Abstract
Pathway-based analysis as an alternative and effective approach to identify disease-related genes or loci has been verified. To decipher the genetic background of plasma adiponectin levels, we performed genome wide pathway-based association studies in extremely obese individuals and normal-weight controls. The modified Gene Set Enrichment Algorithm (GSEA) was used to perform the pathway-based analyses (the GenGen Program) in 746 European American females, which were collected from our previous GWAS in extremely obese (BMI > 35 kg/m2) and never-overweight (BMI<25 kg/m2) controls. Rac1 cell motility signaling pathway was associated with plasma adiponectin after false-discovery rate (FDR) correction (empirical P < 0.001, FDR = 0.008, family-wise error rate = 0.008). Other several Rac1-centered pathways, such as cdc42racPathway (empirical P < 0.001), hsa00603 (empirical P = 0.003) were among the top associations. The RAC1 pathway association was replicated by the ICSNPathway method, yielded a FDR = 0.002. Quantitative pathway analyses yielded similar results (empirical P = 0.001) for the Rac1 pathway, although it failed to pass the multiple test correction (FDR = 0.11). We further replicated our pathway associations in the ADIPOGen Consortium data by the GSA-SNP method. Our results suggest that Rac1 and related cell motility pathways might be associated with plasma adiponectin levels and biological functions of adiponectin.
Adiponectin is an adipocyte-specific secretory protein, which is associated with type 2 diabetes mellitus (T2DM), obesity, dyslipidemia, and other insulin-resistance related phenotypes. With the development of genome-wide association studies (GWAS), more than a dozen genes were found to be associated with adiponectin. The R112C mutation in the third exon of the adiponectin gene was significantly associated with lower plasma adiponectin levels1. Some of those genes were replicated in different populations, including ADIPOQ, ARL15, and CDH132,3,4,5. Adiponectin levels were correlated with body mass index (BMI), yet only a few genes were associated with both BMI and adiponectin. Although, many studies suggested that adiponectin connects obesity, glucose metabolism, and inflammation, the genetic background of that connection remains enigmatic.
During the past two decades, we have been working on genetics of human obesity in families segregating extremely obese and normal-weight siblings6,7,8,9. After plasma adiponectin levels were measured in our subjects, we performed a GWAS for plasma adiponectin in extremely obese cases and in unrelated normal-weight controls.
Although the intensive search for adiponectin-related genes by GWAS was successful in many studies, we still could not explain the heritability of adiponectin by several top candidate genes. On the other hand, an enormous number of genes were only marginally associated with adiponectin. Many those genes might affect major pathways that play important roles in biological functions of adiponectin. To identify the genes and signaling pathways associated with plasma adiponectin, we further performed pathway-based association analyses in extremely obese individuals and normal-weight controls based on our adiponectin GWAS data.
Results
Genome-wide Association Studies
We firstly performed a quantitative GWAS for adiponectin in 737 females. We found that the ADIPOQ gene was associated with BMI-adjusted adiponectin (rs822387, P = 3.77 × 10−7). Weaker associations were found for DLEU1 (rs200220, P = 7.49 × 10−7), ALK (rs13029602, P = 7.48 × 10−6), and CREG2 (rs4352251, P = 1.86 × 10−6). Associations with P < 1 × 10−5 are shown in Table 1. No genome-wide significance was reached (P < 1 × 10−7, given the 550K SNPs in the genotyping panel).
Table 1. Genome-wide association studies for quantitative plasma adiponectin.
| Chr | SNP | BP | MAF | Gene | P (ADI) | P (BMI-ADI) | P(Log-ADI) | ADIPOGen Consortium10(Pvalues) |
|---|---|---|---|---|---|---|---|---|
| 13 | rs200220 | 49858379 | 0.122 | DLEU1 | 7.29 × 10−6 | 7.49 × 10−7 | 1.74 × 10−5 | 0.0120 |
| 13 | rs188014 | 49867924 | 0.116 | DLEU1 | 5.36 × 10−6 | 7.85 × 10−7 | 1.65 × 10−5 | 0.2278 |
| 3 | rs822387 | 188038731 | 0.077 | ADIPOQ | 8.39 × 10−6 | 3.77 × 10−7 | 1.03 × 10−4 | 0 |
| 3 | rs17300539 | 188042154 | 0.076 | ADIPOQ | 1.46 × 10−4 | 6.14 × 10−6 | 9.61 × 10−4 | 0 |
| 2 | rs13029602 | 29428379 | 0.371 | ALK | 4.95 × 10−5 | 7.48 × 10−6 | 5.32 × 10−5 | 0.3367 |
| 2 | rs4352251 | 101350094 | 0.111 | CREG2 | 2.51 × 10−6 | 1.86 × 10−6 | 4.26 × 10−5 | 0.8026 |
| 2 | rs10497399 | 173822385 | 0.075 | ZAK | 2.37 × 10−6 | 4.00 × 10−4 | 3.15 × 10−5 | 0.8006 |
| 2 | rs6731322 | 173825266 | 0.103 | ZAK | 2.28 × 10−5 | 1.24 × 10−2 | 1.07 × 10−4 | 0.6263 |
| 10 | rs2447014 | 14050128 | 0.372 | FRMD4A | 2.26 × 10−5 | 1.32 × 10−3 | 1.72 × 10−4 | 0.4372 |
| 10 | rs4750440 | 14053960 | 0.397 | FRMD4A | 5.60 × 10−6 | 7.57 × 10−4 | 6.17 × 10−5 | 0.5617 |
| 10 | rs10508470 | 14061828 | 0.308 | FRMD4A | 8.82 × 10−5 | 2.98 × 10−3 | 4.63 × 10−4 | 0.9373 |
| 12 | rs11832828 | 42881817 | 0.088 | TMEM117 | 4.46 × 10−5 | 1.66 × 10−4 | 1.88 × 10−4 | 0.4925 |
| 12 | rs7961974 | 42890295 | 0.096 | TMEM117 | 2.26 × 10−5 | 4.90 × 10−5 | 8.36 × 10−5 | 0.5429 |
| 13 | rs7327245 | 23698063 | 0.336 | SPATA13 | 3.24 × 10−5 | 5.66 × 10−5 | 9.70 × 10−5 | 0.5228 |
| 13 | rs9578695 | 23699309 | 0.325 | SPATA13 | 4.17 × 10−5 | 8.00 × 10−5 | 1.94 × 10−4 | 0.3563 |
| 17 | rs2377301 | 74505964 | 0.100 | CANT1 | 8.41 × 10−5 | 5.26 × 10−4 | 8.02 × 10−4 | 0.5259 |
| 17 | rs4789848 | 74514952 | 0.132 | CANT1 | 9.66 × 10−5 | 2.18 × 10−4 | 5.52 × 10−4 | 0.9170 |
| 18 | rs3813086 | 46442790 | 0.253 | MAPK4 | 9.32 × 10−5 | 9.40 × 10−5 | 3.24 × 10−4 | 0.4497 |
| 18 | rs3911593 | 46436326 | 0.265 | MAPK4 | 1.67 × 10−4 | 8.88 × 10−5 | 4.32 × 10−4 | 0.5883 |
| 18 | rs4260159 | 46483873 | 0.130 | MAPK4 | 7.03 × 10−5 | 3.06 × 10−3 | 2.25 × 10−4 | 0.9807 |
MAF, minor allele frequency; ADI, adiponectin; BMI-ADI, BMI-adjusted adiponectin; Log-ADI, log-transformed adiponectin.
Pathway Association Studies
Pathway-based association studies (GenGen) were carried out for plasma adiponectin, in the case/control analysis, 74 pathways achieved a significance of empirical P < 0.05; however, only the Rac1 pathway (empirical P < 0.001, FDR = 0.008, FWER = 0.008) passed correction for multiple testing. RAC1 was present in both Rac1 and Cdc42/Rac pathways (empirical P < 0.001, FDR = 0.188, FWER = 0.249; Table 2).
Table 2. Pathway association study for adiponectin and BMI-adjusted adiponectin.
| Pathway ID | genes | empirical P | FDR | FWER | database |
|---|---|---|---|---|---|
| Adiponectin (binary) | |||||
| Rac1 | 23 | <0.001 | 0.008 | 0.008 | BioCarta |
| Cdc42/Rac | 15 | <0.001 | 0.188 | 0.249 | BioCarta |
| GO0042287 | 13 | <0.001 | 0.884 | 1.000 | GO |
| Log-adiponectin Rac1 | 23 | 0.009 | 0.685 | 0.409 | BioCarta |
| BMI-adjusted adiponectin (quantitative) | |||||
| RAS | 43 | <0.001 | 0.01 | 0.01 | Self-defined* |
*A cluster of RAS pathway genes were selected based on the pathway association study (listed in Table 3).
In the quantitative pathway-based association study, 61 pathways associated with BMI-adjusted adiponectin and 71 were associated with log-adiponectin (empirical P < 0.05). These pathways contained many genes related to T2DM and insulin resistance. Genes of the RAS and downstream PI3K/AKT and MAPK/ERK signaling pathways were found in multiple significant pathways, including TID, RNA, hsa04150, actin y, Par1, and GH pathways. The RAS signaling pathway was associated with adiponectin after FDR correction (empirical P < 0.001, FDR = 0.01; Table 2). RAC1 was also present in the hsa05030 (empirical P = 0.048), MAL (empirical P = 0.040), and actin y (empirical P = 0.019) pathways. Multiple complement-activation-related pathways were also associated with adiponectin levels (empirical P = 0.001).
Seventy-five (75) pathways yielded significant empirical P-values (empirical P < 0.05) in the quantitative pathway-based association study for unadjusted adiponectin. The RAC1 gene was present in the GO0032270 (empirical P = 0.011) and GO0051247 (empirical P = 0.011) pathways, although none of these pathways remained significant after correction for multiple testing.
Seventy pathways were associated with BMI in extremely obese cases and nominal-weight controls (empirical P < 0.05); however, none of them reached FDR < 0.05 after correcting for multiple tests.
The RAC1 pathway association was replicated by the ICSNPathway assay. The RAC1 pathway yielded a FDR = 0.002 (empirical P < 0.001) for binary adiponectin.
Pathway association studies were also performed for body weight related phenotypes, including BMI, %fat, waist and hip circumferences, waist/hip ratio, and A Body Shape Index (ABSI). None of these analyses reached FDR < 0.05 by GenGen. Besides, the Rac1 pathway was not among top pathways (ranked by empirical P values) that associated with body weight related phenotypes.
We further replicated our results in the ADIPOGen Consortium. Using SNP specific P values from the ADIPOGen GWAS, we performed pathway association studies by GSA-SNP. A key pathway found in our study, GO0032990 (centered by the Rac-1 pathway and the CDH13 gene) was among the top associations (corrected P = 1.06 × 10−13, FDR = 0) in the ADIPOGen data (Supplement Information).
Since no phenotype (adiponectin) was available online from the ADIPOGen Consortium data10, we could not test pathway associations for the ADIPOGen data by GenGen. To further verify our results, we have conducted GSA-SNP11 pathway association tests using our GWAS data. Rac-1 and related pathways were still among the most significant associations, while pathways that harbored CDH13 showed significant results as in the ADIPOGen data (Supplement Information). Forty-nine (49) pathways yielded significant associations (corrected P < 0.05, FDR = 0), 12 of 49 pathways were Rac-1 related.
Discussion
In this pathway-based GWASs, we found downstream branches of the RAS signaling pathway, including the PI3K/AKT, MAPK/ERK, and Rac1 cell motility signaling pathways, were associated with plasma adiponectin levels. Among them, the Rac1 cell motility signaling pathway was associated with adiponectin in both case/control and quantitative genome-wide pathway association studies. We found many T2DM susceptibility genes in adiponectin-related pathways, but only a few of them were present in BMI-related pathways. These data suggest that adiponectin may play a role in connecting obesity and T2DM.
So far, more than a dozen GWASs on adiponectin have been carried out, and at least 15 loci have been associated with plasma adiponectin levels12. ADIPOQ, ARL15, and CDH13 are among the most replicated associations2,3,4. Moreover, polymorphisms on FER13, ETV514, and KNG115 have been associated with plasma adiponectin. Dastani et al.10 performed meta-analysis of adiponectin GWAS in 45891 individuals (The ADIPOGen Consortium), ADIPOQ yielded the most significant association. ADIPOQ gene had among the strongest associations in our GWAS. SNPs rs822387 and rs17300539 reached significance (P < 10−5) for both quantitative adiponectin and BMI-adjusted adiponectin. We also found ADIPOQ in the obesity (empirical P = 0.019) and hsa04930 (empirical P = 0.027) pathways associated with BMI-adjusted adiponectin. The interactions among ADIPOQ and other genes in these pathways may contribute to the association with adiponectin. Hivert16 and Schwarz et al.17 found that ADIPOQ polymorphisms were associated with plasma adiponectin levels and T2DM. The promoter region of ADIPOQ has C/EBP and PPARγ binding sites, and the biological interactions between ADIPOQ and C/EBP are well documented18.
CDH13 is another adiponectin-related gene, although CDH13 was not significantly associated with adiponectin in our GWAS, it was present in several pathways that associated with adiponectin, including GO0045296, GO0030100, and GO0007156.
Although “top” associations may explain some of the genetic variation in plasma adiponectin levels, many minor associations contributed the majority of genetic background. Given the relatively limited sample size and genetic relative risk, those gene polymorphisms are unlikely to be identified by GWAS. However, many of those genes are clustered in certain pathways that are jointly associated with adiponectin.
In our study, we used modified GSEA (GenGen) to identify pathways for both binary (case/control) and quantitative adiponectin, and found the Rac1 pathway was the most significant related pathway. In this pathway, RAC1 plays an important role in cell migration via its regulation of actin filament organization19. In the Cdc42/Rac pathway (P < 0.001), N-WASP activates ARP-2/3 for actin polymerization and filopodium formation20. The function of N-WASP is dependent on the formation of the RAC-IRSP53-WAVE2 trimolecular complex. Nakamura et al.21 found that adiponectin increased the phosphorylation of Akt and activated Cdc42 and Rac1 in endothelial progenitor cells (EPCs). Interestingly, in our study the PI3K-Rac1 pathway was associated with plasma adiponectin levels and remained significant after FDR correlation. We need to point out that the “nominal” P values obtained by the GenGen analysis were actually empirical P values: it was calculated by comparing the pathway enrichment score versus 1000 times phenotypes permutation, it was far more conservative than nominal P values in association studies. It was rare to have significant FDR in both GenGen and ICSNPathway.
RAC1 harbored in many pathways that associated with adiponectin and BMI-adjusted adiponectin, which suggests that RAC1 could be a key connector between adiponectin and its biological functions in cell adhesion, migration, and inflammation. Syed et al.22 provided evidence of a direct connection among hyperglycemia, Rac1 activation, and β-cell apoptosis: Rac1 expression was significantly increased in human islets after exposure to 30 mmol/L glucose; Rac1-Nox-ROS signaling led to caspase 3 activity and mitochondrial dysregulation. It worth further studies on whether Rac1 also mediated adiponectin related cell survival and apoptosis.
In our quantitative pathway association study, many downstream pathways of the RAS signal transduction, including PI3K/AKT, MAPK/ER, NF-κB (nuclear factor κB), and mTOR (mammalian target of rapamycin), were associated with adiponectin. The PI3K/AKT pathway is well studied in insulin signal transduction. TNF-α (tumor necrosis factor-α) and IL-6 (interleukin-6) activate the PI3K/AKT pathway to induce cell growth and maintain cell survival, whereas adiponectin inhibits TNF-α and IL-6 to regulate the PI3K/AKT pathway23. Indeed, adiponectin inhibits TNF-α expression in macrophages24. Ouchi et al.25 showed that adiponectin stimulated angiogenesis by promoting cross-talk between AMP-activated protein kinase and Akt signaling.
NF-κB is a nuclear factor that plays key roles in apoptosis, immune response, and inflammation and can be activated by TNF-α and IL-1β. Adiponectin could inhibit TNF-α and thereby suppress the activity of NF-κB and expression of adhesion factors. The anti-inflammatory effect of adiponectin is related to its inhibition of NF-κB through the PI3K/AKT pathway26,27; Kobashi et al.28 indicated that adiponectin inhibits NF-κB via the PI3K/AKT pathway and thereby regulates inflammatory factors in endothelial cells.
mTOR is an important downstream serine/threonine protein kinase of the PI3K/AKT pathway. It has profound biology effects on ribosome syntheses, gene transcription, protein translation, inflammation, apoptosis, and cell proliferation and differentiation. Many studies have demonstrated that adiponectin was the key mediator between mTOR and its biological effects29. Sugiyama et al.30 also found that adiponectin inhibits colorectal cancer cell growth through the AMPK/mTOR pathway.
The MAPK/ERK pathway involved in cell proliferation, differentiation, and apoptosis has profound connections with inflammation and carcinogenesis. Although this study was the first to associate the MAPK/ERK pathway with adiponectin, many other studies have suggested that MAPK/ERK mediates biological effects of adiponectin: adiponectin regulates wound healing by promoting keratinocyte proliferation and migration via the ERK pathway31. Li et al.32 found that adiponectin upregulates prolyl-4-hydroxylase α1 expression in aortic smooth muscle cells via ERK1/2 and Sp1.
All PI3K/AKT, MAPK/ERK, and Rac1 pathways are lower branches of the RAS signal transduction pathway (http://www.biocarta.com/pathfiles/h_rasPathway.asp), it is suggested that the RAS pathway might be associated with adiponectin. We tested a cluster of RAS pathway genes (Table 3) with GenGen and found a significant association for BMI-adjusted adiponectin (empirical P < 0.001, FDR = 0.01). Ample research has indicated that almost all “branches” of the RAS signaling pathway mediate biological effects of adiponectin.
Table 3. RAS pathway genes tested for associations with BMI-adjusted plasma adiponectin.
| RAS pathway genes (empirical P < 0.001, FDR = 0.01) | ||||||||
|---|---|---|---|---|---|---|---|---|
| ACTA1 | BCL2 | CASP8 | FRAP1 | CDK5RAP2 | PIK3CA | PIK3R3 | MAPK3 | VAV1 |
| AKT1 | BCL2L1 | CASP9 | FRAP2 | MAP2K1 | PIK3CB | PIK3R5 | MAPK8 | VAV2 |
| AKT2 | BRAF | CDC42 | HRAS | MAP2K2 | PIK3CD | RALBP1 | NFKB1 | VAV3 |
| AKT3 | CASP1 | SOS2 | LIMK1 | MAP4K2 | PIK3CG | WASF1 | PTPN6 | |
| ARFIP2 | CASP10 | CFL1 | CDK5R1 | NFKBIA | PIK3R1 | RHOA | RAC1 | |
Many T2DM susceptibility genes, including PPARG, CDKN2A/B (ANRIL), and KCNJ11, are found in key adiponectin-related pathways identified in this study. However, only a few of these genes are present in pathways associated with BMI. The connections between adiponectin and T2DM, inflammation, and cancer are well studied; it would be interesting to examine how many T2DM and inflammation-related genes and pathways were actually associated with plasma adiponectin levels in the obese population.
Although pathway association studies may less likely to pick up false positive results than in single-SNP associations, replication in separate cohort is needed. We tested our pathway associations in a much larger data set, the ADIPOGen Consortium. Not to our surprise, CDH13 and ADIPOQ centered pathways showed top associations. Interestingly, a Rac-1 related pathway is among the most significant pathway associations. Similarly, Rac-1 related pathways dominated the GSA-SNP association list in our data set. Actually, Rac-1 pathways were among top associations in both binary and quantitative assays and in all 3 different tests using phenotype or SNP specific P values permutations (GenGen, ICSNPathway, and GSA-SNP). The most important, the Rac-1 pathway association was replicated in different populations.
The Rac-1 gene association was not among the top associations in GWASs, however, Rac-1 related pathways were significantly associated with plasma adiponectin levels. Needless to say, more studies are needed to decipher the connections between these pathway associations and the biological effects of adiponectin.
Materials and Methods
Subjects
We collected 1,071 unrelated European Americans: 526 extremely obese (BMI > 35 kg/m2) and 545 normal-weight controls (BMI < 25 kg/m2)9. Subjects were originally collected and genotyped for a GWAS for obesity9. Of these, we had adiponectin data for 764, 746 of which were females. In this study, we performed our analyses only in females. Adiponectin outliers (>3 SD) were excluded from the data set, 9/746 samples were removed, (see Table 4).
Table 4. Distribution of plasma adiponectin in all subjects (all females).
| N | max | min | mean | SD* | skewness | kurtosis | |
|---|---|---|---|---|---|---|---|
| Adiponectin (mg/L) | 737 | 39.01 | 1.18 | 13.97 | 7.60 | 0.85 | 0.37 |
| Log-adiponectin | 737 | 1.59 | 0.07 | 1.07 | 0.26 | −0.74 | 1.16 |
| BMI-adiponectin** | 737 | 2.97 | −2.12 | 0 | 1.00 | 0.86 | 0.79 |
*Outliers (>±3 SD) were deleted in this study.
**Adiponectin adjusted by BMI.
All subjects gave informed consent, and the protocol was approved by the Committee on Studies Involving Human Beings at the University of Pennsylvania. The study was carried out in accordance with the approved guidelines. Plasma adiponectin levels were detected by radioimmunoassay by the Obesity Unit of the Institute for Diabetes, Obesity and Metabolism at the Perelman School of Medicine, University of Pennsylvania.
Genome-wide Association Study
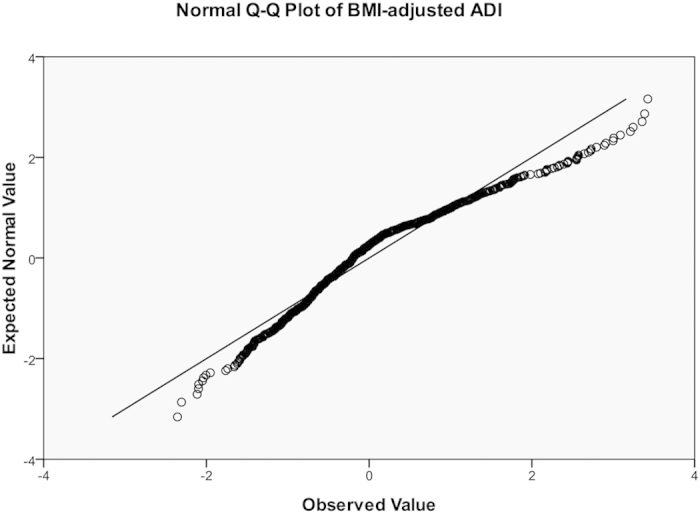
We genotyped 550,000 single-nucleotide polymorphisms (SNPs) by Illumina HumanHap 550 SNP Arrays in our previous GWAS for obesity9. Genome-wide association analyses for adiponectin were carried out by PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/)33, the values of Minor Allele Frequency (MAF) and Hardy-Weinberg equilibrium(HWE) were defaulted. Since subjects were originally recruited for an obese case-control study, plasma adiponectin levels were adjusted by BMI: linear regressions were performed for adiponectin against BMI and standardized residuals were saved as “BMI-adjusted adiponectin” for association studies (mean = 0, standard deviation = 1). Both original, log transformed, and BMI-adjusted adiponectin data were used for quantitative association studies. Outliers (>±3 SD) were deleted before analysis. Distributions of adiponectin, log-transformed, and BMI-adjusted adiponectin were shown as Figs 1, 2, 3 and Table 4.
Figure 1. Q-Q plots for distributions of adiponectin.
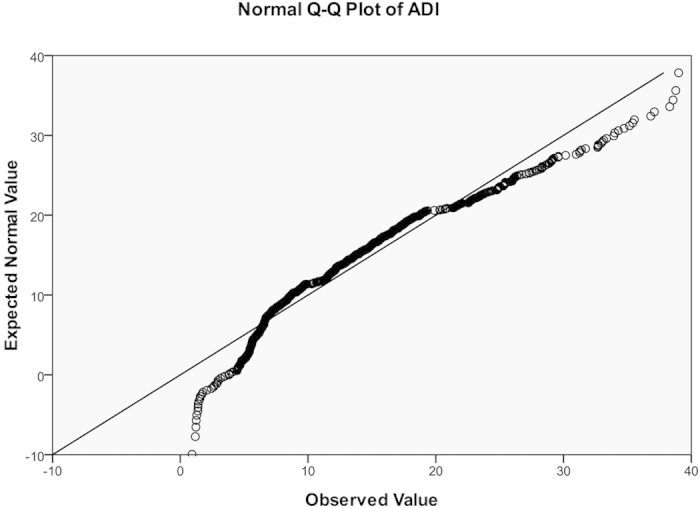
Figure 2. Q-Q plots for distributions of log-transformed adiponectin.
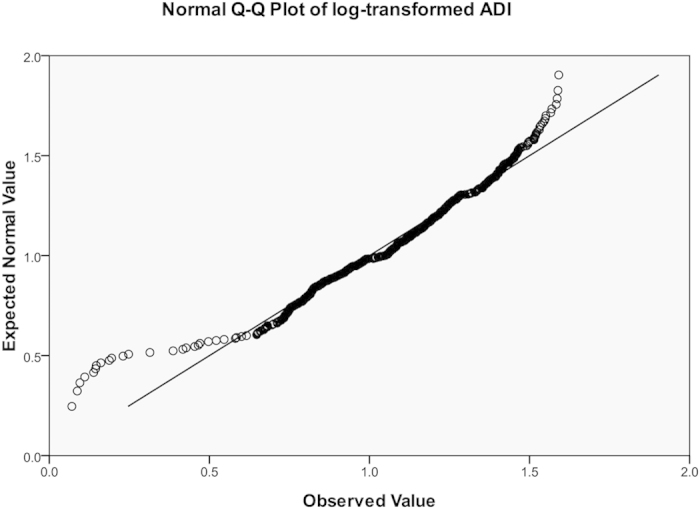
Figure 3. Q-Q plots for distributions of BMI-adjusted adiponectin.
Pathway Association Study
Pathway-based association studies were performed using the GenGen program34, with calculations based on the modified Gene Set Enrichment Algorithm (GSEA)35. Briefly, For each gene, the SNP with the highest test statistic (chi-square detection/F-test) among all SNPs mapped to the gene was selected to represent the gene. All genes were ranked by sorting their statistic values from the largest to smallest, denoted by r(1), …, r(N). For any given gene set S composed of NH genes, an enrichment score (ES) was calculated,
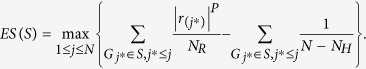 |
Where 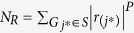 and p is a parameter that gives higher weight to genes with extremes statistic values (default p = 1). Then, in order to adjust the size of different genes, phenotype label permutation with 1000 permutations was performed, and for each permutation, all the above calculations were repeated. We then calculated the normalized enrichment score (NES):
and p is a parameter that gives higher weight to genes with extremes statistic values (default p = 1). Then, in order to adjust the size of different genes, phenotype label permutation with 1000 permutations was performed, and for each permutation, all the above calculations were repeated. We then calculated the normalized enrichment score (NES): 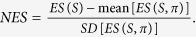
Finally, a false-discovery rate (FDR) procedure was conducted to control the fraction of expected false-positive findings: 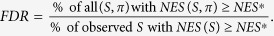
Where NES* denotes the normalized ES in the observed data.
A total of 1,347 pathways compiled from the BioCarta, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO) databases were analyzed. SNPs with minor allele frequencies <0.01 and Hardy-Weinberg equilibrium <0.001 were deleted from the study. Thus, we used 518,230 SNPs in 17,437 genes for GenGen analyses (http://www.openbioinformatics.org/gengen/), 20 k base pairs upstream and downstream of each gene was considered to be a part of the gene.
In dichotomous “case-control” pathway association studies, we defined individuals with plasma adiponectin >10.4 mg/L as “cases” and those with plasma adiponectin <7.4 mg/L as “controls” (Table 5). The “cases” and “controls” were defined by a dichotomous threshold (with a gray area in between) rather than an affection status. In quantitative analyses, we deleted outliers with >±3 SD. Plasma adiponectin levels were also adjusted by BMI: linear regression was performed for adiponectin against BMI, and standardized residuals were saved such that mean = 0 and SD = 1. Quantitative pathway association studies (GenGen) were performed for original, log transformed, and BMI-adjusted adiponectin.
Table 5. Distribution of plasma adiponectin in “cases” and “controls”.
| N | age | adiponectin (mg/L) | BMI > 35 kg/m2 | BMI < 25 kg/m2 | max | min | mean | SD | |
|---|---|---|---|---|---|---|---|---|---|
| Cases | 474 | 43.74 ± 8.49 | >10.4 | 88 | 386 | 51.98 | 10.42 | 18.74 | 7.37 |
| Controls | 165 | 40.68 ± 8.81 | <7.4 | 130 | 35 | 7.30 | 1.18 | 5.47 | 1.59 |
Since pathway association studies showed multiple RAS-pathway-related associations (see Results), we further tested a cluster of genes in the RAS pathway by GenGen for both quantitative and discrete adiponectin. We also conducted pathway association studies for binary BMI in the extremely obese cases and normal-weight controls.
Pathway association studies by ICSNPathway
After we got the results from GenGen, we further tested the pathway associations by ICSNPathway (http://icsnpathway.psych.ac.cn/)36. ICSNPathway took P values of each SNP that obtained from GWAS, however, no phenotype permutation was performed. ICSNPathway analyses were performed for case/control data, SNPs with P < 0.01 (PLINK for binary adiponectin GWAS) and r2 > 0.6 were selected in our study.
Replication of the pathway association results in ADIPOGen Consortium
We further replicated our pathway associations using the ADIPOGen Consortium data10 (http://www.mcgill.ca/genepi/adipogen-consortium). We obtained SNP specific P values from the ADIPOGen GWAS for 45891 individuals, GSA-SNP11 tests were performed to identify pathways that associated with plasma adiponectin levels. GSA-SNP permutated SNP specific P values other than the phenotype (adiponectin) used in GenGen. To better compare ADIPOGen pathway associations with ours, we also conducted GSA-SNP pathway association studies in our binary adiponectin data that used in GenGen.
Conclusions
In this study, we found significant associations between the Rac1 pathway and the plasma adiponectin level in extremely obese individuals and normal-weight controls. Genome-wide pathway association studies (GenGen) yielded similar results in binary and quantitative analyses. The Rac-1 association was further verified using the ICSNPathway method. It is suggest that Rac1 and related cell motility pathways might be associated with plasma adiponectin levels and biological functions of adiponectin.
Additional Information
How to cite this article: Li, W.-D. et al. Pathway-Based Genome-wide Association Studies Reveal That the Rac1 Pathway Is Associated with Plasma Adiponectin Levels. Sci. Rep. 5, 13422; doi: 10.1038/srep13422 (2015).
Supplementary Material
Acknowledgments
We thank all subjects who donated blood samples for genetic research purposes. This work was supported in part by National Institutes of Health grants R01DK44073, R01DK56210, and R01DK076023 to R.A.P. and by a Scientist Development Grant (0630188N) from the American Heart Association, grant 81070576 from the National Natural Science Foundation of China, and grant 12JCZDJC24700 from Tianjin Municipal Science and Technology Commission to W.-D.L. and National Institutes of Health/National Human Genome Research Institute HG006037 to K.W. Genome-wide genotyping was funded in part by an Institutional Development Award to the Center for Applied Genomics (H.H.) from the Children’s Hospital of Philadelphia. Adiponectin radioimmunoassay was supported by a pilot grant from the Institute for Diabetes, Obesity and Metabolism of the University of Pennsylvania School of Medicine (to R.A.P.) and performed by the Obesity Unit led by R.A.
Footnotes
Author Contributions W.-D.L. designed the study, researched data and wrote the manuscript. H.J. and K.W. researched data and edited the manuscript. F.Y. researched data. S.F.A.G. and H.H. generated genotype data and contributed to the discussion. R.A. generated plasma adiponectin data in subjects. R.A.P. designed the study, collected samples, and contributed to the discussion.
References
- Takahashi M. et al. Genomic structure and mutations in adipose-specific gene, adiponectin. International journal of obesity and related metabolic disorders: journal of the International Association for the Study of Obesity 24, 861–868 (2000). [DOI] [PubMed] [Google Scholar]
- Heid I. M. et al. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis 208, 412–420, 10.1016/j.atherosclerosis.2009.11.035 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J. B. et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet 5, e1000768, 10.1371/journal.pgen.1000768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H. et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring) 17, 737–744, 10.1038/oby.2008.625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee S. H. et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet 87, 545–552, 10.1016/j.ajhg.2010.09.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Reed D. R. & Price R. A. Familial risk ratios for extreme obesity: implications for mapping human obesity genes. Int J Obes Relat Metab Disord 21, 935–940 (1997). [DOI] [PubMed] [Google Scholar]
- Lee J. H. et al. Genome scan for human obesity and linkage to markers in 20q13 [published erratum appears in Am J Hum Genet 2000 Apr;66(4):1472]. Am J Hum Genet 64, 196–209 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. D., Dong C., Li D., Zhao H. & Price R. A. An obesity-related locus in chromosome region 12q23-24. Diabetes 53, 812–820 (2004). [DOI] [PubMed] [Google Scholar]
- Wang K. et al. A genome-wide association study on obesity and obesity-related traits. PLoS One 6, e18939, 10.1371/journal.pone.0018939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastani Z. et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet 8, e1002607, 10.1371/journal.pgen.1002607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam D., Kim J., Kim S. Y. & Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Res 38, W749–754, gkq428 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld J., Stumvoll M. & Kovacs P. Genetics of adiponectin. Biochimie 94, 2157–2163, 10.1016/j.biochi.2012.03.004 (2012). [DOI] [PubMed] [Google Scholar]
- Qi L. et al. Novel locus FER is associated with serum HMW adiponectin levels. Diabetes 60, 2197–2201, 10.2337/db10-1645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau-Chonka D. C. et al. Population-specific coding variant underlies genome-wide association with adiponectin level. Hum Mol Genet 21, 463–471, 10.1093/hmg/ddr480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. et al. Genome-wide association study for adiponectin levels in Filipino women identifies CDH13 and a novel uncommon haplotype at KNG1-ADIPOQ. Hum Mol Genet 19, 4955–4964, 10.1093/hmg/ddq423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivert M. F. et al. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 57, 3353–3359, 10.2337/db08-0700 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz P. E. et al. Hypoadiponectinemia is associated with progression toward type 2 diabetes and genetic variation in the ADIPOQ gene promoter. Diabetes Care 29, 1645–1650, 10.2337/dc05-2123 (2006). [DOI] [PubMed] [Google Scholar]
- Saito K. et al. Regulation of gelatin-binding protein 28 (GBP28) gene expression by C/EBP. Biol Pharm Bull 22, 1158–1162 (1999). [DOI] [PubMed] [Google Scholar]
- Rea S. & James D. E. Moving GLUT4: the biogenesis and trafficking of GLUT4 storage vesicles. Diabetes 46, 1667–1677 (1997). [DOI] [PubMed] [Google Scholar]
- Sells M. A. et al. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol 7, 202–210 (1997). [DOI] [PubMed] [Google Scholar]
- Nakamura N. et al. Adiponectin promotes migration activities of endothelial progenitor cells via Cdc42/Rac1. FEBS Lett 583, 2457–2463, 10.1016/j.febslet.2009.07.011 (2009). [DOI] [PubMed] [Google Scholar]
- Syed I. et al. Increased phagocyte-like NADPH oxidase and ROS generation in type 2 diabetic ZDF rat and human islets: role of Rac1-JNK1/2 signaling pathway in mitochondrial dysregulation in the diabetic islet. Diabetes 60, 2843–2852, 10.2337/db11-0809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. F. & Chen J. Z. Obesity, the PI3K/Akt signal pathway and colon cancer. Obes Rev 10, 610–616, 10.1111/j.1467-789X.2009.00607.x (2009). [DOI] [PubMed] [Google Scholar]
- Simons P. J., van den Pangaart P. S., Aerts J. M. & Boon L. Pro-inflammatory delipidizing cytokines reduce adiponectin secretion from human adipocytes without affecting adiponectin oligomerization. J Endocrinol 192, 289–299, 10.1677/JOE-06-0047 (2007). [DOI] [PubMed] [Google Scholar]
- Ouchi N. et al. Adiponectin stimulates angiogenesis by promoting cross-talk between AMP-activated protein kinase and Akt signaling in endothelial cells. J Biol Chem 279, 1304–1309, 10.1074/jbc.M310389200 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi N. et al. Adiponectin, an adipocyte-derived plasma protein, inhibits endothelial NF-kappaB signaling through a cAMP-dependent pathway. Circulation 102, 1296–1301 (2000). [DOI] [PubMed] [Google Scholar]
- Touyz R. M. Endothelial cell IL-8, a new target for adiponectin: implications in vascular protection. Circ Res 97, 1216–1219, 10.1161/01.RES.0000196745.09234.36 (2005). [DOI] [PubMed] [Google Scholar]
- Kobashi C. et al. Adiponectin inhibits endothelial synthesis of interleukin-8. Circ Res 97, 1245–1252, 10.1161/01.RES.0000194328.57164.36 (2005). [DOI] [PubMed] [Google Scholar]
- Song G., Ouyang G. & Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med 9, 59–71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M. et al. Adiponectin inhibits colorectal cancer cell growth through the AMPK/mTOR pathway. Int J Oncol 34, 339–344 (2009). [PubMed] [Google Scholar]
- Shibata S. et al. Adiponectin regulates cutaneous wound healing by promoting keratinocyte proliferation and migration via the ERK signaling pathway. J Immunol 189, 3231–3241, 10.4049/jimmunol.1101739 (2012). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Adiponectin upregulates prolyl-4-hydroxylase alpha1 expression in interleukin 6-stimulated human aortic smooth muscle cells by regulating ERK 1/2 and Sp1. PLoS One 6, e22819, 10.1371/journal.pone.0022819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 81, 1278–1283, 10.1086/522374 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550, 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. et al. ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework. Nucleic Acids Res 39, W437–443, 10.1093/nar/gkr391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


