Abstract
Programs that aim to control vector-borne zoonotic diseases require information on zoonotic hosts and on the feeding behavior of bridging vectors that are capable of transmitting pathogens from those hosts to humans. Here we describe an assay developed to identify bloodmeals in field-collected cat fleas (Ctenocephalides felis Bouché) to assess this species’ potential role as a Yersinia pestis bridging vector in a plague-endemic region of Uganda. Our assay uses a single primer set and SYBR Green I-based real-time polymerase chain reaction to amplify a segment of the 12S mitochondrial ribosomal RNA gene for identification by sequencing. The assay capitalizes on the sensitivity of real-time polymerase chain reaction and the specificity of sequencing and can be used to differentiate vertebrate bloodmeals to the genus or species level without a priori knowledge of the host community. Because real-time assays that detect vertebrate DNA are highly sensitive to human DNA contamination, we analyzed detection in artificially fed and unfed fleas to establish a Ct cutoff that optimized specificity without completely sacrificing sensitivity. Using the established cutoff, our assay detected human, rat, and goat DNA in artificially fed C. felis up to 72 h postfeeding.
Keywords: bloodmeal identification, flea, real-time PCR, threshold cycle, cutoff
Programs that aim to control vector-borne zoonotic diseases require information on zoonotic hosts and on the feeding behavior of bridging vectors capable of transmitting pathogens from those hosts to humans (Gratz 1999, Kirstein and Gray 1999, Kilpatrick et al. 2005, Steuber et al. 2005). To better understand the feeding behavior of bridging vector species, researchers have developed various methodologies to identify bloodmeals in hematophagous arthropods.
For decades, researchers relied on serologic techniques, including precipitin tests, passive hemagglutination inhibition tests, and enzyme linked immunosorbent assays, to identify bloodmeals in arthropod vectors (Tempelis 1975, Washino and Tempelis 1983, Pant et al. 1987). These antigen-based methods have provided invaluable insights into the feeding behavior of many disease vectors, but they require the production of antisera against each potential bloodmeal source (Washino and Tempelis 1983). Serologic tests thus require a priori knowledge of the host community; they cannot detect “unpredicted” hosts. In addition, their specificity is limited by the potential for cross-reactivity between related animal antisera (Rurangirwa et al. 1986, Van den Hurk et al. 2007).
Over the last two decades, researchers have adapted polymerase chain reaction (PCR)-based techniques to identify arthropod bloodmeals. These assays have been used to identify bloodmeal sources from a variety of disease vectors, including mosquitoes (Chow–Shaffer et al. 2000, Kent and Norris 2005), ticks (Humair et al. 2007), sand flies (Haouas et al. 2007), tsetse flies (Steuber et al. 2005), kissing bugs (Mota et al. 2007), biting midges (Bartsch et al. 2009), and fleas (Woods et al. 2009, Franklin et al. 2010). Molecular methods for bloodmeal identification include amplification and analysis of microsatellites, minisatellites, short and long interspersed elements, heteroduplex analysis, reverse line-blot hybridization, restriction fragment length polymorphism, and terminal restriction fragment-length polymorphism analysis (Mukabana et al. 2002a, Pizarro and Stevens 2008, Kent 2009). PCR-based assays can be more specific than serologic tests, allowing identification to the species or even the individual level (Chow–Shaffer et al. 2000, Steuber et al. 2005). Some researchers have combined molecular methods with serologic techniques, using PCR, for example, to identify species that an enzyme linked immunosorbent assay cannot reliably detect or differentiate (Van den Hurk et al. 2007). Others have developed highly specific stand-alone assays using conventional PCR with species-specific primers, probes or restriction fragment-length polymorphism analysis (Kent and Norris 2005, Steuber et al. 2005, Humair et al. 2007).
Real-time PCR is more sensitive than conventional PCR (Lanciotti et al. 2000). A highly sensitive assay is advantageous for detecting DNA from tiny bloodmeals that may be degraded because of digestion or storage conditions (Mukabana et al. 2002a, Kent 2009). Researchers have developed probe-based real-time assays to identify bloodmeals in mosquitoes and fleas, but this technique requires knowledge of potential host species (Van den Hurk et al. 2007, Woods et al. 2009). Using “universal primers” to amplify a molecular target from a group of species (e.g., vertebrates or mammals) and then sequencing the amplicon to identify the bloodmeal source allows for specific identification of both predicted and unpredicted hosts (Haouas et al. 2007, Kent et al. 2009). This is useful in situations when there is little or no data available on host abundance and diversity (Haouas et al. 2007). Here, we describe a SYBR Green I-based real-time PCR assay using a single primer set to amplify vertebrate DNA for identification by sequencing.
Our assay capitalizes on the sensitivity of real-time PCR and the specificity of sequencing. Like other sensitive molecular assays that amplify vertebrate DNA, real-time PCR is subject to human DNA contamination (Urban et al. 2000). Here, we discuss the utility of establishing an appropriate threshold cycle (Ct) cutoff value to dichotomize positive and negative results and thus reduce the risk of false positives. Having established an appropriate cutoff, we tested the assay’s ability to detect vertebrate DNA in artificially fed fleas up to 98 h postfeeding.
We developed this method specifically to identify bloodmeals in field-collected cat fleas (Ctenocephalides felis Bouché) to assess this species’ potential role as a bridging vector in the West Nile region of Uganda. Within this region, cat fleas are the predominant off-host species collected from human habitations, the presumed sites of human exposure to the plague bacterium (Eisen et al. 2008c). Previous studies have demonstrated that C. felis is a competent vector of Yersinia pestis (Eisen et al. 2008c). However, cat flea feeding preferences in this region are largely unknown. Our aim was to develop an assay that would allow us to determine what proportion of all cat flea bloodmeals come from small mammals that are susceptible to Y. pestis and might serve as a source of infection in this region, and what proportion of cat flea bloodmeals come from humans that could become infected by feeding cat fleas. We required an assay that could be used to identify bloodmeals from all vertebrate hosts to establish the proportion of bloodmeals that could be attributed to any given species. Existing bloodmeal assays were deemed inadequate to assess the role of cat fleas as bridging vectors because they were either not sensitive enough to detect the very small amounts of DNA that are often observed in field-collected fleas or unable to capture the wide range of potential vertebrate hosts in this region.
Materials and Methods
Primer Selection
Mitochondrial DNA is a commonly used and effective target for bloodmeal assays because of its high copy number and rapid rate of evolution relative to nuclear DNA (Kent 2009). Optimal amplicon size for real-time assays using SYBR Green I as a fluorescent reporter is between 75 and 200 base pairs (bp) (Bio-Rad Laboratories 2006), and the ability to detect partially digested bloodmeals via PCR appears to be inversely related to amplicon size (Kirstein and Gray 1996, Kent and Norris 2005, Davey et al. 2007). Therefore, we sought to identify primers that would amplify a relatively small mitochondrial gene fragment from vertebrates without amplifying flea DNA. Additional criteria for our molecular target included low intra-species variability, sufficient inter-species variability to allow for differentiation at the species or genus level, and minimal primer degeneracy as this increases the risk of mispriming and may thus generate unintended amplification products and false positives in SYBR Green I-based assays (Bustin et al. 2009). We found the mitochondrial gene encoding 12S ribosomal RNA best suited to our purposes.
Humair et al. (2007) identified two highly conserved regions flanking a variable region within the 12S rRNA gene. We retrieved the corresponding nucleotide sequences from the GenBank database for C. felis (U73741.1) and for six species believed to be potential hosts for C. felis in the West Nile region of Uganda: cat (Felis catus; NC_001700.1), chicken (Gallus gallus; NC_001323.1), human (Homo sapiens; NC_012920.1), black rat (Rattus rattus; AJ005780.1), domestic dog (Canis lupus familiaris; NC_002008.4), and goat (Capra hircus; NC_005044.2). We aligned the sequences using ClustalW2 and designed a two-fold degenerate forward primer, 12S_425F (5′ – TGT AAA ACG ACG GCC AGT GGG ATT AGA TAC CCY ACT ATG C – 3′), that can be paired with the 12S_9R primer described by Humair et al. (2007) (5′ – CAG GAA ACA GCT ATG ACA GAA CAG GCT CCT CTA G – 3′) to amplify vertebrate DNA without amplifying C. felis DNA. Each primer included an M13 tag (shown in italics in primer sequences) to facilitate sequencing (Ivanova et al. 2007). When we compared real-time amplification of DNA from three different vertebrate species using primers with and without these tags, we found that they behaved similarly, although the tagged primers benefited from an increased annealing temperature (data not shown). Using the tags allowed us to achieve longer, cleaner sequences, so we used the tagged primers for all experiments described here. The target amplicon length was ≈ 175 bp.
DNA Extraction
Before extracting DNA from individual fleas, we removed surface contaminants by soaking each flea in 50% bleach (3.08% sodium hypochlorite) and rinsing in calcium- and magnesium-free Dulbecco’s Phosphate Buffered Saline (DPBS). Each flea was then homogenized in 100 μl DPBS using 3-mm glass beads and a Mixer Mill (model MM300, Retsch, Hann, Germany) set at 20 beats per second for 10 min. We extracted DNA from the homogenate using the QIAamp DNA Micro Kit (Qiagen, Valencia, CA) per the manufacturer’s instructions for purification of genomic DNA from small volumes of blood and eluted the DNA with 70 μl PCR-grade water (Roche Diagnostics, Indianapolis, IN). We extracted DNA from whole, citrated Rattus norvegicus Wistar Hannover blood (Bioreclamation, Westbury, NY) using the DNeasy Blood and Tissue Kit (Qiagen) and following the standard kit protocol. All DNA was stored at −80°C until PCR analysis.
Real-Time PCR
Each 25-μl real-time reaction contained 12.5 μl iQ SYBR Green Supermix (2X; Bio-Rad Laboratories, Hercules, CA), 100 nM forward and reverse primers, and 10 μl DNA template. We performed all reactions in an Mx3005P thermal cycler (Stratagene, LA, Jolla, CA). A 5-min initial denaturation at 95°C was followed by 38 – 40 amplification cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s. Bound SYBR Green I fluorescence was measured after each amplification cycle. The software set the background fluorescence for all samples at a common starting point (baseline correction), then plotted relative fluorescence (dR) versus cycle number to generate an amplification plot for each sample. The final amplification cycle was immediately followed by a melting analysis cycle during which all products were melted at 95°C for 1 min, annealed at 55°C for 30 s, and then subjected to a gradual rise in temperature to 95°C. The PCR software collected fluorescence data after each incremental temperature increase. The software then plotted the negative first derivative of the raw fluorescence data versus temperature to generate a melting peak. A single peak indicates the presence of a single PCR product in the corresponding well, and similar amplicons peak at similar temperatures (Ririe et al. 1997).
Each real-time run included at least two no-template-control (NTC) wells (water in place of template DNA), and control DNA isolated from R. norvegicus whole blood, diluted to generate a single stock, and stored at −80°C in single-use aliquots. The R. norvegicus DNA served as a positive control and an inter-run calibrator (IRC). We ran the IRC in the same position on all plates. After each run, we manually adjusted the threshold value such that the IRC amplification plot crossed the threshold at the same cycle. By keeping the IRC Ct value constant, we ensured equivalent threshold settings across runs. After we set the threshold, the software determined a Ct value for each sample.
We ran all samples, including the IRC, in duplicate. Following each run, we examined the results to ensure that replicates had similar Ct values and melting peaks. The software calculated a collective Ct value from each pair of replicates by averaging the fluorescence of both replicate wells at every cycle to generate a common amplification plot for the sample. We considered a sample positive at a given Ct cutoff value if the collective Ct value was less than or equal to the cutoff, at least one replicate crossed the threshold below the cutoff and the second replicate crossed no > 2 cycles above the cutoff. We repeated any sample with dissimilar replicates. If a sample repeatedly yielded inconsistent replicates, or if a sample repeatedly generated melting curves with multiple peaks, we concluded that the assay could not reliably detect a bloodmeal in that sample.
Sequencing
Following PCR, all amplicons were stored promptly at 4°C or −20°C. We chose a single replicate from each positive sample for sequencing and purified the amplicon using the QIAquick PCR Purification Kit (Qiagen). We eluted the purified product with 20 μl nuclease-free H2O and estimated its concentration using a NanoDrop 2000 Spectrophotometer (Thermo Fisher, Waltham, MA). Purified products were used immediately or stored at −20°C until sequencing. We prepared forward and reverse sequencing reactions for each sample. Each 20-μl sequencing reaction contained ≈ 6 ng purified amplicon, 6.6 pmol M13 forward or reverse sequencing primer (Ivanova et al. 2007), and 8.0 μl BigDye Terminator v3.1 Ready Reaction Mix (Applied Biosystems, Foster City, CA). Cycle sequencing comprised a 1 min denaturation at 96°C followed by 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. We used the BigDye Xterminator Kit (Applied Biosystems) to remove unincorporated dyes before analyzing the samples on an ABI 3130XL genetic analyzer. Using Lasergene software (DNASTAR, Inc., Madison, WI), we analyzed forward and reverse sequence traces and assembled a single sequence for each sample. We then manually removed the primer sequences from either end of the sequence and identified the source of the DNA using Basic Local Alignment Search Tool (BLAST) to find matching mitochondrial sequences in the nucleotide collection database.
To minimize the risk of cross-contamination, we conducted DNA isolation, PCR set-up, and amplicon processing in three separate rooms. We used certified DNA-free pipet tips and tubes for DNA isolation and PCR set-up. All surfaces and equipment were treated with 0.62– 0.99% sodium hypochlorite or DNA Away (Molecular BioProducts, San Diego, CA) and rinsed with 70% reagent alcohol (Ricca Chemical Company, Arlington, TX) before and after use.
Establishing Ct Cutoff Value
Blood
We established a Ct cutoff value using rat and human blood because the assay was developed to assess the proportion of vertebrate C. felis bloodmeals taken from humans and the proportion taken from small mammals that might serve as a source of Y. pestis infection in the West Nile region of Uganda. The black rat (R. rattus) is the predominant small mammal species in human habitations in this region, is highly susceptible to Y. pestis, and develops the very high bacterial burden required to infect feeding fleas (Pollitzer 1954, Lorange et al. 2005, Borchert et al. 2007, Amatre et al. 2009, Tollenaere et al. 2010). We selected R. norvegicus as our rat blood source for this experiment because it is commercially available and the complete R. norvegicus 12S sequence is available from the GenBank database. We determined that the R. norvegicus sequence flanked by our primers differs from the R. rattus sequence by only three nucleotides. Therefore, we concluded that R. norvegicus blood could serve as a reasonable proxy for R. rattus blood.
Fleas
To determine the assay’s sensitivity and specificity at different Ct cutoff values, we used Xenopsylla cheopis Rothschild reared in colonies maintained by the Centers for Disease Control and Prevention Division of Vector-Borne Diseases (Fort Collins, CO). We starved adult fleas for 6 d before offering them a bloodmeal using artificial feeders containing citrated whole blood that had been collected 2 d earlier from either human or rat (Bioreclamation). The artificial feeding system was developed previously for transmission studies using Y. pestis-infected blood (Eisen et al. 2006, 2007, 2008a, c; Wilder et al. 2008). Blood was added to a glass reservoir to which a shaved mouse skin had been secured. Fleas were placed in a 50-ml syringe barrel secured to the other side of the membrane. A plunger was then inserted into the barrel, forcing the fleas to within a few centimeters of the membrane. The feeder was suspended with the membrane facing down, and water circulating through a jacket around the reservoir kept the blood at 37°C. After 1 h, fleas were removed from the feeders, immobilized by chilling on ice and examined by light microscopy for the presence of an obvious red bloodmeal in the proventriculus or midgut. We stored fed fleas alive for 24 h in flasks containing filter paper at 23°C and ~85% relative humidity (RH). We then transferred all surviving fed fleas to petri dishes containing the solution used to trap off-host fleas in our field studies: 2% NaCl with two drops per liter Tween 80. The fleas died in the trap solution, and we transferred them by hand to 95% ethanol. Because fleas are usually removed from traps in Uganda by hand without wearing gloves, we used the same method in this experiment so that our results would reflect any impact this might have on vertebrate DNA detection using our assay. We then stored individual fleas in tubes of 95% ethanol at −20°C until DNA isolation.
Unfed X. cheopis were collected by sifting pupae from the colony medium and holding the pupae in a conical tube for 2 wk. We then shook the tube gently to encourage emergence and harvested teneral fleas by vacuum aspiration. We transferred the fleas to trap solution and processed them using the same method described above for fed fleas.
Calculating Optimal Ct Cutoff Value
We established a Ct cutoff value for our real-time assay using a modification of the “epidemiological approach” described by Caraguel et al. (2011). We expected DNA isolated from fed fleas to contain either human or rat DNA and DNA isolated from unfed fleas to be negative for vertebrate DNA. Using the real-time protocol described above with 40 amplification cycles, we tested each sample in duplicate and recorded either its collective Ct value or that it had no detectable blood-meal. To ensure that any differences between real-time runs would similarly affect all sample types, each run included rat blood-fed flea DNA, human blood-fed flea DNA, unfed flea DNA, and NTCs. We then determined whether each human and rat blood-fed flea would be classified as having a detectable blood-meal at each Ct cutoff value between 34 and 40 cycles. From this we calculated assay sensitivity (the percentage of blood-fed fleas with a detectable blood-meal) at each Ct cutoff value. We followed the same procedure using the unfed flea data to determine assay specificity (the percentage of negative samples in which we could not detect vertebrate DNA). We sequenced amplicons from all false-positive samples to identify the source of contaminating vertebrate DNA. We then calculated the positive predictive value (PPV, the percentage of samples with detectable rat DNA that came from rat blood-fed fleas and the percentage of samples with detectable human DNA that came from human blood-fed fleas) at each Ct cutoff value. We used these calculations to identify the Ct cutoff value that maximized PPV. That is, we selected the Ct cutoff that maximized the chances that a flea that tested positive for rat or human DNA had actually consumed a rat or human bloodmeal, respectively. We sequenced all samples with a Ct value below that cutoff to verify that we had amplified the expected sequence from human and rat blood-fed fleas.
Determining Real-Time Assay Linear Dynamic Range, Limit of Detection, and Variability
To verify that the established Ct cutoff fell within the dynamic range over which the real-time reaction is linear, we diluted R. norvegicus DNA 10-fold over six logs and ran five replicates of each dilution. We defined the limit of detection (LOD) for our assay as the lowest serial concentration at which all five replicates crossed the threshold before cycle 40 (Caraguel et al. 2011). We defined linear dynamic range (LDR) as the range of concentrations (highest to LOD) over which data from all replicates could be fit to a standard curve plot with an R2 ≥ 0.985. We analyzed intra-assay variation by calculating the standard deviation of Ct values at each concentration within the LDR (Bustin et al. 2009).
Assessing Assay Sensitivity to Bloodmeal DNA Over Time
To determine the effect of C. felis digestion on assay sensitivity over time, we allowed unfed, colony-reared C. felis (Heska Corporation, Loveland, CO) to feed on 1-d-old, citrated human, rat, or goat blood (Bioreclamation) using artificial feeders. We included goat blood in this analysis because a previous survey found that this domesticated species was present in or around >75% of huts in the West Nile region of Uganda (Centers for Disease Control and Prevention, unpublished data), and others have reported that C. felis readily infests goats (Fagbemi 1982, Kaal et al. 2006). Given that field-collected fleas from Uganda were likely to have fed on this domesticated species, we sought to determine if the assay showed similar sensitivity to goat, human, and rat blood in artificially fed cat fleas. Fed fleas were identified by light microscopy and held alive as described above. Surviving fleas were collected by vacuum aspiration and stored at −20°C in 95% ethanol at seven time points postfeed: 2, 4, 12, 24, 48, 72, and 98 h. We chose 98 h as the cutoff because C. felis generally die within 4 d of leaving their host (Dryden 1989). We tested each flea using our real-time assay with 38 amplification cycles, and a sample was considered positive only if it had a collective Ct value ≤36. Each real-time run included an IRC, NTCs, and negative control DNA from unfed C. felis. Amplicons from all positive samples were sequenced to verify that we had amplified the correct vertebrate DNA (human, rat, or goat). We compared the percentage of positive samples between time points for each blood type and between blood types for each time point using contingency table analysis and Fisher Exact tests, with a P < 0.05 indicating statistical significance. We conducted all statistical comparisons using JMP statistical software (SAS Institute, Cary, NC).
Results
Ct Cutoff Value
We isolated DNA from individual, artificially fed X. cheopis collected 24 h postfeed (n = 50 rat blood-fed and 50 human blood-fed fleas), and from unfed X. cheopis adults (n = 50). Using our real-time assay, we determined the Ct value for each sample. The DNA samples from 5 (10%) of the human blood-fed fleas, 6 (12%) of the rat blood-fed fleas, and 44 (88%) of the unfed fleas had a Ct value >40 or repeatedly yielded dissimilar replicates. Two additional rat blood-fed flea samples (4%) repeatedly yielded melting curves with multiple peaks. We classified all of these samples as having no detectable bloodmeal at any Ct cutoff value. All other samples were considered positive at a given Ct cutoff value if both replicates amplified no >2 cycles above the cutoff and had a collective Ct value less than or equal to the cutoff. Sensitivity to vertebrate DNA in rat blood-fed fleas and human blood-fed fleas decreased from 84 to 68% and from 90 to 20%, respectively, as we decreased the Ct cutoff from 40 to 34 (Table 1). Assay specificity (the proportion of unfed fleas correctly identified as having no detectable vertebrate blood-meal), increased by 8%, from 88 to 96%, over the same range (Table 1). Via sequencing, we identified the DNA in all six false-positive samples (unfed flea isolates with detectable vertebrate DNA) as human. Because none of the false positives was identified as R. norvegicus DNA, the effective PPV of a sample identified as rat-fed was 100% regardless of the Ct cutoff value (100% of fleas identified as rat blood-fed were rat-blood fed; Table 1). Therefore, we focused on the PPV associated with samples containing human DNA. Specifically, we calculated the PPV associated with human blood-fed versus unfed fleas (the percentage of samples in which the assay detected human DNA that came from human blood-fed fleas). The maximum PPV occurs at a Ct cutoff value of 36 (Table 1). Using this cutoff value, our assay is 94% specific; we expect a 6% false positive rate.
Table 1.
Assay performance using Ct cutoff values between 34 and 40
| Ct cutoff value | Se (%): RF (n = 50 fleas) | Se (%): HF (n = 50 fleas) | Sp (%)(n = 50 fleas) | PPV (%):RF vs UF | PPV (%):HF vs UF | % NTCs with no detectable vertebrate DNA (n = 110 wells) |
|---|---|---|---|---|---|---|
| 40 | 84 | 90 | 88 | 100 | ≤88.2a | ≥52.7b |
| 39 | 84 | 90 | 90 | 100 | ≤90.0a | ≥66.4b |
| 38 | 84 | 90 | 90 | 100 | ≤90.0a | ≥80.9b |
| 37 | 78 | 76 | 92 | 100 | ≤90.5a | ≥95.5b |
| 36 | 74 | 64 | 94 | 100 | 91.4 | 100 |
| 35 | 72 | 40 | 96 | 100 | 90.9 | 100 |
| 34 | 68 | 20 | 96 | 100 | 83.3 | 100 |
Se, sensitivity, the percentage of rat blood-fed (RF) fleas or human blood-fed (HF) fleas with a detectable vertebrate blood meal; Sp, specificity, the percentage of unfed fleas without detectable vertebrate DNA; PPV, positive predictive value, the percentage of fleas with detectable rat DNA that had consumed rat blood (RF vs unfed [UF]) or the percentage of fleas with detectable human DNA that consumed human blood (HF vs UF); NTC, no template control.
Sequencing indicated that all unfed flea DNA isolates with Ct ≤ 40 contained human DNA. We sequenced amplicons from fed flea samples only if they had a Ct value ≤ 36. We calculated PPV assuming that all rat blood-fed fleas and human blood-fed fleas with a detectable vertebrate blood meal and a Ct value >36 contained rat or human DNA, respectively. The true PPV for with Ct cutoff values > 36 could therefore be lower.
We did not sequence amplicons from NTCs with Ct > 36. Sequencing could lead us to re-classify some false-positive NTCs as negative, increasing the percent of NTCs with no detectable vertebrate DNA.
Using a Ct cutoff value of 36, we did not detect a significant difference between the percentage of rat blood-fed fleas with detectable rat DNA (74.0%) and the percentage of human blood-fed fleas with detectable human DNA (64.0%; Fisher Exact, P = 0.39). This indicates that, using this cutoff, the assay is no more or less sensitive to rat DNA than to human DNA in artificially fed X. cheopis collected 24 h after feeding. We sequenced amplicons from all of our positive samples with a collective Ct value ≤ 36. Amplicon sequences from all positive samples isolated from human blood-fed fleas were most similar to the expected region of human 12S rDNA. Likewise, amplicon sequences from all positive samples isolated from rat blood-fed fleas were most similar to the expected region of R. norvegicus 12S rDNA.
We included a total of 110 NTCs in our real-time runs. Using a Ct cutoff value of 40, our assay yielded 52 (52.7%) false positives (Table 1). Only one NTC well had a Ct value ≤ 36, however, and repeated attempts to sequence the amplicon failed. Therefore, we classified this NTC as containing no detectable vertebrate DNA. Thus, using a Ct cutoff value of 36, 100% of our NTCs were effectively negative (Table 1).
Linear Dynamic Range, Limit of Detection, and Variability
Using 10-fold dilutions of R. norvegicus DNA, we established 100 fg as the LOD for our real-time assay. The corresponding collective Ct value was 36.43. The highest concentration we tested had a collective Ct value of 21.73. All replicates between 100 fg and 1 ng could be fit to a linear standard curve plot (R2 = 0.990). Thus, the Ct cutoff value we selected, 36, falls within the assay’s LDR (21.73 ≤ Ct ≤ 36.43). As is typical in real-time assays, intra-assay variation increased as DNA concentration decreased (Bustin 2009). At the highest concentration tested, Ct standard deviation over five replicates was 0.09. At the limit of detection, the standard deviation for Ct variance over five replicates was 1.19.
Assay Sensitivity to Bloodmeal DNA Over Time
To determine the effect of digestion on assay sensitivity in C. felis, we artificially fed C. felis either human, rat (R. norvegicus), or goat blood and collected a subset of fed fleas at each of seven time points. We considered a sample positive for vertebrate DNA only if it yielded similar replicates with a collective Ct value ≤ 36. Amplicon sequences from all positive samples isolated from human blood-fed fleas were most similar to the expected region of human 12S rDNA. One rat blood-fed flea collected 98 h after feeding and one goat blood-fed flea collected 72 h after feeding also tested positive for human DNA. All other amplicon sequences from positive rat and goat blood-fed fleas were most similar to the expected region of R. norvegicus and C. hircus 12S rDNA, respectively.
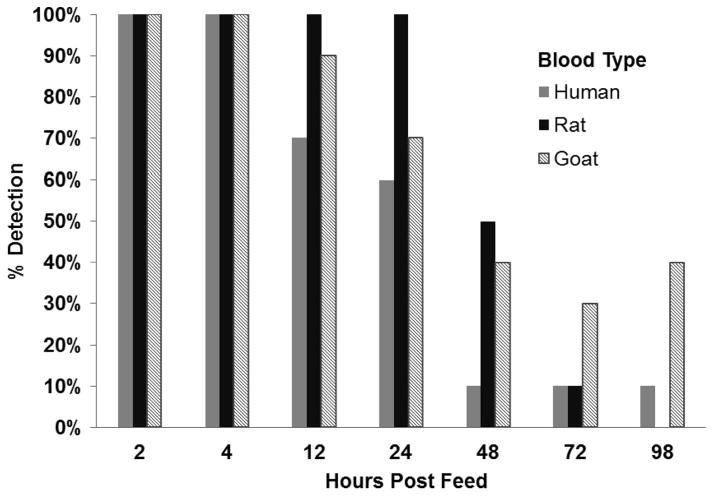
We determined the percentage of fed fleas in which the assay detected and correctly identified a blood-meal at each time point. The results are summarized in Fig. 1. Based on pair-wise comparisons of each blood type, we did not detect a significant difference between the proportion of human blood-fed, rat blood-fed, and goat blood-fed fleas with an identifiable bloodmeal overall or at any single time point (two-tailed Fisher Exact, P ≥ 0.09). As expected, detection of all blood types decreased over time. Comparing detection at the first time point (2 h postfeeding; 100% detection) to detection at each subsequent time point for each blood type, we found a significant decrease in the percentage of fleas with a detectable human bloodmeal beginning at 24 h (one-tailed Fisher Exact, P = 0.04), and a significant decrease in the percentage of fleas with a detectable rat or goat bloodmeal beginning at 48 h (one-tailed Fisher Exact, P ≤0.02). We detected human, rat, and goat DNA in fed fleas out to 72 h. We detected human DNA in a single human blood-fed flea and in four goat blood-fed fleas at 98 h. We did not detect rat DNA in any of the rat blood-fed fleas at this final time point, but there was not a significant difference between the proportion of human, rat, and goat-blood-fed fleas with an identifiable bloodmeal at 98 h postfeeding (two-tailed Fisher Exact, P ≥0.09).
Fig. 1.
Percentage of bloodmeals detected and correctly identified in artificially fed C. felis by time point and blood type (n = 10 fleas per blood type per time point).
Discussion
Since the precipitin test was first adapted almost a century ago to identify bloodmeals in vectors of malaria parasites, researchers have recognized vector feeding habits as a key factor in vector-borne disease transmission dynamics and bloodmeal assays as a powerful tool to help elucidate those feeding habits (King and Bull 1923). Since then, researchers have developed and refined a range of serologic and molecular tools to improve bloodmeal assay sensitivity and specificity. Here we describe an assay that combines real-time PCR using SYBR Green I detection and sequencing to identify vertebrate bloodmeals in fleas. This method takes advantage of the sensitivity afforded by real-time PCR to overcome some of the challenges inherent in detecting tiny, often degraded bloodmeals in vectors. Because real-time assays that detect vertebrate DNA are highly sensitive to human DNA contamination, we analyzed detection in artificially fed and unfed fleas to establish a Ct cutoff that optimized specificity without completely sacrificing sensitivity. Our assay uses sequencing to allow for specific identification of a wide range of vertebrate bloodmeal sources; it can be used to differentiate vertebrate bloodmeals to the genus or species level without a priori knowledge of the host community.
One of the primary challenges in developing a molecular bloodmeal assay is achieving sufficient sensitivity to detect DNA in tiny bloodmeals, often from species with enucleated erythrocytes (Kent 2009). Franklin et al. (2010) used species-specific primers in a conventional PCR assay to detect rodent DNA in field-collected fleas. Though they only tested fleas with a visible bloodmeal in the abdomen, they did not detect a vertebrate bloodmeal in any of 230 off-host fleas. This suggests that bloodmeal identification in fleas collected off of hosts may demand a more sensitive detection method than conventional PCR, perhaps because some flea species take relatively small bloodmeals. While some studies have found that there is no association between bloodmeal size and the rate of DNA amplification (Ali Oshaghi et al. 2006), Mota et al. (2007) reported that the amount of blood consumed was the principle limiting factor for amplification of triatomine bloodmeal DNA by PCR. Haouas et al. (2007) also suggested that the difficulty in detecting bloodmeals in sand flies may stem, in part, from their tiny size (0.5–1 μl). Flea bloodmeal size varies within and between species. C. felis tend to feed frequently, and actively reproducing females can consume an average of 13.6 μl per day when allowed to feed on a cat for 48 h (Dryden and Gaafar 1991). It is not clear, however, exactly how much blood the C. felis gut contains at any given point. Adult fleas, particularly C. felis, begin to defecate soon after they begin feeding, and they excrete large amounts of fecal material (Dryden and Gaafar 1991, Silverman and Appel 1994). Adult cat flea feces is an essential component of the larval flea diet, and studies of the protein content in adult cat flea feces suggest that adult fleas may digest blood less completely than other hematophagous arthropods to excrete more nutrients for larval development (Hinkle et al. 1991, Silverman and Appel 1994). Digestion may therefore have less impact on the ability to detect a bloodmeal in fleas (particularly C. felis) than in some other hematophagous arthropods, but fleas may also excrete many components of the bloodmeal (including DNA) more quickly than other vectors.
To optimize assay sensitivity, we used SYBR Green-I-based real-time PCR. Given that real-time PCR is exquisitely sensitive to contamination, however, we recognized the potential for our assay to yield false positive results, particularly because it detects human DNA. Forensic and anthropological studies highlight the risk of detecting contaminating human DNA when using highly sensitive molecular techniques to amplify DNA from tiny or degraded samples (von Wurmb–Schwark et al. 2008, Deguilloux et al. 2011). We took measures to eliminate contamination, and we sought to establish a Ct cutoff value for our real-time assay that would maximize specificity (minimize false positives) without completely compromising sensitivity. To this end, we conducted a laboratory study to determine the Ct cutoff value that would maximize PPV. We identified 36 as the Ct cutoff value below which real-time amplification of DNA isolated from a flea is most likely to indicate the presence of DNA from a bloodmeal (as opposed to contaminating human DNA). Significantly, selection of a Ct cutoff value based on maximum PPV is not prevalence dependent (Caraguel et al. 2011). Therefore, 36 should be an appropriate Ct cutoff regardless of the actual proportion of field-collected fleas that contain a detectable vertebrate bloodmeal.
Ideally, we would have established the Ct cutoff using field-collected fleas from our target population, but this was not possible given that we were unable to verify the true status (fed vs. unfed) of our field-collected fleas from Uganda. Available flea samples were stored in ethanol and developed cloudy, gray midguts. This made it impossible to reliably distinguish blood-fed from unfed specimens via microscopy. One limitation associated with using a laboratory experiment to establish an appropriate Ct cutoff value is that it does not account for possible contamination associated with phoretic hosts. It is possible that skin cells from such a host might contaminate the exoskeleton. We tried to mitigate the effects of external contamination by soaking each flea in 50% bleach before the DNA isolation process. Previous studies showed that treatment with bleach eliminated surface contamination from maggots (Linville and Wells 2002) and ancient bones (Kemp and Smith 2005) without compromising subsequent amplification of DNA from the crop contents or bone, respectively. Notably, we did not identify mouse DNA in any of the fleas we tested, though all fleas that received an artificial bloodmeal for this experiment fed through mouse membranes, and the colony-reared X. cheopis used to determine an appropriate Ct cutoff value had been maintained on live mice. Four of the rat blood-fed X. cheopis were classified as having an “undetectable bloodmeal” because melting peak analysis of the real-time results indicated the presence of contaminants, which may have included mouse DNA, in some or all of the replicate wells, but such samples were rare. We suspect that we continued to observe human DNA contamination in our bleach-treated samples because human DNA is ubiquitous in the lab and may be inadvertently introduced during the DNA isolation or PCR set-up (Urban et al. 2000). Ultimately, however, we cannot rule out the possibility of detecting contaminating DNA from phoretic hosts using this assay.
Using a Ct cutoff of 36, we detected contaminating human DNA in 6% of our unfed X. cheopis. Care must therefore be taken when interpreting the results from field-collected fleas that test positive for human DNA using this assay. Notably, we did not detect human DNA in any of the rat blood-fed X. cheopis with a Ct value less than or equal to 36, although the four rat blood-fed X. cheopis classified as having an undetectable bloodmeal because melting peak analysis indicated the presence of contaminants may have contained human DNA. Only one of the 70 rat blood-fed and one of the 70 goat blood-fed C. felis we tested in the time-course experiment were misclassified as positive for human DNA. We conclude that most false positives are likely to result from detecting contaminating human DNA in unfed fleas. Therefore, it should be possible to reduce the number of false positives by only testing vectors known to have taken a bloodmeal.
Having established a Ct cutoff of 36, we tested the assay’s ability to detect rat, human, and goat DNA in artificially fed C. felis out to 98 h postfeeding. Our assay accurately classified all blood-fed fleas held up to 4 h after feeding as positive for vertebrate DNA. As expected, our ability to detect human, rat, and goat DNA was inversely related to how long the fleas were held alive after feeding. Numerous studies in a variety of vectors have found a similar decline in sensitivity as the age of the bloodmeal increases (Mukabana et al. 2002b, Pichon et al. 2003, Steuber et al. 2005, Ali Oshaghi et al. 2006, Van den Hurk et al. 2007). It should be noted that because we used an artificial feedings system, assay sensitivity to host DNA in fleas fed under natural conditions might differ. Using an artificial feeding system, however, allowed us to examine sensitivity to different blood types ingested under similar conditions. Because our aim was to develop an assay that would allow us to accurately determine the proportion of bloodmeals from humans and small mammals in field-collected C. felis, we sought to achieve similar sensitivity to blood from different species. Indeed, we did not detect significant differences between the assay’s ability to detect rat, human, and goat blood at any time point. Using a Ct cutoff of 36, we also found no significant difference between the assay’s ability to detect vertebrate DNA in rat blood-fed versus human blood-fed X. cheopis held for 24 h after feeding.
Given that we only tested fleas that had fed on blood from three species, we cannot conclude that the assay has similar sensitivity to blood from all vertebrate species. It might, for example, be more sensitive to avian species that, unlike mammals, have nucleated erythrocytes (Lee et al. 2002). In addition, we found a significant decrease in detection beginning at 24 h postfeed for human blood-fed fleas, while detection in rat and goat blood-fed fleas did not decrease significantly until 48 h postfeed. This may indicate that C. felis excretes or digests rat blood more slowly than human and goat blood, at least under our laboratory conditions. Other studies have demonstrated that host species can have a significant effect on the rate of blood digestion in fleas (Krasnov et al. 2003, 2007), and a difference in digestion rate might explain the impact of blood type on the ability of fleas to maintain a Y. pestis infection (Eisen et al. 2008b). Given these caveats, care must be taken when extrapolating flea feeding behavior based on the proportion of detected bloodmeals taken from any given species. Our results strongly suggest, however, that our assay is sufficiently sensitive to detect and accurately identify vertebrate DNA in fleas that have taken a recent bloodmeal from a rat, human or goat. This will allow us to confidently determine if a proportion of field-collected C. felis have fed on R. rattus or other small mammals that might serve as a source of Y. pestis infection and if this flea species might therefore serve as a bridging vector to humans in the West Nile region of Uganda.
We acknowledge several additional limitations associated with this type of assay. This method requires specialized equipment that may not be available in laboratories in developing countries. In addition, the actual Ct value associated with any sample is likely to vary based on the specific real-time equipment and reagents used as well as on the calibrator used to set the threshold for each run. Any lab wishing to use this assay would therefore have to conduct its own experiment to set an appropriate Ct cutoff value. Because storage conditions can impact the efficiency of blood-meal detection (Coulson et al. 1990, Chow–Shaffer et al. 2000), other laboratories would also need to consider storage conditions when adapting this assay. Other studies describing PCR-based bloodmeal identification have used frozen (Kirstein and Gray 1996, Ngo and Kramer 2003, Van den Hurk et al. 2007) or dried (Kent and Norris 2005, Steuber et al. 2005) samples, or samples stored in ethanol (Haouas et al. 2007, Bartsch et al. 2009, Allan et al. 2010). Linville et al. (2004) compared eight preservation conditions and reported successful amplification of mitochondrial DNA from the crop contents of maggots stored up to 6 mo at −70°C or in ethanol (70 or 95%), although efforts to amplify nuclear DNA indicated that there is some initial decline in DNA quality when samples are stored in ethanol. Others have reported successful amplification of host DNA from bloodmeals preserved by squashing mosquito abdomens on filter paper and then storing the paper dry or in lysis buffer at room temperature (Chow–Shaffer et al. 2000). We elected to assess assay sensitivity and specificity using fleas that had been stored in ethanol because this method is feasible under field conditions, even in remote areas where it is often impossible to keep samples frozen. Given that this method may results in some initial decline in DNA quality (Linville et al. 2004), the assay might be more sensitive to host DNA in frozen samples. Ethanol preservation also allows for subsequent arthropod identification based on morphological features. Laboratories that identify arthropods using molecular means, however, might consider assessing assay performance using dried bloodmeals on filter paper.
Unlike a probe-based real-time assay, the method described here requires postreaction processing, and it does not allow for rapid identification of mixed bloodmeals. Woods et al. (2009) described a probe-based real-time assay that was successfully used to identify bloodmeals in artificially fed fleas and in pools of field-collected fleas from the Democratic Republic of Congo. This probe-based assay showed similar sensitivity to bloodmeals in artificially fed fleas at 1 and 5 h postfeeding (compared with 2 and 4 h postfeeding using the universal assay). The probe-based assay was more sensitive than our universal assay at 24, 48, and 72 h (two-tailed Fisher Exact, P ≤ 0.02). It should be noted, however, that artificially fed fleas tested using the probe-based assay were stored frozen rather than in ethanol. As noted above, storage in ethanol may negatively impact assay sensitivity, but it is a more realistic storage method for field-collected samples. The probe-based assay was also highly specific; the only DNA the assay detected in artificially fed fleas was from the experimental blood source, and it did not detect DNA in mouse-fed fleas. Assay validation did not include testing unfed fleas, however, so it is not known whether the probe-based assay may have detected contaminating human DNA in a proportion of unfed samples.
Overall, such a probe-based real-time PCR assay has some advantages in situations where it is possible to predict potential bloodmeal sources, although there was some variation in the sensitivity of different primer-probe sets. The assay was significantly less likely to detect dog DNA than human DNA, for example, in artificially fed fleas at 24, 48, and 60 h postfeeding (two-tailed Fisher Exact, ≤ 0.02). In addition, a stated weakness of the assay was that given that a flea did not contain an identifiable bloodmeal, the bloodmeal might be absent, degraded, or the flea might have fed on a species not include in the test panels. Mastomys natalensis, which has been implicated as a potentially important source of Y. pestis infection in the West Nile region and other parts of East Africa (Gratz 1999, Borchert et al. 2007, Eisen et al. 2008c), was absent from the panels. When using a probe-based assay, it is necessary to run a separate real-time reaction using each panel for each sample, so increasing the number of panels to include additional species would increase the amount of time and reagent required for processing. Adding an additional species to a multiplexed panel can compromise assay sensitivity or result in the formation of nonspecific products (Gunson et al. 2008). Our SYBR Green real-time PCR-based assay has the advantage that each sample must be run with only one primer set. More importantly, it allows us to be more confident that we will not miss potentially important host species that are not included in the panel. Thus, it enables us to conclude with more confidence that if we detect DNA from a particular species in a very small proportion of field-collected fleas containing detectable vertebrate DNA, the fleas in that population rarely feed on that species.
Despite its limitations, the assay described here will be useful for studies of flea-feeding behavior and plague ecology in the West Nile region of Uganda. With appropriate modifications, this assay could also be applied to research of other vector-borne diseases. Emerging zoonotic infectious diseases with a wildlife origin represent a significant and growing threat to global health (Jones et al. 2008). As vector-borne zoonotic diseases emerge and reemerge, they are most likely to originate in “hotspots” like tropical Africa, Latin America, and Asia, precisely those regions where surveillance efforts are relatively weak (Jones et al. 2008). Control of these diseases will require identification and understanding of potential reservoir hosts and bridging vectors in regions where surveillance data may be absent or out of date. An assay combining real-time PCR with sequencing is sensitive enough to detect tiny, partially digested bloodmeals, specific enough to identify the bloodmeal source to genus or species, and does not rely on a priori knowledge of host communities. This type of assay may therefore be particularly useful for investigating disease dynamics in the regions at greatest risk.
Acknowledgments
We thank J. Stovall and K. Boroughs and for technical assistance, K. MacMillan for comments on the manuscript, and J. M. Petersen for helpful discussions.
References Cited
- Ali Oshaghi M, Chavshin AR, Vatandoost H, Yaaghoobi F, Mohtarami F, Noorjah N. Effects of post-ingestion and physical conditions on PCR amplification of host blood meal DNA in mosquitoes. Exp Parasitol. 2006;112:232–236. doi: 10.1016/j.exppara.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Allan BF, Goessling LS, Storch GA, Thach RE. Blood meal analysis to identify reservoir hosts for Amblyomma americanum ticks. Emerg Infect Dis. 2010;16:433–440. doi: 10.3201/eid1603.090911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatre G, Babi N, Enscore RE, Ogen–Odoi A, Atiku LA, Akol A, Gage KL, Eisen RJ. Flea diversity and infestation prevalence on rodents in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2009;81:718–724. doi: 10.4269/ajtmh.2009.09-0104. [DOI] [PubMed] [Google Scholar]
- Bartsch S, Bauer B, Wiemann A, Clausen PH, Steuber S. Feeding patterns of biting midges of the Culicoides obsoletus and Culicoides pulicaris groups on selected farms in Brandenburg, Germany. Parasitol Res. 2009;105:373–380. doi: 10.1007/s00436-009-1408-y. [DOI] [PubMed] [Google Scholar]
- Bio-Rad Laboratories. Real-time PCR applications guide. Bio-Rad Laboratories; Hercules, CA: 2006. [Google Scholar]
- Borchert JN, Mach JJ, Linder TJ, Ogen–Odoi A, Angualia S. In: Witmer GW, Pitt WC, Fagerstone A, editors. Invasive rats and bubonic plague in Northwest Uganda; Managing Vertebrate Invasive Species: Proceedings of an International Symposium; Fort Collins, CO: U.S. Dep. Agric./APHIS Wildlife Services, National Wildlife Research Center; 2007. pp. 283–293. [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Caraguel CGB, Stryhn H, Gagne N, Dohoo IR, Hammell KL. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: analytical and epidemiologic approaches. J Vet Diagn Invest. 2011;23:2–15. doi: 10.1177/104063871102300102. [DOI] [PubMed] [Google Scholar]
- Chow–Shaffer E, Sina B, Hawley WA, De Benedictis J, Scott TW. Laboratory and field evaluation of polymerase chain reaction-based forensic DNA profiling for use in identification of human blood meal sources of Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2000;37:492–502. doi: 10.1603/0022-2585-37.4.492. [DOI] [PubMed] [Google Scholar]
- Coulson RMR, Curtis CF, Ready PD, Hill N, Smith DF. Amplification and analysis of human DNA present in mosquito bloodmeals. Med Vet Entomol. 1990;4:357–366. doi: 10.1111/j.1365-2915.1990.tb00452.x. [DOI] [PubMed] [Google Scholar]
- Davey JS, Casey CS, Burgess IF, Cable J. DNA detection rates of host mtDNA in bloodmeals of human body lice (Pediculus humanus L., 1758) Med Vet Entomol. 2007;21:293–296. doi: 10.1111/j.1365-2915.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- Deguilloux MF, Ricaud S, Leahy R, Pemonge MH. Analysis of ancient human DNA and primer contamination: one step backward one step forward. Forensic Sci Int. 2011;210:102–109. doi: 10.1016/j.forsciint.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Dryden MW. Host association, on-host longevity and egg production of Ctenocephalides felis felis. Vet Parasitol. 1989;34:117–122. doi: 10.1016/0304-4017(89)90171-4. [DOI] [PubMed] [Google Scholar]
- Dryden MW, Gaafar SM. Blood consumption by the cat flea, Ctenocephalides felis (Siphonaptera: Pulicidae) J Med Entomol. 1991;28:394–400. doi: 10.1093/jmedent/28.3.394. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Wilder AP, Bearden SW, Montenieri JA, Gage KL. Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J Med Entomol. 2007;44:678–682. doi: 10.1603/0022-2585(2007)44[678:etoypb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Holmes JL, Schotthoefer AM, Vetter SM, Montenieri JA, Gage KL. Demonstration of early-phase transmission of Yersinia pestis by the mouse flea, Aetheca wagneri (Siphonaptera: Ceratophylidae), and implications for the role of deer mice as enzootic reservoirs. J Med Entomol. 2008a;45:1160–1164. doi: 10.1603/0022-2585(2008)45[1160:doetoy]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Vetter SM, Holmes JL, Bearden SW, Montenieri JA, Gage KL. Source of host blood affects prevalence of infection and bacterial loads of Yersinia pestis in fleas. J Med Entomol. 2008b;45:933–938. doi: 10.1603/0022-2585(2008)45[933:sohbap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Borchert JN, Holmes JL, Amatre G, Van Wyk K, Enscore RE, Babi N, Atiku LA, Wilder AP, Vetter SM, Bearden SW, Montenieri JA, Gage KL. Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am J Trop Med Hyg. 2008c;78:949–956. [PubMed] [Google Scholar]
- Fagbemi BO. Effect of Ctenocephalides felis strongylus infestation on the performance of West African dwarf sheep and goats. Vet Quart. 1982;4:92–95. doi: 10.1080/01652176.1982.9693846. [DOI] [PubMed] [Google Scholar]
- Franklin HA, Stapp P, Cohen A. Polymerase chain reaction (PCR) identification of rodent blood meals confirms host sharing by flea vectors of plague. J Vector Ecol. 2010;35:363–371. doi: 10.1111/j.1948-7134.2010.00095.x. [DOI] [PubMed] [Google Scholar]
- Gratz NG. Rodent reservoirs and flea vectors of natural foci of plague. In: Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Plague Manual: Epidemiology, Distribution, Surveillance and Control. World Health Organization; Geneva, Switzerland: 1999. pp. 63–96. [Google Scholar]
- Gunson RN, Bennett S, Maclean A, Carman WF. Using multiplex real time PCR in order to streamline a routine diagnostic service. J Clin Virol. 2008;43:372–375. doi: 10.1016/j.jcv.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouas N, Pesson B, Boudabous R, Dedet JP, Babba H, Ravel C. Development of a molecular tool for the identification of leishmania reservoir hosts by blood meal analysis in the insect vectors. Am J Trop Med Hyg. 2007;77:1054–1059. [PubMed] [Google Scholar]
- Hinkle NC, Koehler PG, Kern WH, Patterson RS. Hematophagous strategies of the cat flea (Siphonaptera: Pulicidae) Fla Entomol. 1991;74:377–385. [Google Scholar]
- Humair PF, Douet V, Moran Cadenas F, Schouls LM, Van De Pol I, Gern L. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J Med Entomol. 2007;44:869–880. doi: 10.1603/0022-2585(2007)44[869:miobsi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 2007;7:544–548. [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:U990–U994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaal JF, Baker K, Torgerson PR. Epidemiology of flea infestation of ruminants in Libya. Vet Parasitol. 2006;141:313–318. doi: 10.1016/j.vetpar.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Kemp BM, Smith DG. Use of bleach to eliminate contaminating DNA from the surface of bones and teeth. Forensic Sci Int. 2005;154:53–61. doi: 10.1016/j.forsciint.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kent R, Juliusson L, Weissmann M, Evans S, Komar N. Seasonal blood-feeding behavior of Culex tarsalis (Diptera: Culicidae) in Weld County, Colorado, 2007. J Med Entomol. 2009;46:380–390. doi: 10.1603/033.046.0226. [DOI] [PubMed] [Google Scholar]
- Kent RJ. Molecular methods for arthropod blood-meal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour. 2009;9:4–18. doi: 10.1111/j.1755-0998.2008.02469.x. [DOI] [PubMed] [Google Scholar]
- Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome b. Am J Trop Med Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Campbell SR, Alleyne EO, Dobson AP, Daszak P. West Nile virus risk assessment and the bridge vector paradigm. Emerg Infect Dis. 2005;11:425–429. doi: 10.3201/eid1103.040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King WV, Bull CG. The blood feeding habits of malaria-carrying mosquitoes. Am J Hyg. 1923;3:497–513. [Google Scholar]
- Kirstein F, Gray JS. A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector Ixodes ricinus. Appl Environ Microb. 1996;62:4060–4065. doi: 10.1128/aem.62.11.4060-4065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein F, Gray JS. Blood meal identification in ticks: a promising tool in ecological research on tick-borne diseases. Zentralbl Bakteriol. 1999;289:760–764. [Google Scholar]
- Krasnov BR, Korine C, Burdelova NV, Khokhlova IS, Pinshow B. Between-host phylogenetic distance and feeding efficiency in hematophagous ectoparasites: rodent fleas and a bat host. Parasitol Res. 2007;101:365–371. doi: 10.1007/s00436-007-0480-4. [DOI] [PubMed] [Google Scholar]
- Krasnov BR, Sarfati M, Arakelyan MS, Khokhlova IS, Burdelova NV, Degen AA. Host specificity and foraging efficiency in blood-sucking parasite: feeding patterns of the flea Parapulex chephrenis on two species of desert rodents. Parasitol Res. 2003;90:393–399. doi: 10.1007/s00436-003-0873-y. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol. 2000;38:4066–4071. doi: 10.1128/jcm.38.11.4066-4071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Hassan H, Hill G, Cupp EW, Higazi TB, Mitchell CJ, Godsey MS, Unnasch TR. Identification of mosquito avian-derived blood meals by polymerase chain reaction-heteroduplex analysis. Am J Trop Med Hyg. 2002;66:599–604. doi: 10.4269/ajtmh.2002.66.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linville JG, Wells JD. Surface sterilization of a maggot using bleach does not interfere with mitochondrial DNA analysis of crop contents. J Forensic Sci. 2002;47:1055–1059. [PubMed] [Google Scholar]
- Linville JG, Hayes J, Wells JD. Mitochondrial DNA and STR analyses of maggot crop contents: effect of specimen preservation technique. J Forensic Sci. 2004;49:341–344. [PubMed] [Google Scholar]
- Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis. 2005;191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- Mota J, Chacon JC, Gutiterrez–Cabrera AE, Sanchez–Cordero V, Wirtz RA, Ordoez R, Panzera F, Ramsey JM. Identification of blood meal source and infection with Trypanosoma cruzi of Chagas disease vectors using a multiplex cytochrome b polymerase chain reaction assay. Vector-Borne Zoon Dis. 2007;7:617–627. doi: 10.1089/vbz.2007.0106. [DOI] [PubMed] [Google Scholar]
- Mukabana WR, Takken W, Knols BGJ. Analysis of arthropod bloodmeals using molecular genetic markers. Trends Parasitol. 2002a;18:505–509. doi: 10.1016/s1471-4922(02)02364-4. [DOI] [PubMed] [Google Scholar]
- Mukabana WR, Takken W, Seda P, Killeen GF, Hawley WA, Knols BGJ. Extent of digestion affects the success of amplifying human DNA from blood meals of Anopheles gambiae (Diptera: Culicidae) Bull Entomol Res. 2002b;92:233–239. doi: 10.1079/BER2002164. [DOI] [PubMed] [Google Scholar]
- Ngo KA, Kramer LD. Identification of mosquito bloodmeals using polymerase chain reaction (PCR) with order-specific primers. J Med Entomol. 2003;40:215–222. doi: 10.1603/0022-2585-40.2.215. [DOI] [PubMed] [Google Scholar]
- Pant CP, Houba V, Engers HD. Bloodmeal identification in vectors. Parasitol Today. 1987;3:324–326. [Google Scholar]
- Pichon B, Egan D, Rogers M, Gray J. Detection and identification of pathogens and host DNA in unfed host-seeking Ixodes ricinus L. (Acari: Ixodidae) J Med Entomol. 2003;40:723–731. doi: 10.1603/0022-2585-40.5.723. [DOI] [PubMed] [Google Scholar]
- Pizarro JC, Stevens L. A new method for forensic DNA analysis of the blood meal in Chagas disease vectors demonstrated using Triatoma infestans from Chuquisaca, Bolivia. PLoS One. 2008;3:1–8. doi: 10.1371/journal.pone.0003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitzer R. Plague, World Health Organization Monograph Series. World Health Organization; Geneva, Switzerland: 1954. [Google Scholar]
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Rurangirwa FR, Minja SH, Musoke AJ, Nantulya VM, Grootenhuis J, Moloo SK. Production and evaluation of specific antisera against sera of various vertebrate species for identification of bloodmeals of Glossina morsitans centralis. Acta Tropica. 1986;43:379–389. [PubMed] [Google Scholar]
- Silverman J, Appel AG. Adult cat flea (Siphonaptera: Pulicidae) excretion of host blood proteins in relation to larval nutrition. J Med Entomol. 1994;31:265–271. doi: 10.1093/jmedent/31.2.265. [DOI] [PubMed] [Google Scholar]
- Steuber S, Abdel–Rady A, Clausen PH. PCR-RFLP analysis: a promising technique for host species identification of blood meals from tsetse flies (Diptera: Glossinidae) Parasitol Res. 2005;97:247–254. doi: 10.1007/s00436-005-1410-y. [DOI] [PubMed] [Google Scholar]
- Tempelis CH. Host-feeding patterns of mosquitoes, with a review of advances in analysis of blood meals by serology. J Med Entomol. 1975;11:635–653. doi: 10.1093/jmedent/11.6.635. [DOI] [PubMed] [Google Scholar]
- Tollenaere C, Rahalison L, Ranjalahy M, Duplantier JM, Rahelinirina S, Telfer S, Brouat C. Susceptibility to Yersinia pestis experimental infection in wild Rattus rattus, reservoir of plague in Madagascar. Eco-health. 2010;7:242–247. doi: 10.1007/s10393-010-0312-3. [DOI] [PubMed] [Google Scholar]
- Urban C, Gruber F, Kundi M, Falkner FG, Dorner F, Hammerle T. A systematic and quantitative analysis of PCR template contamination. J Forensic Sci. 2000;45:1307–1311. [PubMed] [Google Scholar]
- Van den Hurk AF, I, Smith L, Smith GA. Development and evaluation of real-time polymerase chain reaction assays to identify mosquito (Diptera: Culicidae) bloodmeals originating from native Australian mammals. J Med Entomol. 2007;44:85–92. doi: 10.1603/0022-2585(2007)44[85:daeorp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- von Wurmb–Schwark N, Heinrich A, Freudenberg M, Gebuhr M, Schwark T. The impact of DNA contamination of bone samples in forensic case analysis and anthropological research. Legal Med (Tokyo, Japan) 2008;10:125–130. doi: 10.1016/j.legalmed.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Washino RK, Tempelis CH. Mosquito host bloodmeal identification: methodology and data analysis. Annu Rev Entomol. 1983;28:179–201. doi: 10.1146/annurev.en.28.010183.001143. [DOI] [PubMed] [Google Scholar]
- Wilder AP, Eisen RJ, Bearden SW, Montenieri JA, Gage KL, Antolin MF. Oropsylla hirsuta (Siphonaptera: Ceratophyllidae) can support plague epizootics in black-tailed prairie dogs (Cynomys ludovicianus) by early-phase transmission of Yersinia pestis. Vector-Borne Zoon Dis. 2008;8:359–367. doi: 10.1089/vbz.2007.0181. [DOI] [PubMed] [Google Scholar]
- Woods ME, Montenieri JA, Eisen RJ, Zeidner NS, Borchert JN, Laudisoit A, Babi N, Atiku LA, Enscore RE, Gage KL. Identification of flea blood meals using multiplexed real-time polymerase chain reaction targeting mitochondrial gene fragments. Am J Trop Med Hyg. 2009;80:998–1003. [PubMed] [Google Scholar]