.4%), polyfunctional (0.28%, 0.56% and 0.28%) and bi-functional (2.5, 1.7% and 3.3%) CD8+ T-cell responses were comparably detected across peptide sets, respectively, (Fig. 3C); and were uniformly distributed across the majority (14/15) of the functional subsets, Fig. 3D.Taken together, these data demonstrate similar ability for ConM, ConA and ConD Gag peptide sets to detect HIV-specific T-cell subsets in this population.
Fig. 3.
Functional Gag-specific T-cell subsets were detected at similar frequencies across peptide sets. Consensus group M, ConA and ConD Gag peptide sets were assessed for the detection of HIV-specific T-cell functions (IFN-γ, IL-2, TNF-α and Perforin) using intracellular cytokine staining assay. Virus-specific T-cell polyfunctionality was defined as the simultaneous detection of three or more functions. The pie charts in (A) show the average T-cell functionality stratified by peptide set. Pie slices represent proportions of CD3+CD8−CD4+ T-cells that detected 4 (light blue), 3 (dark blue), 2 (light brown) and 1 function (dark brown). Pie arcs represent proportions of the total CD3+CD4+ T-cell response that contains Perforin (orange arcs), TNF-α (blue arcs), IFN-γ (purple arcs) and IL-2 (green arcs). (B) illustrates the CD3+CD4+ T-cell responses detected by ConM (blue bars), ConA (red bars) and ConD peptides (green bars), and their overall distribution across 15 different functional subsets (combinations of IFN-γ, IL-2, Perforin and TNF-α functions). Bars represent means and error bars indicate standard deviations. The Y-axis represents proportions of total CD3+CD4+ T-cell response contributing a given functional profile. The X-axis represents the number of positive and negative responses contributing to a given functional profile. Similarly, proportions (C), and relative contributions of CD3+CD8+ functional T-cell responses (D) are illustrated. Significant differences between peptide sets are highlighted; * denotes p-value ≤0·05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Monitoring virus-specific T cell responses in populations infected with HIV-1 clades A and D has been partially limited by a paucity of clade-specific reagents. We previously showed that consensus group M (ConM) peptides detect high frequencies of virus-specific IFN-γ in clade A1 and D infected populations [20]. It remained unclear how detection of T cell responses to the locally circulating A1 and D strains compares between ConM peptides with peptides based on consensus clades. In this study, we found that ConM peptides detected both Gag- and Nef-specific T cell responses in comparable proportions of subjects; and at comparable magnitudes as did clade-based consensus reagents. Detection was most improved when the reagent was based on the infecting clade. Consensus Clade D peptides detected greater breadths of Gag-specific IFN-γ across clades. Clade-based consensus peptides detected Nefspecific IFN-γ breadths at similar frequencies across clades, while Con M underestimated the breadths of both Gag and Nef IFN-γ. Cross-recognition by ConM peptides was predominantly attributed to targeting of highly conserved regions with gag p24 and the Nef core regions. Sustained cross-recognition of peptides with substantial amino acid dissimilarities was observed suggesting that epitope recognition tolerated that level of amino acid substitution, as previously described by others [28].
Underestimation of breadths by ConM is not unexpected; ConM reagents target the highly conserved regions across clades, potentially favoring cross-clade recognition. Equally, peptides based on infecting clades match the circulating strains better, and are putatively better able to detect responses to both the conserved and the relatively variable regions. Even studies in clade C-infected populations revealed that despite the extensive presence of cross-reactive clades A, B and D Gag epitopes, clade C-infected individuals recognized consensus peptides based on infecting clade variants with greater frequency [24]. Others also demonstrated substantial relative reduction in Gag breadths to non-infecting clades in clade B-infected populations [29]. Our findings are consistent with earlier ConM studies that reported lower Env breadths in clade B-infected US subjects [8]; and lower Gag breadths in clade C-infected Brazilians [17].
In one subject, Gag-specific IFN-γ responses were distinctively detected by only ConD, and by only ConM in another; similar situations were also observed for Nef responses. These findings support others that demonstrated that use of a single reagent inevitably underrepresents the frequency of detected responses [8,24,25]. The data also underscore the need to evaluate approaches designed to better detect breadths. Strategies to improve N-mer coverage in order to expand breadths of T cell detection and to ensure uniform coverage across the major clades have been evaluated [26,30]. Other demonstrated considerable improvement in detection of Nef-specific responses by PTE peptides compared to consensus peptides in clade B-infected populations [31]. It will be necessary to evaluate such approaches in other diversely infected populations.
Our findings contrast others that reported similar detection of breadths by consensus group M, consensus B or consensus C peptides in subtype B- and C-infected US and Zambian patients, respectively [16]. Inconsistent detection of ConM breadths in the Ugandan, Zambian and Brazilian populations are unexpected especially to the highly conserved Gag protein. Based on the Gag protein distances of 9% to ConM, and 5.6% to the consensus infecting clade in this population, we anticipated comparable breadths detection across the three reagents sets. Differing outcomes across the two studies are likely attributed to variations in the intrinsic design of the reagent sets. In our population, detection of virus-specific breadth by the three Gag peptide sets ranked as ConD > ConA > ConM. This hierarchy was consistent with the coverage of conserved and unique regions in the respective reagent set. Consensus clade D reagents had a broader coverage of highly conserved; as well as unique group M, Clade A and Clade D sequences. This likely contributed to the better cross-clade detection of virus-specific responses by ConD. In contrast, ConA and D ConM reagents designs were likely to better target highly conserved regions (73% and 77% respectively).
Disparity between the Ugandan and Brazilian findings might also be attributed to differences in study methods. While we assessed responses to the complete Gag and Nef proteins, Cortes et al. [17] evaluated only the Nef core and portions of Gag that had been predetermined to detect high frequencies of virus-specific response in the Brazilian population. In this clade A- and D-infected population, it is apparent that clade-based consensus sequences were able to detect responses to both the core and terminal regions of Nef; and that ConM peptides recognized only the Nef core. Probably, breadths across peptide sets would have been equivalent to those seen in the Brazilian population if we had also assessed just the Nef core. Comparable detection of breadths by group and clade-based consensus peptides as reported in the clade C-infected Zambians [32] was not seen in our population. These contrasting findings are also possibly attributed to variations in study methods. We defined breadth as number of mapped peptides recognized while Bansal et al. [32] defined breadth as the number of pools recognized. Because each pool contained 22 peptides, there was increased likelihood of detecting a positive response to a pool compared to designs where individual peptides were considered.
Breadths of CD4 and CD8 Gag T-cell response have been linked improved HIV-1 disease outcome [28,33]. This study was not designed to define clinical correlates of protection. Nonetheless, clade A1-infected subjects targeted significantly more Gag peptides than clade D subjects; consistent with reported associations of Gag breadth with protection [34] and of clade A infection with better disease outcome [35–37]. This study had some limitations. First, group M and clade-based consensus peptides were found to detect similar proportions and frequencies of HIV-specific of functional T-cell subsets; interpretations of the quantitative data should be treated with caution. Although ELISpot and Intracellular cytokine staining assays provide incredibly useful qualitative data, it is been difficult to compare such data quantitatively. Our study was not adequately powered to detect real quantitative differences. The study was also not designed to control for critical confounding factors like differences in host HLA alleles [38], disease progression status [39,40] and variability in reagent design that can quantitatively influence HIV-specific T cell responses. Also, the magnitudes of response may substantially depend on the ability to target highly conserved, immunodominant regions, as seen for ConM. We have shown that the peptides we evaluated are pre-designed with different abilities to detect T cell responses to the highly conserved immunodominant regions; hence quantitative comparisons using hence reagents would be inequitable. Secondly, the reagent sets we compared contained varying numbers of N-mers peptides. This is a potential limitation, but a necessary one given that these reagents are available for use as is. Our findings will be relevant when using these reagents to monitor virus-specific T cell responses in the relevant populations. The clade of the infecting virus was determined by sequencing of the dominant virus. Possibly, other co-circulating variants, not determined in this study may have contributed to the variant cross-recognition observed in this population.
Overall, these data show that despite differences in genetic distance the evaluated ConM peptides detect comparable antigenic potency as consensus peptides based on circulating clade A and D strains. The data also underscore the limitations associated with use of group M consensus peptides to detect breadths of clade A1 and D-specific response. The data also highlight the importance of reagent selection when monitoring HIV-specific T cell responses in various populations. Overall, the data support the use of Consensus group M peptides to screen for Gag- and Nef-specific T-cell responses in clade A and D infected populations.
Supplementary Material
Acknowledgements
We thank all the study participants and the dedicated clinical and field staff of The AIDS Support Organization (TASO) clinic in Entebbe. We thank the Medical Research Council (MRC UK) for funding this study and enabling access to the study cohort. The peptides were obtained through the NIH AIDS Research and Reference reagent program, Division of AIDS, NIAID, NIH. Flowcytometry antibodies were obtained using funds from “Training Health Researchers into Vocational Excellence in East Africa” (THRiVE), grant number 087540. Study participant recruitment was funded by the International Atomic Energy Agency, project code RAF6/029 through a grant to the WHO/African AIDS Vaccine Program.
Footnotes
This manuscript has not been published in its current form or a substantially similar form. There are no financial, consultant, institutional and other relationships that might lead to bias or a conflict of interest. Part of this work was presented as an abstract at the South African Immunology Society Conference, 5th–8th December 2010 with a title “Comparison of cross reactive epitopes using clade A, D and group M consensus peptides in an HIV-1 infected Ugandan population”.
Conflict of interest: None declared.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2014.05.021.
References
- [1].Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- [3].Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–72. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS. 2011;25:679–89. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liang B, Luo M, Scott-Herridge J, Semeniuk C, Mendoza M, Capina R, et al. A comparison of parallel pyrosequencing and sanger clone-based sequencing and its impact on the characterization of the genetic diversity of HIV-1. PLoS ONE. 2011;6:e26745. doi: 10.1371/journal.pone.0026745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Altfeld M, Addo MM, Shankarappa R, Lee PK, Allen TM, Yu XG, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–40. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Doroudchi M, Yegorov O, Baumgartner T, Kernaleguen AE, Breton G, Ndongala ML, et al. Autologous HIV-1 clade-B Nef peptides elicit increased frequency, breadth and function of CD8+ T-cells compared to consensus peptides. PLoS ONE. 2012;7:e49562. doi: 10.1371/journal.pone.0049562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rutebemberwa A, Currier JR, Jagodzinski L, McCutchan F, Birx D, Marovich M, et al. HIV-1 MN Env 15-mer peptides better detect HIV-1 specific CD8T cell responses compared with consensus subtypes B and M group 15-mer peptides. AIDS. 2005;19:1165–72. doi: 10.1097/01.aids.0000176216.02743.98. [DOI] [PubMed] [Google Scholar]
- [9].Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–60. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- [10].Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, et al. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78:4463–77. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–92. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feeney ME, Roosevelt KA, Tang Y, Pfafferott KJ, McIntosh K, Burchett SK, et al. Comprehensive screening reveals strong and broadly directed human immunodeficiency virus type 1-specific CD8 responses in perinatally infected children. J Virol. 2003;77:7492–501. doi: 10.1128/JVI.77.13.7492-7501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ramduth D, Chetty P, Mngquandaniso NC, Nene N, Harlow JD, Honeyborne I, et al. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J Infect Dis. 2005;192:1588–96. doi: 10.1086/496894. [DOI] [PubMed] [Google Scholar]
- [14].Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathog. 2007;3:e157. doi: 10.1371/journal.ppat.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, et al. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–51. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bansal A, Gough E, Ritter D, Wilson C, Mulenga J, Allen S, et al. Group M-based HIV-1 Gag peptides are frequently targeted by T cells in chronically infected US and Zambian patients. AIDS. 2006;20:353–60. doi: 10.1097/01.aids.0000206501.16783.67. [DOI] [PubMed] [Google Scholar]
- [17].Cortes FH, Bello G, Vorsatz C, Pilotto JH, Guimaraes ML, Grinsztejn B, et al. Higher cross-subtype IFN-gamma ELISpot responses to Gag and Nef peptides in Brazilian HIV-1 subtype B- and F1- than in C-infected subjects. Vaccine. 2013;31:1106–12. doi: 10.1016/j.vaccine.2012.12.023. [DOI] [PubMed] [Google Scholar]
- [18].Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, et al. HIV Sequence Compendium. Published by Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, NM, LA-UR 13-26007; 2013. [Google Scholar]
- [19].Kaleebu P, Whitworth J, Hamilton L, Rutebemberwa A, Lyagoba F, Morgan D, et al. Molecular epidemiology of HIV type 1 in a rural community in southwest Uganda. AIDS Res Hum Retroviruses. 2000;16:393–401. doi: 10.1089/088922200309052. [DOI] [PubMed] [Google Scholar]
- [20].Serwanga J, Mugaba S, Pimego E, Nanteza B, Lyagoba F, Nakubulwa S, et al. Profile of T cell recognition of HIV type 1 consensus group M gag and Nef peptides in a clade A1- and D-infected ugandan population. AIDS Res Hum Retroviruses. 2012;28:384–92. doi: 10.1089/aid.2011.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fu TM, Dubey SA, Mehrotra DV, Freed DC, Trigona WL, Adams-Muhler L, et al. Evaluation of cellular immune responses in subjects chronically infected with HIV type 1. AIDS Res Hum Retroviruses. 2007;23:67–76. doi: 10.1089/aid.2006.0114. [DOI] [PubMed] [Google Scholar]
- [22].Novitsky V, Cao H, Rybak N, Gilbert P, McLane MF, Gaolekwe S, et al. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J Virol. 2002;76:10155–68. doi: 10.1128/JVI.76.20.10155-10168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Frahm N, Korber BT, Adams CM, Szinger JJ, Draenert R, Addo MM, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78:2187–200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zembe L, Burgers WA, Jaspan HB, Bekker LG, Bredell H, Stevens G, et al. Intra- and inter-clade cross-reactivity by HIV-1 Gag specific T-cells reveals exclusive and commonly targeted regions: implications for current vaccine trials. PLoS ONE. 2011;6:e26096. doi: 10.1371/journal.pone.0026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Currier JR, Visawapoka U, Tovanabutra S, Mason CJ, Birx DL, McCutchan FE, et al. CTL epitope distribution patterns in the Gag and Nef proteins of HIV-1 from subtype A infected subjects in Kenya: use of multiple peptide sets increases the detectable breadth of the CTL response. BMC Immunol. 2006;7:8. doi: 10.1186/1471-2172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Finnefrock AC, Liu X, Opalka DW, Shiver JW, Casimiro DR, Condra JH. HIV type 1 vaccines for worldwide use: predicting in-clade and cross-clade breadth of immune responses. AIDS Res Hum Retroviruses. 2007;23:1283–92. doi: 10.1089/aid.2007.0098. [DOI] [PubMed] [Google Scholar]
- [27].Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytom A: J Int Soc Anal Cytol. 2011;79:167–74. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, et al. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007;81:2440–8. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yu XG, Lichterfeld M, Perkins B, Kalife E, Mui S, Chen J, et al. High degree of inter-clade cross-reactivity of HIV-1-specific T cell responses at the single peptide level. AIDS. 2005;19:1449–56. doi: 10.1097/01.aids.0000183126.32077.c8. [DOI] [PubMed] [Google Scholar]
- [30].Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, McElrath JM, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24:6893–904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- [31].Malhotra U, Li F, Nolin J, Allison M, Zhao H, Mullins JI, et al. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J Virol. 2007;81:5225–37. doi: 10.1128/JVI.02564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bansal A, Yue L, Conway J, Yusim K, Tang J, Kappes J, et al. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS. 2007;21:2387–97. doi: 10.1097/QAD.0b013e3282f13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- [34].Julg B, Williams KL, Reddy S, Bishop K, Qi Y, Carrington M, et al. Enhanced anti-HIV functional activity associated with Gag-specific CD8 T-cell responses. J Virol. 2010;84:5540–9. doi: 10.1128/JVI.02031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kiwanuka N, Laeyendecker O, Robb M, Kigozi G, Arroyo M, McCutchan F, et al. Effect of human immunodeficiency virus type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197(5):707–13. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- [36].Kaleebu P, French N, Mahe C, Yirrell D, Watera C, Lyagoba F, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–50. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- [37].Easterbrook PJ, Smith M, Mullen J, O’Shea S, Chrystie I, de Ruiter A, et al. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc. 2010;13:4. doi: 10.1186/1758-2652-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- [39].Streeck H, Schweighardt B, Jessen H, Allgaier RL, Wrin T, Stawiski EW, et al. Loss of HIV-1-specific T-cell responses associated with very rapid HIV-1 disease progression. AIDS. 2007;21:889–91. doi: 10.1097/QAD.0b013e3280f77439. [DOI] [PubMed] [Google Scholar]
- [40].Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.