Abstract
Alternative NFκB signaling is crucial for B cell activation and immunoglobulin production, and is mainly regulated by the IKK regulatory complex. Dysregulation of alternative NFκB signaling in B cells could therefore lead to hyperactive B cells and immunoglobulin overproduction. In our previous study we found that DBC1 is a suppressor of the alternative NFκB pathway to attenuate B cell activation. In this study, we further report that loss of DBC1 results in spontaneous overproduction of immunoglobulin in mice after 10 months of age. Using a double mutant genetic model, we confirm that DBC1 suppresses B cell activation through RelB inhibition. At the molecular level, we show that DBC1 interacts with alternative NFκB members RelB and p52 through its LZ domain. In addition, phosphorylation of DBC1 at its C terminus by IKKα facilitates its interaction with RelB and IKKα, indicating that DBC1-mediated suppression of alternative NFκB is regulated by IKKα. Our results define the molecular mechanism of DBC1 inhibition of alternative NFkB activation in suppressing B cell activation.
Introduction
The NFκB pathway is critical in many processes such as cell survival, inflammatory cytokine signaling and apoptosis (1–3). While the canonical signaling pathway is highly expressed in most cell types, the alternative NFκB pathway is central to a much smaller subset of cell types, namely bone, dendritic cells and B cells (1, 4–6). In addition, the canonical and alternative NFκB pathways have distinct functions (3). In B cells, the alternative NFκB pathway regulates development from transitional to mature B cells, cell cycle entry, isotype switching and differentiation, and plasma cell survival (5, 7–13). As a result, either reduced or increased alternative NFκB activation is associated with defective B cell response or B cell-mediated autoimmune diseases respectively (5, 12, 14–17).
One of the main regulatory mechanisms of the NFκB pathway is through the Inhibitor of kappa B Kinase (IKK) regulatory complex, which consists of at least IKKα, IKKβ and or IKKγ (NEMO) (18). IKKα and IKKβ are both about 750 amino acids in size, whereas IKKγ is 300 amino acids smaller in size, and has a distinct structure from the former two (18, 19). Although IKKα and β structurally very similar, knock out studies show that IKKα and IKKβ differentially regulate the alternative and canonical NFκB pathways respectively (18–21). IKKβ preferentially binds to and phosphorylates IκBα, which is then degraded to release RelA: p50 heterodimers into the nucleus (18). On the other hand, IKKα phosphorylates the regulatory domain of p100, a necessary step to yield transcriptionally active RelB:p52 dimers (18, 21).
Deleted in Breast Cancer 1 (DBC1) was found to interact with IKKβ through a mass spectrometry screen (22). Although initially identified as a tumor suppressor, the role of DBC1 is cell-type dependent, as upregulated DBC1 levels are found in various other cancers (23–26). In addition to IKKβ, DBC1 has been shown to interact with various nuclear proteins, such as ER, AR, BRCA1, HDAC3, SUV39H1, and Sirt1, thus implicating its role in regulating transcription and epigenetic modification (27–33). We previously identified a novel role of DBC1 as a suppressor of B cell activation (34). In addition, through a microarray screen, we showed that in DBC1 KO B cells, RelB activity was significantly upregulated (34). In this study we further report that loss of DBC1 in mice leads to spontaneous dysregulation of B cells at 10 months of age, leading to increased production of autoreactive immunoglobulin. Furthermore, we confirm that DBC1 suppresses B cell regulation through RelB using a double mutant genetic mouse model. At the molecular level, we show that DBC1 interacts with both alternative NFκB members RelB and p52, as well as its regulator IKKα. Lastly, while the N-terminus Leucine Zipper (LZ) domain of DBC1 is required for its interaction with RelB, phosphorylation of DBC1 at its C terminus by IKKα is required for its interaction with both RelB and IKKα Our study further defines the molecular mechanism of DBC1 suppression of NFκB, and its role in B cell regulation.
Materials and Methods
Mouse and Cell Lines
HEK293T and NIH3T3 cells lines were maintained in DMEM (Gibco) supplemented with 10% Fetal Calf Serum (FCS) and 1% Penicillin/Streptomycin. EL4 cell line was maintained in RPMI supplemented with 10% FCS and 1% penicillin/streptomycin. Dbc1−/− mice were gifted by the Chini lab, and further backcrossed to C57BL/6 background for more than 5 generations. Relbshep/shep mice were purchased from Jackson Laboratories, and further crossed with DBC1 KO mice to generate Dbc1−/− Relbshep/shep double mutant mice with mixed genetic background. For all experiments, littermates were used as controls. All mice used in this study were maintained and used at the Northwestern University mouse facility under pathogen-free conditions according to institutional guidelines and animal study proposals approved by the Institutional Animal Care and Use Committee.
Plasmids, Antibodies and Reagents
PcDNA-Myc-DBC1 plasmid was purchased from Addgene. Truncation mutants were subcloned into pCMV-Myc (Clontech), and Myc-DBC1-SA mutant was generated by site directed mutagenesis using Advantage GC Rich PCR kit (Clontech) using standard protocol. RelA-cFlag pcDNA3, RelB-cFlag pcDNA3, C-Rel RHD-cFlag-pcDNA3, p50-cFlag-pcDNA3, p52-cFlag pcDNA3, pCR-Flag-IKKα, pCR-Flag-IKKβ plasmids were purchased from Addgene. Antibodies used for immunoblotting and co-immunoprecipitation were anti-p30/DBC1 (Bethyl laboratories), anti-RelB (C-19) (D-4), p100/p52 (C-5), Myc (A-14), (9E10), IKKα/β (H-470) (Santa Cruz), anti-Flag (F7425) (F1804) (Sigma), anti-Tubulin (DM1A) (Calbiochem) anti-phosphoserine (AB1603), phosphothreonine (AB1607) (Millipore).
Primary B cell Isolation and Culture
Primary B cells were negatively isolated from 8–12 week old mice using Dynabeads® Mouse CD43 (Untouched B cells) (Life Technologies) per manufacturer’s instructions. Primary B cells were maintained at 106/mL in RPMI (Dibco) supplemented with 10% Fetal Bovine Serum, 50μM β-mercaptoethanol, 100mM sodium pyruvate, 100mM HEPES buffer, and 1% penicillin/streptomycin. B cells were activated with goat f(ab)2 anti- mouse IgM (10μg/ml; Jackson ImmunoResearch), anti-CD40 (1μg/ml; eBioscience) supplemented with IL4 (10ng/ml;), LPS (500ng/ml;), BAFF (100ng/ml; Peprotech) as indicated.
Carboxyfluorescein diacetatesuccinimidyl ester (CFSE) proliferation assay
For cell proliferation and immunoglobulin production assays, purified B cells were stained with Cell Trace Carboxyfluorescein diacetatesuccinimidyl ester (5μm; Life Technologies), and cultured at 106 cells/ml for 5 days with indicated stimuli. After 5 days, cells were subjected to flow cytometry and analysis.
Flow cytometry
Single-cell suspensions were Fc-blocked with anti-CD16/32 antibody (eBioscience), stained with the appropriate fluorophore-conjugated antibodies then collected by an Accuri C6 Flow Cytometer or FACSCanto (BD Biosciences). Fluoroscence labeled-antibodies used include fluorescein-isothiocyanate-conjugated anti-mouse IgA, phycoerythrin-conjugated anti-CD138, allophyocyanin-conjugated anti-IgG1 (BD Biosciences), Peridinin-chlorophyll Cy5.5-conjugated anti-B220 (Biolegend). For intracellular staining of immunoglobulin, cells were fixed and permeabIlized using the CytoFix/Perm Kit (BD Biosciences) per manufacturer’s instructions, then incubated with 1:400x dilution of isolated serum in 1% Bovine Serum Albumin, followed by staining with the appropriate fluorophore-conjugated antibodies.
Immunofluorescence Staining
NIH3T3 Cells were seeded on Poly-L-Lysine coated cover slips in 6-well plates. Cells were fixed with 4% Formaldehyde, permeabilized with 0.1% Triton-X100, then incubated with 1:400x diluted serum in 1% Bovine Serum Albumin for 1 hr at RT. Cells were washed and incubated with Alexafluor 594-conjugated anti mouse IgG (Invitrogen) and biotin-labeled anti-mouse IgA for 1 hr at RT, followed by Alexafluor-488 avidin (Invitrogen) for 1 hr at RT. Cells were stained with DAPI before visualized using the Nikon eclipse Ti Fluorescence microscope at 40x, 2s exposure.
ELISA
96-well flat bottom plates were coated overnight at 4°C with 50ng purified anti-mouse antibodies (Biolegend) for the specific isotypes. Wells were then blocked with 1% BSA at RT for 2 hours, then incubated with mouse sera at 1:200 to 1:10000 dilutions overnight at 4°C. Wells were then incubated with biotin-coated secondary antibodies against specific immunoglobulin for 1 hour at RT, followed by avidin-HRP, and TMB substrate (Thermo Scientific). Absorbance at 450nm was detected using FilterMax F5 microplate reader (Molecular Devices).
Real Time PCR
106 cells were lysed in Trizol (Invitrogen), and RNA isolated per manufacturer’s instructions. 1μg of isolated RNA was then reversed transcribed using qScript cDNA synthesis kit (Quanta BioSciences). Real Time PCR was performed in duplicate wells using the iCycler Sequence and SsoFast SYBR Green Supermix (BioRad).
Statistical analysis
Student’s t-test was used to calculate statistical significance for ELISA, flow cytometry, and western blot densitometry analysis. For CFSE, ELISA and qPCR analysis of DKO mice, the Tukey-Kramer method was used for calculating statistical significance. A value of p ≤ 0.05 was considered significant.
Results
Loss of DBC1 leads to increased immunoglobulin production in 10 month-old mice
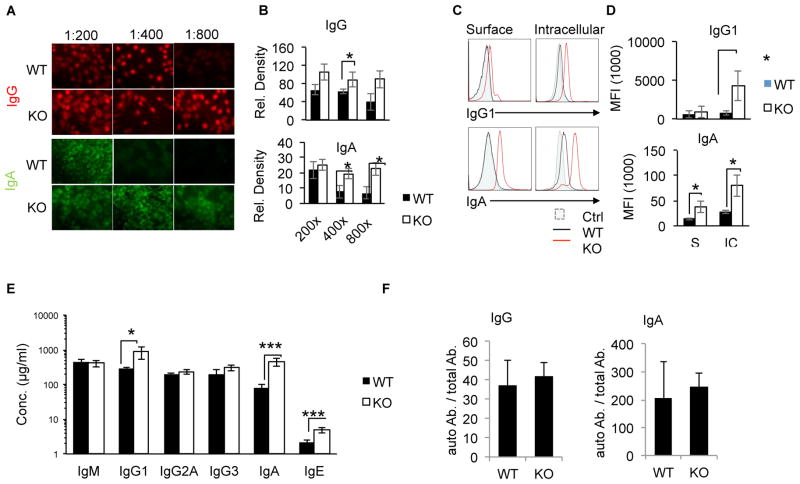
In our previous study, we showed that loss of DBC1 in mice leads to increased autoantibody production when induced with Experimental Autoimmune Myasthenia Gravis (34). Through our study we proposed that the tight regulation of B cell activation is lost in DBC1 KO mice. Interestingly, we now find that at 10 months of age, DBC1 KO mice spontaneously produce increased levels of autoreactive immunoglobulin. Using an indirect IF staining method, serum from 10 month-old DBC1 KO mice revealed higher levels of autoreactive IgG and IgA towards the murine fibroblast cell line NIH 3T3 (Fig. 1A & B). Specifically, autoreactive IgG was undetectable from WT serum at 1:800 dilution. In contrast, serum from DBC1 KO mice had persistently high levels of autoreactive IgG across all dilutions (Fig. 1A top panel, 1B left graph). Likewise, for the IgA isotype, WT serum had barely detectable levels of fibroblast-bound IgA at 1:400 dilution, whereas autoreactive IgA from DBC1 KO mice remained at high levels even at 1:800 dilution (Fig. 1A bottom panel, Fig. 1B right graph).
Figure 1. DBC1 KO mice spontaneously increase production of serum Ig at 10 months.
(A) Autoreactive IgG (red) and IgA (green) from the sera of 10 month-old WT and DBC1 KO mice was detected by indirect Immunofluorescent (IF) staining using NIH 3T3 cell line. (B) Mean fluorescence density from (A). (C) Serum from WT and DBC1 KO mice were tested for IgG1 and IgA reactivity towards surface (top) and intracellular (bottom) autoantigens in EL4 murine T lymphoma cell line, and detected by flow cytometry. (D) Mean fluorescence intensity (MFI) of EL4- bound immunoglobulin as detected in (C). (E) Blood serum was isolated from 10 month-old WT and DBC1 KO mice, and serum immunoglobulin levels for the indicated isotypes were measured by ELISA. (F) Autoreactive IgG and IgA as measured in (A) normalized to total IgG and IgA levels (fluorescence density (A.U)/μg). N=5, error bars represent SEM. * p<0.05, ** p<0.01, ***P<0.001.
We confirmed the increase of autoreactive immunoglobulin in DBC1 KO mouse serum by testing reactivity towards the murine T cell lymphoma cell line EL4. Similarly, significantly higher levels of autoreactive IgG and IgA levels in DBC1 KO serum were detected when analyzed by flow cytometry. Furthermore, DBC1 KO immunoglobulin could bind to both surface and intracellular autoantigens, as shown by flow analysis of differential surface staining and intracellular staining of EL4 cell line (Fig. 1C & D). The increased levels of autoreactive immunoglobulin correlated with significantly higher levels of total serum immunoglobulin, specifically IgG1, IgA and IgE isotypes (Fig. 1E). However, due to increased total immunoglobulin levels in DBC1 KO mice, DBC1 KO mice autoantibody levels normalized to total antibody levels (fluorescence relative density (A.U.)/total Ab. Conc.) are not significantly different from WT littermates (Fig. 1F). Taken together, the data indicate that loss of DBC1 in mice leads to loss of tight regulation of B cell activation, and spontaneous increased production of autoreactive and total immunoglobulin upon 10 months of age.
DBC1 interacts with RelB and P52
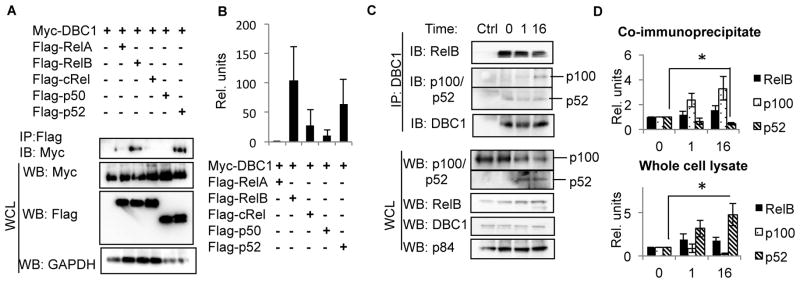
Our findings prompted us to determine the molecular basis for the dysregulation of B cell activation in DBC1 KO mice. In our previous study, microarray data and Chromatin Immunoprecipitation (ChIP) experiments suggest that RelB activity is increased in DBC1 KO B cells (34). We further tested if DBC1 suppresses RelB activity through binding with RelB. We co-transfected Myc-tagged DBC1 with each of the Flag-tagged NFκB members into HEK293T cells, and tested for interaction by co-immunoprecipitation (Co-IP). We observed that DBC1 co-immunoprecipitated with RelB and p52 (Fig. 2A & B). In contrast, we did not detect an interaction between DBC1 and RelA, c-Rel or p50 (Fig. 2A & B). In addition, the interaction of DBC1 with RelB and p52 was confirmed in mouse primary splenic B cells by immunoprecipitating DBC1 (Fig. 2C & D). RelB interacted with DBC1 in primary splenic B cells, and this interaction was reduced upon 16 hours of CD40 stimulation (Fig. 2C, 3rd panel & D). Similarly, we detected an interaction between DBC1 and p52 in naïve primary B cells (Fig. 2C, 2nd panel & D). Interestingly, upon CD40 activation, DBC1 partially reduces its interaction with p52, and increases its interaction with the inactive precursor p100 instead (Fig. 2C, 2nd panel & D). These results thus indicate that DBC1 interacts with the alternative NFκB members RelB and p52, supporting our hypothesis that DBC1 regulates B cell function by selectively modulating the transcriptional activity of the alternative NFκB pathway.
Figure 2. DBC1 interacts with RelB and p52.
(A) Flag-tagged RelA, RelB, c-Rel, p50 and p52 were each co-transfected with myc-tagged DBC1 into HEK293T cells, cell lysates were then immunoprecipitated using anti-Flag antibody. Interaction with DBC1 was detected by immunoblotting with anti-Myc antibody. (B) Densitometry analysis of (A) based on 5 independent trials. (C) Splenic B cells from WT mice were activated with anti-CD40 for 0, 1 and 16 hours, then cell lysates were immunoprecipitated with anti-DBC1 antibody. Interaction between endogenous DBC1 and RelB and p52 were detected by immunoblotting. Normal rabbit IgG serves as negative control. (D) Densitometry analysis of (C) based on 6 independent trials. * p<0.05.
The Leucine Zipper (LZ) domain of DBC1 is necessary and sufficient for interaction with RelB
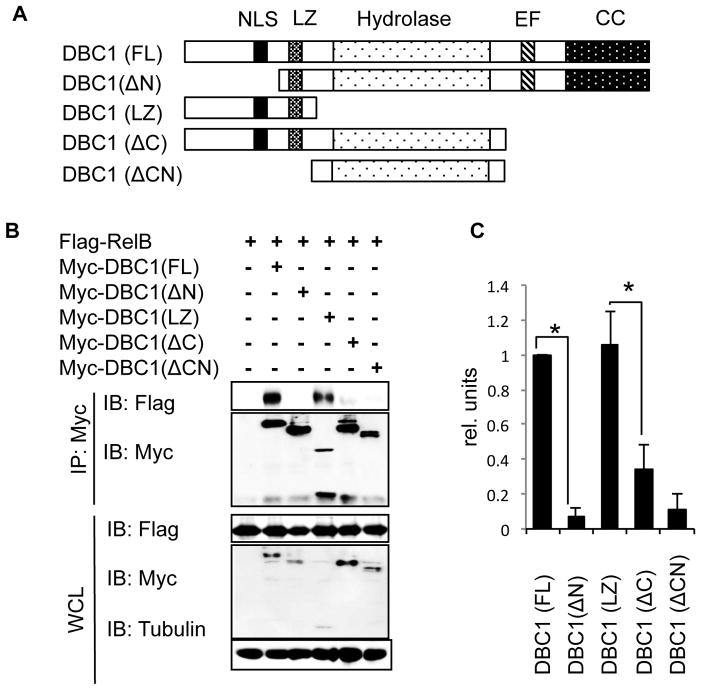
To characterize the molecular mechanism of DBC1 suppression of RelB, we mapped the interaction domains of DBC1 and RelB. DBC1 protein has a Leucine Zipper (LZ), putative hydrolase domain, inactive EF hand, Nuclear Localization Sequence (NLS) and coiled-coil domain (Fig. 3A)(31, 34). We generated five truncated mutants of DBC1 (Fig. 3A), then co-transfected Flag-tagged RelB with either Myc-tagged full-length DBC1 (FL), or each of the five Myc-tagged truncated mutants of DBC1 into HEK293T cells (Fig 3B). Co-IP results show that the LZ domain of DBC1 is required and sufficient for interaction with RelB (Fig. 3B). Specifically, deletion of the N terminus led to loss of DBC1 interaction with RelB, whereas expression of the N terminus of DBC1 alone was sufficient for interaction with RelB (Fig. 3B, lanes 3 and 4). Interestingly, we observed that when the center hydrolase domain was expressed along with the LZ domain, interaction between DBC1 and RelB was significantly reduced compared to interaction with the LZ domain of DBC1 alone, suggesting that a regulatory domain is present C-terminus to the LZ domain of DBC1 (Fig. 3B & C).
Figure 3. The LZ of DBC1 is necessary and sufficient for interaction with RelB.
(A) Schematic of structure of full length DBC1 and generated truncated mutants. NLS: Nuclear Localization Signal, LZ: Leucine Zipper, Hydrolase: Putative hydrolase domain, EF: Inactive EF hand, CC: Coiled-coil. (B) Myc-tagged DBC1 and its truncated mutants were co-transfected with Flag-tagged RelB into HEK293T cells, and cell lysates were immunoprecipitated with anti-Myc antibody. Interaction with RelB was detected by Western Blot using anti-Flag antibody (top panel). Whole cell lysate (WCL) (bottom panel) serves as loading control. (C) Densitometry analysis of (B). RelB co-immunoprecipitated with full length DBC1 (lane 2 of (B)) was used to normalize RelB co-immunoprecipitate levels with truncated mutants. N=3, error bars represent SEM. *p<0.05.
Increased proliferation and Ig production of DBC1 KO mice is abrogated by deletion of RelB
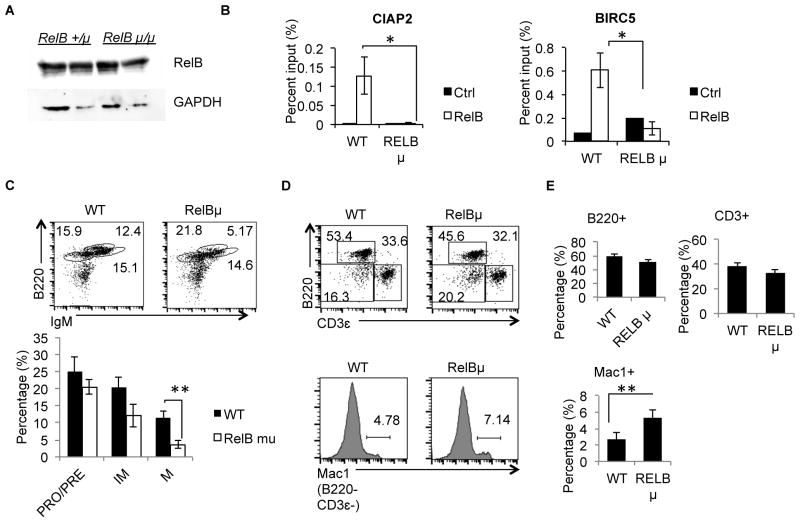
To confirm that hyper-activation of DBC1 KO B cells is due to loss of DBC1 suppression of RelB in B cells, we crossbred DBC1 KO mice with RelBshep/shep (RelB μ) mice, which bear a spontaneous loss-of-function point mutation of (35). Although RelB protein level was unaffected in the mutant (Fig. 4A), RelB function was significantly diminished in B cells from RelB shep/shep mice, since we could not detect mutant RelB DNA-binding activity using Chromatin Immunoprecipitation (ChIP) assay (Fig. 4B). As a result, at 6 to 8 weeks of age, RelBshep/shep mice exhibit a similar but milder phenotype than RelB-null mice in the hematopoietic compartment (12), specifically a slight reduction of mature B cell compartment in the bone marrow (Fig. 4C) and spleen (Fig. 4D), and increase in B220− CD3ε− Mac1+ cells (Fig. 4D& E).
Figure 4. RelB shep/shep mutation leads to loss-of-function RelB protein.
(A) Western Blot analysis of RelB protein levels from 2 pairs of RelB +/shep and RelB shep/shep mice. GAPDH levels serves as loading control. (B) B cells from RelB +/shep (WT) and RelB shep/shep (RelB mu) mice were activated with anti-CD40 overnight and subjected to ChIP analysis. RelB-bound target gene promoters were quantified by qPCR. (C) Bone marrow cells were isolated from WT and RelBshep/shep mice and B220LO IgM−, B220LO IgM+, and B220HI IgM+ cells analyzed by flow cytometry (top panel). Mean percentages of B cell subpopulations of WT and RelBshep/shep mice are shown (bar graph at bottom panel) (D) Splenocytes were isolated WT and RelBshep/shep mice and analyzed for B220+, CD3ε+ and B220−CD3ε− Mac1+ cells. (E) Mean percentages of B cells, T cells and macrophages/monocytes as analyzed in (D). For (A) and (B) N=5 from 2 independent experiments. For (C) – (E), N= 5. Error bars indicate S.E.M. *P<0.05.
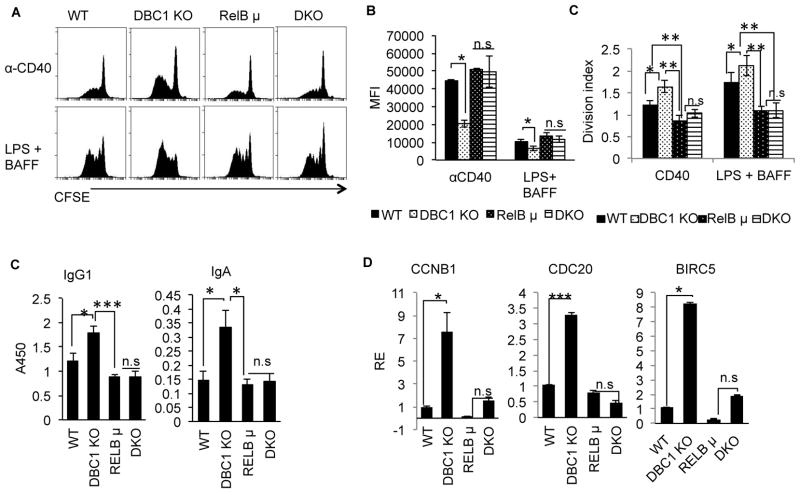
Naïve B cells were then isolated from WT, Dbc1−/− (DBC1 KO), RelB shep/shep (RelB μ) and Dbc1−/−RelBshep/shep (DKO) mice and analyzed for proliferation and immunoglobulin production. Consistent with our hypothesis, when assayed for proliferation by CFSE, DBC1 KO B cells proliferated at a faster rate compared to WT when stimulated with RelB agonists anti-CD40 or BAFF (Fig. 5A & B). In contrast, RelB μ B cells had reduced proliferation compared to WT as expected. Importantly, proliferation of Dbc1−/− RelBshep/shep (DKO) B cells were identical to RelB μ B cells, indicating that loss of RelB function abrogates the hyper-proliferative phenotype observed in DBC1 KO B cells. (Fig. 5A–C). Consistent with the proliferation data, ELISA using culture supernatants reveal that IgG1 and IgA production was increased in DBC1 KO B cells, but not in RelB μ and DKO B cells (Fig. 5D). In addition, we previously found that NFκB target genes involved in proliferation were upregulated in CD40-activated DBC1 KO B cells. In agreement with our previous findings, we observed increased expression of the NFκB target genes CCNB1, CDC20 and BIRC5 in DBC1 KO B cells (Fig. 5E). However, RelB μ B cells have basal expression of all three genes, which could not be rescued by the DKO B cells (Fig. 5E). In sum, these experiments show that DBC1 negatively regulates B cell proliferation, immunoglobulin production, and NFκB target gene expression through a RelB-dependent mechanism.
Figure 5. Increased proliferation and Ig production in DBC1 KO B cells is abrogated by deletion of RelB.
(A) Splenic B cells were isolated from WT, DBC1 KO, RelBshep/shep (RelB μ) and Dbc1−/− RelBshep/shep (DKO) mice. CFSE-stained B cells were stimulated with anti-CD40 or LPS+ BAFF for 5 days then subjected to flow cytometry for detection of proliferation. (B) Mean Fluorescence Intensity (MFI) of CFSE from (A). Reduction of MFI indicates increased proliferation. (C) Division index calculated from CFSE experiments as performed in (A). (D) ELISA measuring IgG1 and IgA levels in culture supernatants of WT, DBC1 KO, RelB μ, and DKO B cells stimulated as in (A). (E) Real Time PCR (qPCR) detection of NFκB target genes from WT, DBC1 KO, RelB μ and DKO B cells activated overnight with anti-CD40. N=5, error bars represent SEM. *p<0.05, ** p<0.01, *** p<0.001, n.s= not significant.
DBC1 interacts with IKKα and IKKβ in B cells
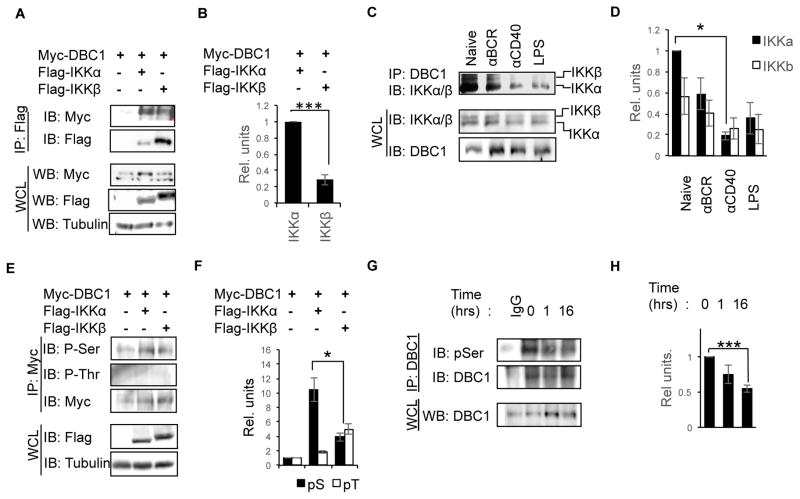
Through a mass spectrometry screen, DBC1 was previously identified as one of the proteins which interact with the IKKβ (22). IKKs are key components of the NFκB regulatory complex, and can activate NFκB by phosphorylating IκBs to promote its degradation (18). However, unlike IKKβ, which regulates classical NFκB signaling through phosphorylation of IκBs, IKKα is required for alternative NFκB signaling by promoting processing of p100 to active p52 (1, 21). Based on published literature and our Co-IP studies, we hypothesized that DBC1 could be regulated by upstream components of NFκB signaling, i.e. the IKK regulatory complex. We found that in addition to IKKβ, DBC1 interacts with greater affinity to the alternative NFκB regulator IKKα, both in HEK293T cells (Fig. 6A & B) and in primary B cells (Fig 6C &D). Interestingly, DBC1 interaction with IKKα and β is reduced in B cells upon stimulation (Fig. 6C & D), suggesting that CD40 stimulation triggers the release of DBC1 from the IKK regulatory complex. Together with our observation that DBC1 suppresses the alternative NFκB pathway, these results suggest that the regulatory function of DBC1 is regulated by IKKα.
Figure 6. DBC1 interacts with IKKα and IKKβ and is Serine phosphorylated by IKKα.
(A) Flag-tagged IKKα and IKKβ were each co-transfected with Myc-tagged DBC1, then cell lysates were immunoprecipitated with anti-Flag antibody. Interaction with DBC1 was detected by anti-Myc antibody. (B) Densitometry analysis of IKKα and IKKβ precipitates relative to IKKα in lane 2. (C) Primary B cells from WT mice were activated with the indicated stimuli for 1 hour. Cell lysates were then immunoprecipitated with anti-DBC1 antibody, and interaction with IKKα and β were detected by immunoblotting with antibody recognizing both IKKα and IKKβ. (D) Densitometry analysis of IKKα and IKKβ protein in the precipitates relative to IKKα levels at naïve state. (E) Myc-tagged DBC1 was co-transfected with Flag-tagged IKKα and β into HEK293T, cell lysates were then immunoprecipitated with anti-Myc, and phosphorylation of DBC1 detected using anti-phosphoSerine (α-pS) and anti-phosphoThreonine (α-pT) antibodies. (F) Densitometry analysis of pS and pT levels from (E) relative to lane 1 (overexpression of Myc-DBC1 alone). (G) Primary B cells were stimulated with α-CD40 for the indicated times, DBC1 was immunoprecipitated from cell lysates and phosphorylation of DBC1 was detected using α-pS antibody. Normal rabbit IgG serves as negative control, and whole cell lysate (WCL) as loading control. (H) Densitometry analysis of p-DBC1 as detected in (G) are normalized to lane 2 (naïve state). N=5, error bars represent SEM. *P<0.05, ** P<0.01, ***P<0.0001.
DBC1 is Serine-phosphorylated by IKKα
Since IKKs function as Serine/Threonine kinases, we speculated that DBC1 could be phosphorylation substrates of IKKs. Indeed, using anti-phosphorylated Serine (α-pS), but not anti-phosphorylated Threonine (α-pT), antibodies, we detected weak phosphorylation of DBC1, which was further enhanced by over-expression of either IKKα or IKKβ (Fig. 6E & F). In agreement with our observation that IKKα interacts with greater affinity with DBC1 than IKKβ, IKKα expression induced higher levels of DBC1 Serine phosphorylation (Fig. 6E & F). Similarly, we detected phosphorylated Serine residues on DBC1 in primary B cells (Fig. 6G & H). Interestingly, DBC1 phosphorylation was reduced 1 hour upon CD40 stimulation, similar to its interaction with IKKα/β, and further reduced at 16 hours upon stimulation. These results suggest that IKKs, in particular IKKα, is a Serine kinase of DBC1.
Serine phosphorylation of DBC1 at its C-terminus is required for interaction with IKKα and RelB
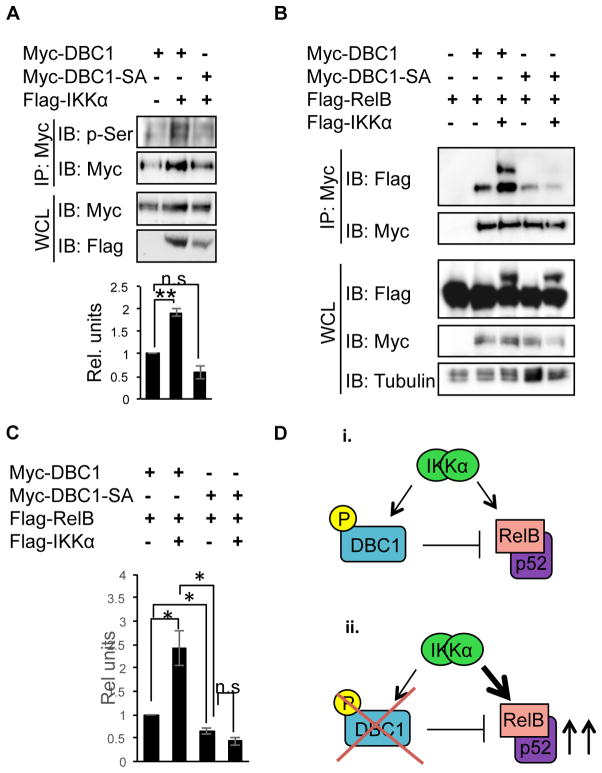
DBC1 was found through Mass Spectrometry to have a cluster of phosphorylable Serine residues at the C-terminus (phosphosite.org). Indeed, replacing all six Serine residues at the C- terminus of DBC1 with Alanine (DBC1-SA) abrogated Serine phosphorylation of DBC1 (Fig. 7A). To elucidate the functional consequence of IKKs-mediated DBC1 phosphorylation, we compared the ability of WT and phosphorylation-deficient DBC1-SA mutant to interact with RelB. Indeed, co-transfecting IKKα enhanced the interaction between DBC1 and RelB. In contrast, the interaction between DBC1 and RelB and IKKα was largely diminished when the Serine residues on DBC1 were replaced with alanine, indicating that phosphorylation at the C-terminus Serine cluster of DBC1 is required for its interaction with RelB (Fig. 7B & C). Therefore, based on our Co-IP experiments, we conclude that DBC1 interaction with RelB is regulated by the alternative NFκB regulatory protein IKKα, and phosphorylation by IKKα promotes the interaction between DBC1 and IKKα, as well as DBC1 and RelB.
Figure 7. DBC1 Serine phosphorylation by IKKα is required for interaction with RelB and IKKα.
(A) Myc-tagged DBC1 and Myc-tagged DBC1-SA mutant were each co-transfected with Flag-tagged IKKα. Both myc-DBC1 and myc-DBC1-SA were then immunoprecipitated using anti-Myc, and phosphorylation levels detected using α-pS antibody. Densitometry analysis of pS levels in the precipitate (bottom bar graph) is shown; pS-levels are relative to lane 1 (overexpression of WT DBC1 alone). (B) Myc-tagged DBC1 or DBC1-SA mutant was co-transfected with Flag-tagged RelB with or without flag-tagged IKKα. Cell lysates were then immunoprecipitated with anti-Myc antibody. Interaction between DBC1 and RelB or IKKα were detected by immunoblotting for Flag. (C) Densitometry analysis of precipitated RelB from (B) relative lane 2 (overexpression of WT DBC1 and RelB). (D) Schematic of proposed molecular mechanism of DBC1 suppression of B cells. i) DBC1 interacts with both IKKα and RelB in B cells and maintains the balance of active and inactive NFκB. ii) Loss of DBC1 suppression of RelB leads to imbalanced increase of alternative NFκB activity, leading to increased B cell proliferation and Ig production. N=5. Error bars represent SEM. *=P<0.05, **=P<0.01, ***=P<0.001.
In summary, our data indicates that DBC1 is a crucial regulator of B cell activation and immunoglobulin production by suppressing RelB activity. Firstly, loss of DBC1 in mice leads to a spontaneous increase in immunoglobulin production in the serum at 10 months of age. Second, we further determined that DBC1 selectively interacts with alternative NFκB members RelB and p52, and the Leucine Zipper (LZ) domain of DBC1 is required and sufficient for this interaction. Third, the increased proliferation and immunoglobulin production in DBC1KO B cells is abrogated by deletion of RelB. Fourth, DBC1 interacts with both IKKα and β, with significantly higher affinity with the former. Lastly, we found that DBC1 is Serine phosphorylated by IKKα, and its phosphorylation is required for its interaction with both RelB and IKKα. We previously identified a novel function of DBC1 as a suppressor of B cell activation. Furthering our previous work, we now determine that DBC1 suppresses B cell activation through RelB, and identify a novel regulatory mechanism of DBC1 through its interaction with IKKα and RelB.
Discussion
In this study we show that loss of DBC1 leads to spontaneous overproduction of immunoglobulin in mice at 10 months of age, and confirm that DBC1 suppresses B cell activation through RelB using double knockout genetic model. In terms of the molecular mechanism of DBC1, we have showed that DBC1 preferentially interacts with alternative NFκB members RelB and p52, and the Leucine Zipper (LZ) domain is necessary and sufficient for its interaction. In addition, DBC1 interacts with the NFκB regulators IKKα and IKKβ. Lastly, DBC1 is Serine phosphorylated by IKKα, and its phosphorylation regulates its interaction with both IKKα and RelB. Based on our findings, we propose that DBC1 regulates B cell activation by binding to RelB:p52 as well as the IKK regulatory complex. Upon B cell stimulation, loss of DBC1 phosphorylation, and its release from IKKα and RelB, allows for full RelB-mediated B cell activation. In WT B cells, through IKKα-mediated phosphorylation, DBC1 maintains the balance between active and inactive alternative NFκB. Loss of DBC1 function or regulation in B cells thus leads to increased RelB activity, and hyperproliferation, increased immunoglobulin production in mice (Fig. 7D).
Loss of integrity of B cell regulation, and polyclonal expansion of auto-reactive B cells especially increases with age (36–38). In our experimental model, the loss of DBC1 leads to production of self-reactive antibodies spontaneously with age. The alternative NFκB signaling pathway, which is induced by CD40L and BAFF in B cells, plays a central role in peripheral B cell regulation (15, 39–42). Specifically, increased BAFF levels or overexpression of CD40 or BAFF are associated with B cell-mediated autoimmunity (14, 16, 42–44). Based on its suppressive function on the alternative NFκB pathway, it is possible that the loss of DBC1 in mice leads to increased CD40 and BAFFR-induced alternative NFκB signaling, resulting in breakdown of B cell regulation and presence of autoantibodies. The alternative NFκB signaling pathway has also been shown to be important in IgG and IgA class switching (12, 45, 46), consistent with our observation that IgG and IgA isotypes are increased in the sera of aged DBC1 KO mice. However, we did not detect spontaneous onset of clinical symptoms of autoimmunity such as glomurelonephritis in DBC1-deficient mice (data not shown). It is possible that an additional genetic predisposition or antigenic challenge is required to induce further autoimmune symptoms. In support of our speculation, we have previously shown that in an Experimental Autoimmune Myasthenia Gravis (EAMG) setting, DBC1-deficient mice have elevated anti-Acetylcholine autoantibody, and exhibit increased disease severity as a result (34).
Using a microarray approach, we previously identified NFκB target genes to be dysregulated by loss of DBC1. ChIP experiments further show that the activity of alternative NFκB member RelB in particular was increased in DBC1 KO B cells (34). Since NFκB plays a critical role in proliferation and isotype switching of activated B cells, we proposed that the hyper-proliferative and increased IgG and IgA phyenotype in DBC1 KO mice is dependent on increased RelB activity. To confirm that loss of DBC1 leads to increased B cell activation through RelB, we used a Dbc1−/− RelBshep/shep double mutant strain and tested for B cell activation and function. Consistent with our hypothesis, loss of RelB function abrogated the hyper-proliferative phenotype of DBC1 KO mice, affirming that DBC1 suppresses B cell proliferation and immunoglobulin production by inhibiting RelB function. Furthermore, through our Co-IP experiments, we show that DBC1 preferentially interacts with RelB and p52, but not other NFκB members RelA, cRel and p50, consistent with our hypothesis that DBC1 suppresses B cell activation through the alternative NFκB pathway.
NFκB activity is regulated by the IKK regulatory complex (18). While IKKα and IKKβ are found within the same complex in vivo, genetic studies have shown that IKKα and β regulate alternative and classical NFκB activity differentially (18, 47). DBC1 was previously identified as an interacting protein with IKKβ through a mass spectrometry screen (22). In our study, we confirmed that in addition to RelB:p52, DBC1 interacts with IKKα and β in B cells. Moreover, in agreement with our finding that DBC1 regulates the alternative NFκB pathway, DBC1 interacts with a much higher affinity with IKKα, which is required for regulation of alternative NFκB signaling, rather than IKKβ(1, 21). Similarly, IKKα is more effective in increasing Serine phosphorylation of DBC1 compared to IKKβ. Interestingly, upon CD40 activation in B cells, DBC1 interaction with IKKα/β is reduced, and correlates with reduced DBC1 phosphorylation, especially after 16 hours. Moreover, in primary B cells, loss of DBC1 phosphorylation at 16 hours leads to reduced interaction between DBC1 and RelB, as well as shift from its interaction with p52 to its inactive precursor p100 (Fig. 2B). Based on our data, we propose that DBC1 phosphorylation by IKKα enables DBC1 suppression of RelB, and the later loss of phosphorylation on DBC1 reduces its suppressive function, leading to full RelB activity. In support of this, replacing six Serines on DBC1 at its C-terminus, reduced phosphorylation levels of DBC1 and also its interaction with RelB. Basal IKK kinase activity has been reported to serve important metabolic functions, and is actively maintained during cell homeostasis (48–50). Our data thus suggests that in addition to promoting the activation of alternative NFκB by phosphorylating p100 for processing, IKKα also negatively regulates alternative NFκB activity by phosphorylating DBC1 and maintaining its suppression of RelB:p52 activity. In addition, DBC1 was reported to be phosphorylated at T454 by ATM during the DNA damage response (51). In our experiments, using α-pT antibody, we could not detect any threonine phosphorylation of DBC1. Instead, Serine phosphorylation regulates DBC1 function on the alternative NFκB pathway. Therefore, DBC1 appears to regulate B cell activation independent of DNA damage responses.
Thus, our results further define the role of DBC1 as an endogenous suppressor of B cell proliferation and immunoglobulin production, as loss of DBC1 in mice leads to spontaneous production of autoreactive immunoglobulin at 10 months of age. In addition, using a double mutant genetic model, we confirm that DBC1 regulates B cell activation by suppressing the alternative NFκB pathway. At a molecular level, DBC1 suppresses B cell activation selectively binding to alternative NFκB members RelB and p52. In addition, DBC1 preferentially binds to IKKα, and is Serine-phosphorylated at the C terminus. Lastly, phopshorylation of DBC1 by IKKα is crucial for its interaction with IKKα and RelB. DBC1 has been implicated in the regulation of proliferation in solid tumors, but the study of DBC1 in the immune response has been limited. As the alternative NFκB pathway is a crucial regulator of B cell activation and autoimmunity, defining the role of DBC1 in regulating the alternative NFκB pathway provides a rationale for further study of its role in the hematopoietic system, in B cell immunoglobulin production and in autoimmune regulation.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants (AI079056, DK083050 and their supplemental grants) to D.F and (DK-084055/DK/NIDDK NIH HHS/United States) to E.N.C.
Footnotes
Disclosure of Conflicts of Interest. The authors declare no competing financial interests.
Author contributions: SK performed the experiments and analyzed the data; CCSC contributed to development of reagents and animal models; SK, ENC & DF wrote the manuscript.
References
- 1.Bonizzi G, Karin M. The two NF-[kappa]B activation pathways and their role in innate and adaptive immunity. Trends in Immunology. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Grossmann M, Metcalf D, Merryfull J, Beg A, Baltimore D, Gerondakis S. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proceedings of the National Academy of Sciences. 1999;96:11848–11853. doi: 10.1073/pnas.96.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. The EMBO Journal. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao Z, Li Y, Yin X, Dong Y, Xing L, Boyce BF. NF-kappaB RelB negatively regulates osteoblast differentiation and bone formation. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2014;29:866–877. doi: 10.1002/jbmr.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, Grinberg A, Tran T, Scharton-Kersten T, Anver M, Love P, Brown K, Siebenlist U. Mice Deficient in Nuclear Factor (NF)-κB/p52 Present with Defects in Humoral Responses, Germinal Center Reactions, and Splenic Microarchitecture. The Journal of Experimental Medicine. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shih VF, Davis-Turak J, Macal M, Huang JQ, Ponomarenko J, Kearns JD, Yu T, Fagerlund R, Asagiri M, Zuniga EI, Hoffmann A. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-kappaB pathways. Nat Immunol. 2012;13:1162–1170. doi: 10.1038/ni.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormier F, Monjanel H, Fabre C, Billot K, Sapharikas E, Chereau F, Bordereaux D, Molina TJ, Avet-Loiseau H, Baud V. Frequent engagement of RelB activation is critical for cell survival in multiple myeloma. PLoS One. 2013;8:e59127. doi: 10.1371/journal.pone.0059127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Tänzer S, Busslinger M, Weih F. Lack of nuclear factor-κB2/p100 causes a RelB-dependent block in early B lymphopoiesis. Blood. 2008;112:551–559. doi: 10.1182/blood-2007-11-125930. [DOI] [PubMed] [Google Scholar]
- 9.Guo F, Weih D, Meier E, Weih F. Constitutive alternative NF-κB signaling promotes marginal zone B-cell development but disrupts the marginal sinus and induces HEV-like structures in the spleen. Blood. 2007;110:2381–2389. doi: 10.1182/blood-2007-02-075143. [DOI] [PubMed] [Google Scholar]
- 10.Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM. IKK controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 2006;2006:3801–3812. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liou HC, Sha WC, Scott ML, Baltimore D. Sequential induction of NF-kappa B/Rel family proteins during B-cell terminal differentiation. Molecular and Cellular Biology. 1994;14:5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck RP, Lira SA, Bravo R. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 13.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-[kappa]B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 14.Kim JY, Yang Y, Moon JS, Lee EY, So SH, Lee HS, Park KD, Choi YC. Serum BAFF expression in patients with myasthenia gravis. J Neuroimmunol. 2008;199:151–154. doi: 10.1016/j.jneuroim.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Davidson A. BAFF and selection of autoreactive B cells. Trends in Immunology. 2011;32:388–394. doi: 10.1016/j.it.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushita T, Hasegawa M, Yanaba K, Kodera M, Takehara K, Sato S. Elevated serum BAFF levels in patients with systemic sclerosis: enhanced BAFF signaling in systemic sclerosis B lymphocytes. Arthritis and rheumatism. 2006;54:192–201. doi: 10.1002/art.21526. [DOI] [PubMed] [Google Scholar]
- 17.Warnatz K, Salzer U, Rizzi M, Fischer B, Gutenberger S, Bohm J, Kienzler AK, Pan-Hammarstrom Q, Hammarstrom L, Rakhmanov M, Schlesier M, Grimbacher B, Peter HH, Eibel H. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106:13945–13950. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Israel A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harbor perspectives in biology. 2010;2:a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB Kinase Complex (IKK) Contains Two Kinase Subunits, IKKα and IKKβ, Necessary for IκB Phosphorylation and NF-κB Activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV. Embryonic Lethality, Liver Degeneration, and Impaired NF-κB Activation in IKK-β-Deficient Mice. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- 21.Senftleben U, Cao Y, Xiao G, Greten FR, Krähn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Activation by IKKα of a Second, Evolutionary Conserved, NF-κB Signaling Pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 22.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-[alpha]/NF-[kappa]B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 23.Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proceedings of the National Academy of Sciences. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH, Park HS, Lee H, Chung MJ, Kang MJ, Lee DG, Moon WS, Jang KY. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009;15:4453–4459. doi: 10.1158/1078-0432.CCR-08-3329. [DOI] [PubMed] [Google Scholar]
- 25.Kim SH, Kim JH, Yu EJ, Lee KW, Park CK. The overexpression of DBC1 in esophageal squamous cell carcinoma correlates with poor prognosis. Histol Histopathol. 2012;27:49–58. doi: 10.14670/HH-27.49. [DOI] [PubMed] [Google Scholar]
- 26.Kim JE, Chen J, Lou Z. p30 DBC is a potential regulator of tumorigenesis. Cell Cycle. 2009;8:2932–2935. doi: 10.4161/cc.8.18.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyama S, Wada-Hiraike O, Nakagawa S, Tanikawa M, Hiraike H, Miyamoto Y, Sone K, Oda K, Fukuhara H, Nakagawa K, Kato S, Yano T, Taketani Y. Repression of estrogen receptor [beta] function by putative tumor suppressor DBC1. Biochemical and Biophysical Research Communications. 2010;392:357–362. doi: 10.1016/j.bbrc.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Hiraike H, Wada-Hiraike O, Nakagawa S, Koyama S, Miyamoto Y, Sone K, Tanikawa M, Tsuruga T, Nagasaka K, Matsumoto Y, Oda K, Shoji K, Fukuhara H, Saji S, Nakagawa K, Kato S, Yano T, Taketani Y. Identification of DBC1 as a transcriptional repressor for BRCA1. Br J Cancer. 2010;102:1061–1067. doi: 10.1038/sj.bjc.6605577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chini CCS, Escande C, Nin V, Chini EN. HDAC3 Is Negatively Regulated by the Nuclear Protein DBC1. Journal of Biological Chemistry. 2010;285:40830–40837. doi: 10.1074/jbc.M110.153270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. Inhibition of SUV39H1 Methyltransferase Activity by DBC1. Journal of Biological Chemistry. 2009;284:10361–10366. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu J, Jiang J, Li J, Wang S, Shi G, Feng Q, White E, Qin J, Wong J. Deleted in Breast Cancer 1, a Novel Androgen Receptor (AR) Coactivator That Promotes AR DNA-binding Activity. Journal of Biological Chemistry. 2009;284:6832–6840. doi: 10.1074/jbc.M808988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong S, Thiruppathi M, Qiu Q, Lin Z, Dong H, Chini EN, Prabhakar BS, Fang D. DBC1 Is a Suppressor of B Cell Activation by Negatively Regulating Alternative NF-kappaB Transcriptional Activity. J Immunol. 2014 doi: 10.4049/jimmunol.1401798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairfield H, Gilbert GJ, Barter M, Corrigan RR, Curtain M, Ding Y, D’Ascenzo M, Gerhardt DJ, He C, Huang W, Richmond T, Rowe L, Probst FJ, Bergstrom DE, Murray SA, Bult C, Richardson J, Kile BT, Gut I, Hager J, Sigurdsson S, Mauceli E, Di Palma F, Lindblad-Toh K, Cunningham ML, Cox TC, Justice MJ, Spector MS, Lowe SW, Albert T, Donahue LR, Jeddeloh J, Shendure J, Reinholdt LG. Mutation discovery in mice by whole exome sequencing. Genome Biol. 2011;12:R86. doi: 10.1186/gb-2011-12-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szabo P, Li F, Mathew J, Lillvis J, Weksler ME. Evolution of B-cell clonal expansions with age. Cellular immunology. 2004;231:158–167. doi: 10.1016/j.cellimm.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Weksler ME. Changes in the B-cell repertoire with age. Vaccine. 2000;18:1624–1628. doi: 10.1016/s0264-410x(99)00497-1. [DOI] [PubMed] [Google Scholar]
- 38.Goidl EA, Michelis MA, Siskind GW, Weksler ME. Effect of age on the induction of autoantibodies. Clin Exp Immunol. 1981;44:24–30. [PMC free article] [PubMed] [Google Scholar]
- 39.Hsing Y, Hostager BS, Bishop GA. Characterization of CD40 signaling determinants regulating nuclear factor-kappa B activation in B lymphocytes. J Immunol. 1997;159:4898–4906. [PubMed] [Google Scholar]
- 40.Rickert RC, Jellusova J, Miletic AV. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol Rev. 2011;244:115–133. doi: 10.1111/j.1600-065X.2011.01067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatada EN, Do RKG, Orlofsky A, Liou HC, Prystowsky M, MacLennan ICM, Caamano J, Chen-Kiang S. NF-κB1 p50 Is Required for BLyS Attenuation of Apoptosis but Dispensable for Processing of NF-κB2 p100 to p52 in Quiescent Mature B Cells. The Journal of Immunology. 2003;171:761–768. doi: 10.4049/jimmunol.171.2.761. [DOI] [PubMed] [Google Scholar]
- 42.Meffre E. The establishment of early B cell tolerance in humans: lessons from primary immunodeficiency diseases. Annals of the New York Academy of Sciences. 2011;1246:1–10. doi: 10.1111/j.1749-6632.2011.06347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lougaris V, Badolato R, Ferrari S, Plebani A. Hyper immunoglobulin M syndrome due to CD40 deficiency: clinical, molecular, and immunological features. Immunol Rev. 2005;203:48–66. doi: 10.1111/j.0105-2896.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- 44.Higuchi T, Aiba Y, Nomura T, Matsuda J, Mochida K, Suzuki M, Kikutani H, Honjo T, Nishioka K, Tsubata T. Cutting Edge: Ectopic expression of CD40 ligand on B cells induces lupus-like autoimmune disease. J Immunol. 2002;168:9–12. doi: 10.4049/jimmunol.168.1.9. [DOI] [PubMed] [Google Scholar]
- 45.Enzler T, Bonizzi G, Silverman Gregg J, Otero DC, Widhopf George F, Anzelon-Mills A, Rickert Robert C, Karin M. Alternative and Classical NF-κB Signaling Retain Autoreactive B Cells in the Splenic Marginal Zone and Result in Lupus-like Disease. Immunity. 2006;25:403–415. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Jin J, Xiao Y, Chang JH, Yu J, Hu H, Starr R, Brittain GC, Chang M, Cheng X, Sun SC. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-kappaB signaling. Nat Immunol. 2012;13:1101–1109. doi: 10.1038/ni.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallabhapurapu S, Karin M. Regulation and Function of NF-κB Transcription Factors in the Immune System. Annual Review of Immunology. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 48.Buss H, Dorrie A, Schmitz ML, Hoffmann E, Resch K, Kracht M. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J Biol Chem. 2004;279:55633–55643. doi: 10.1074/jbc.M409825200. [DOI] [PubMed] [Google Scholar]
- 49.Palma L, Crinelli R, Bianchi M, Magnani M. De-ubiquitylation is the most critical step in the ubiquitin-mediated homeostatic control of the NF-kappaB/IKK basal activity. Mol Cell Biochem. 2009;331:69–80. doi: 10.1007/s11010-009-0146-x. [DOI] [PubMed] [Google Scholar]
- 50.Tam AB, Mercado EL, Hoffmann A, Niwa M. ER stress activates NF-kappaB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS One. 2012;7:e45078. doi: 10.1371/journal.pone.0045078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]