Abstract
The prognosis of pancreatic cancer patients is very poor, with a 5-year survival of less than 6%. Previous studies demonstrated that the loss of function of CDKN2A is mainly caused by the hypermethylation of CDKN2A gene promoter; however, whether or not it is associated with the incidence of pancreatic cancer still remains unclear. In this study, we systematically reviewed the association between CDKN2A promoter methylation and pancreatic cancer using meta-analysis methods. The pooled data were analyzed by Review Manager 5.2. Fourteen studies eligible studies, including 418 pancreatic cancer, 155 pancreatic intraepithelial neoplasia (PanINs) and 45 chronic pancreatitis (CP) patients were analyzed. We observed that the frequency of CDKN2A methylation was significantly higher in pancreatic cancer patients than in normal healthy controls, the pooled OR = 17.19, 95% CI = 8.72–33.86, P < 0.00001. The frequency of CDKN2A methylation was also significantly higher in PanINs patients than that in normal individual controls, OR = 12.35, 95% CI = 1.70–89.89, P = 0.01. In addition, CDKN2A methylation was associated with worse survival in pancreatic cancer, HR = 4.46, 95% CI = 1.37–14.53, P = 0.01. The results strongly suggest that CDKN2A methylation is correlated with an increased risk of pancreatic cancer. CDKN2A methylation plays a critical role in pancreatic carcinogenesis and may serve as a prognostic marker.
Pancreatic cancer has a high mortality rate and is the 7th most frequent cause of cancer death1. It is estimated that 43,920 people in the United States have suffered from pancreatic cancer, and 37,390 people died from pancreatic cancer in 20122. Pancreatic cancer is a devastating disease with poor survival at advanced stages. Over the last two decades, the 5-year overall survival for pancreatic cancer only slightly improved despite the death rates of most cancers have decreased due to improvements in early diagnosis and efficient treatments3 .The insidious onset, lack of effective screening and early biomarker detection methods, as well as few efficient therapies (due to the complicated cellular and molecular makeup of the pancreatic tumors and their surrounding microenvironment) contribute to the unsatisfied clinical outcomes4. Therefore, biomarkers for early diagnosis and new/effective treatments are urgently needed.
In exocrine origin, pancreatic malignancy display heterogeneous glandular and duct-like structure with infiltration of the most pancreatic parenchyma and partial desmoplastic stroma. This typical pancreatic ductal adenocarcinoma (PDA) is a highly invasive metastatic disease evolved from a premalignant lesion5. Although PDA shows histological and clinical heterogeneity, the studies suggest that the majority of PDA expresses a successive accumulation of highly penetrant genetic changes at genetic genes such as K-ras, p53, CDKN2A and smad4/DPC46 as well as epigenetic alterations1. In endocrine origin, pancreatic neuroendocrine neoplasms (PENs) account for 5% of all pancreatic malignancies which include both cystic and solid PENs7. Cystic PENs is estimated to account for up to 11.5% of all PENs8,9, cystic PENs had less aggressive behavior compared to the solid PENs7. Cystic PENs are thought to be developed from solid counterparts as a result of degeneration, necrosis and hemorrhage of tumors10.
Tumor suppressor CDKN2A gene is located on chromosome 9p21, which is one of the crucial defenses against cancer development. A large body of evidence suggests that CDKN2A is a target of inactivation in pancreatic cancer11. In addition to homozygous deletions and mutation, frequent 5′-CpG island methylation of CDKN2A gene promoter resulting in transcriptional silencing of this gene is noted as an important event in the development of pancreatic cancer. Improved understanding of the role of CDKN2A in pancreatic cancer may offer a tool to refine diagnosis and therapeutic management of pancreatic cancer patients.
The aim of this study was to review the available publications and to summarize the data using meta-analysis to characterize the clinical significance of CDKN2A gene promoter methylation in the pancreatic tumorigenesis.
Results
Identification of relevant studies
Seventy publications were identified by the search method as described above. Fifty-six of those were excluded due to laboratory studies, non-original articles (review), lacking of matched controls or studies irrelevant to the current analysis. Eventually, there were 14 studies included in final meta-analysis (Fig. 1).
Figure 1. Flow chart of study selection.

Study characteristics
Fourteen studies published from 2002 to 2012 were eligible for meta-analysis12,13,14,15,16,17,18,19,20,21,22,23,24,25. A total of 418 pancreatic cancer, 155 PanINs and 45 chronic pancreatitis (CP) patients from China, Singapore, Japan, Germany, England, and United States (USA) were enrolled. Their basic characteristics were summarized in Table 1.
Table 1. Basic characteristics of the included studies.
| Study/Country | Patients/samples | Methods | Primary Aim | Methylation site |
|---|---|---|---|---|
| Dauksa 2012 12/Germany | 26/blood | MSP, SIRPH | Study whether methylation changes in blood could provide a method for early detection of PDA | Promoter, CpG islands |
| Li 201213/China | 5/tissue | MSP | Investigate whether epigenetic modification via hypermethylation represents a mechanism for the inactivation of CDKN2A gene in PDA | Promoter, CpG islands |
| Tan 2007 14/Singapore | 2/blood | MSP | Detect whether serum methylation of CDKN2A is a marker for PDA | Promoter, CpG islands |
| Li 200715/USA | 57/blood, 9/tissue | MSP | Detect whether plasma DNA might be a useful surrogate in epigenetic alterations of PDA | Promoter, CpG islands |
| Matsubayashi 200616/USA | 11/PJ | MSP | Examine whether aberrantly methylated DNA in PJ is an approach for diagnosis of PDA | Promoter, CpG islands |
| Peng 200617/Japan | 56/tissue | MSP | Study whether accumulation of DNA methylation of multiple tumor-related genes is involved in multistage carcinogenesis of pancreas | Promoter, CpG islands |
| Liu 200518/USA | 16/tissue | MSP | Detect whether epigenetic changes in PENs vary in age, histopathologic type and metastasis | Promoter, CpG islands |
| Chan 200319/USA | 10/tissue | MSP | Determine whether methylation profile of PENs differs from carcinoid tumors | Promoter, CpG islands |
| Yan 200520/England | 42/PJ | MSP | Utilize molecular analysis to detect PDA in high-risk groups | Promoter, CpG islands |
| Klump 200321/Germany | 37/PJ | MSP | Determine a role for CDNK2A as a diagnostic marker in differentiation of benign and malignant pancreatic disease | Promoter, CpG islands |
| House 200322/USA | 48/tissue | MSP | Study whether methylation of TSG was an independent predictor of early PENs recurrence and OS following surgical resection | Promoter, CpG islands |
| Ohtsubo 200324/Japan | 60/tissue | MSP | Detest expression of CDKN2A protein and the clinicopathological parameters | Promoter, CpG islands |
| Fukushima 200325/USA | 33/tissue, 45/PJ | MSP | Examine whether CDKN2A in PJ can be a diagnostic approach for PDA | Promoter, CpG islands |
| Fukushima 200223/USA | 15/tissue | MSP | Examine whether CDKN2A can be an indicator of the potential malignancy of epithelial cells of the pancreas | Promoter, CpG islands |
PJ: pancreatic juice, tissue: pancreatic tissue, MSP: methylation-specific PCR, SIRPH: sNuPE with IP-RP-HPLC, PENs: pancreatic neuroendocrine neoplasm, TSG: tumor suppressor gene, OS: overall survival.
CDKN2A methylation and clinicopathological features
The inactivation of CDKN2A through methylation in chronic pancreatitis (CP)
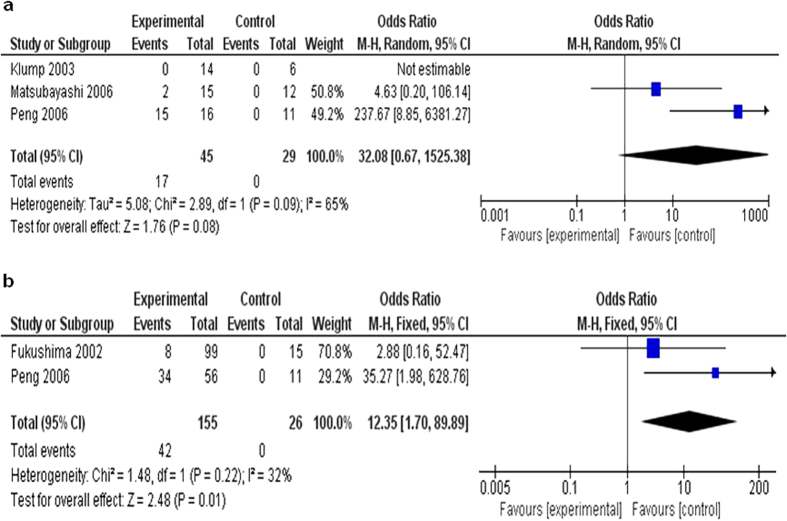
We observed that frequency of CDKN2A methylation was higher in chronic pancreatitis than that in normal individual controls, but it did not reach significant difference. The pooled OR from 3 studies including 45 patients with chronic pancreatitis and 29 healthy individuals is shown in Fig. 2a (OR = 32.08, 95% CI = 0.67–1525.38, P = 0.08). These findings indicate that although an increased risk was identified in CP patients for the development of pancreatic cancer, CDKN2A gene methylation is not the only determinant for its malignancy. Another study showed that CDKN2A methylation was detected in 10% of CP. We excluded the article because no healthy individual controls were available26.
Figure 2.
The studies included to investigate CDKN2A methylation status between (a) 45 chronic pancreatitis patients and 29 normal individuals with the combined OR being 32.08 (95% CI: 0.67–1525.38; Z = 1.76; p = 0.08), and (b) 155 PanINs patients and 26 normal healthy controls with the pooled OR being 12.35 (95% CI: 1.70–89.89; Z = 2.48; p = 0.01).
The inactivation of CDKN2A through methylation in pancreatic intraepithelial neoplasia (PanINs)
We observed that the frequency of CDKN2A methylation was significantly higher in PanINs patients than that in normal individual controls. The pooled OR from 2 studies including 155 patients with PanINs and 26 healthy individuals is shown in Fig. 2b (OR = 12.35, 95% CI = 1.70–89.89, P = 0.01). PanIN is considered the precursor lesion of invasive pancreatic cancer27. Our findings indicate that CDKN2A gene methylation could be one of the determinants for its malignancy.
The inactivation of CDKN2A through methylation in pancreatic cancer
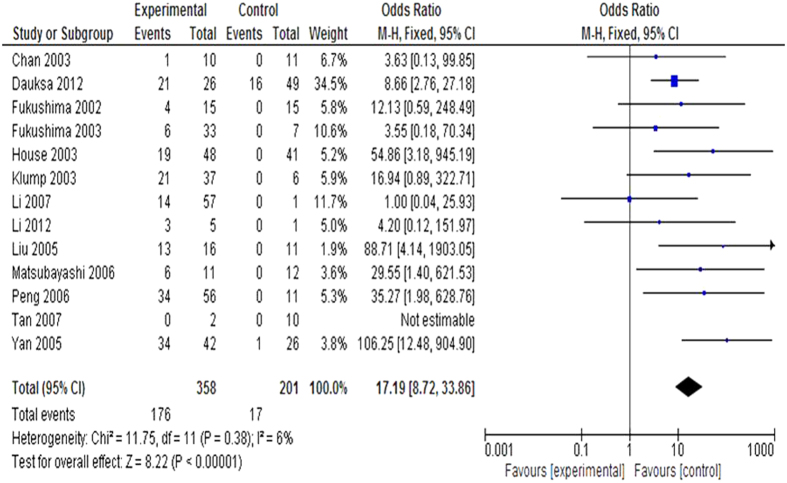
We observed that the frequency of CDKN2A methylation was significantly higher in pancreatic cancer patients than in normal healthy controls. The pooled OR from 13 studies including 358 pancreatic cancer patients and 201 normal individuals is shown in Fig. 3 (OR = 17.19, 95% CI = 8.72–33.86, P < 0.00001), indicating that CDKN2A inactivation through methylation plays an important role in the pathogenesis of pancreatic cancer.
Figure 3. The studies included to investigate CDKN2A methylation status between 358 patients with pancreatic cancer and 201 normal individuals.
The combined OR was 17.19 (95% CI: 8.72–33.86; Z = 5.26; p < 0.00001).
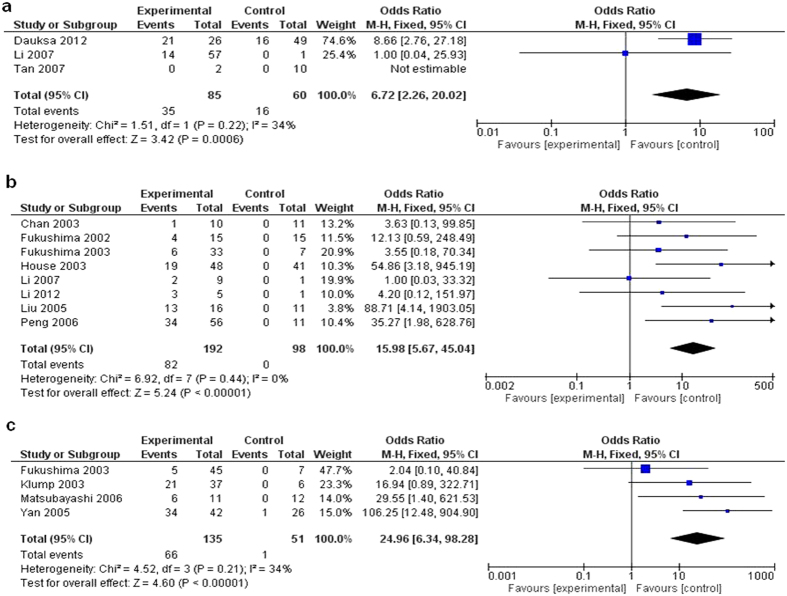
Previously, diagnostic accuracy of some biomarkers such as K-ras analysis for pancreatic carcinoma has been shown to be diverse in different samples;28 therefore, we stratified the analysis of CDKN2A methylation by specimen types (blood, pancreatic tissue and pancreatic juice) for pancreatic carcinoma. The pooled OR for blood analysis from 3 studies including 85 pancreatic cancer patients and 60 healthy controls is shown in Fig. 4a (OR = 6.72, 95% CI = 2.26–20.02, P = 0.0006). We further calculated the sensitivity, specificity of CDKN2A methylation and other parameters in blood samples from pancreatic cancer patients as described in published literature29. Based on Table 2 which is derived from Fig. 4a the results are as follows: sensitivity is 41.0% (a/a + c = 0.41/0.41 + 0.59), specificity is 73.0% (d/d + b = 0.73/0.27 + 0.73), positive predictive value (PPV) is 57.6% (a/a + b = 0.41/0.41 + 0.27), and negative predictive value (NPV) is 57.6% (0.73/0.59 + 0.73).
Figure 4. Pooled results of methylation analysis of CDKN2A gene in different samples in pancreatic cancer.
The pooled OR for blood analysis is shown in Fig. 4a (OR = 6.72, 95% CI = 2.26–20.02, P = 0.0006). As shown in Fig. 4b,c, the pooled OR for pancreatic tissue is 15.98 (95% CI = 5.67–45.04, Z = 5.24, P < 0.00001) and for pancreatic juice is 24.96 (95% CI = 6.34–98.28, Z = 4.60, P < 0.00001).
Table 2. Calculation sensitivity and specificity in blood samples from pancreatic cancer patients.
| Pancreatic cancer | Non-pancreatic cancer | |
|---|---|---|
| CDKN2 positive | 35/85 = 0.41 (a, TP) | 16/60 = 0.27 (b, FP) |
| CDKN2 negative | 50/85 = 0.59 (c, FN) | 44/60 = 0.73 (d, TN) |
TP: true positive, FP: false positive, FN: false negative, TN: true negative.
As shown in Fig. 4b,c, the pooled OR for pancreatic tissues from 8 studies having 192 pancreatic cancer patients and 98 healthy controls was 15.98 (95% CI = 5.67–45.04, Z = 5.24, P < 0.00001) and for pancreatic juice from 4 studies including 135 pancreatic cancer patients and 51 normal individual controls was 24.96 (95% CI = 6.34–98.28, Z = 4.60, P < 0.00001). The overall methylation frequency of CDKN2A in blood, pancreatic tissue and juice for pancreatic carcinoma were more than that in normal controls, suggesting a potential role of CDKN2A methylation analysis in diagnosing pancreatic cancer.
PDA remains one of the most deadly malignancies worldwide with extremely poor overall survival4. While PENs, an indolent neuroendocrine tumor which may secret neuropeptides causing clinical manifestations, is rare, the estimated incidence of PENs in USA has increased by almost 10 times over the past decades7,11. The prognosis of PENs cannot be reliably predicted from histopathological assessment because of neuropeptides secretion19. This is the justification for us to make meta-analysis of CDKN2A methylation in PDA and PENs separately. As shown in Fig. 5a, PENs patients display significantly enhanced CDKN2A methylation frequency (OR = 34.96, 95% CI: 6.27–194.87, Z = 4.05, P < 0.0001) compared to controls. As shown in Fig. 5b, the same observation was obtained for PDA patients when compared to corresponding controls (OR = 14.33, 95% CI = 6.83–30.07, Z = 7.04, P < 0.00001). These data indicated that loss of CDKN2A gene expression through epigenetic modification correlated with both types of aforementioned pancreatic cancers.
Figure 5. Pooled results of methylation analysis of CDKN2A gene PENs and PDA.
The pooled OR for PENs analysis is shown in Fig. 5a (OR = 34.96, 95% CI = 6.27–194.87, Z = 4.05, P < 0.0001). As shown in Fig. 5b, the pooled OR for PDA is 14.33 (95% CI = 6.83–30.07, Z = 7.04, P < 0.00001).
Prognostic values of CDKN2A gene methylation in PENs/pancreatic cancers (PCs)
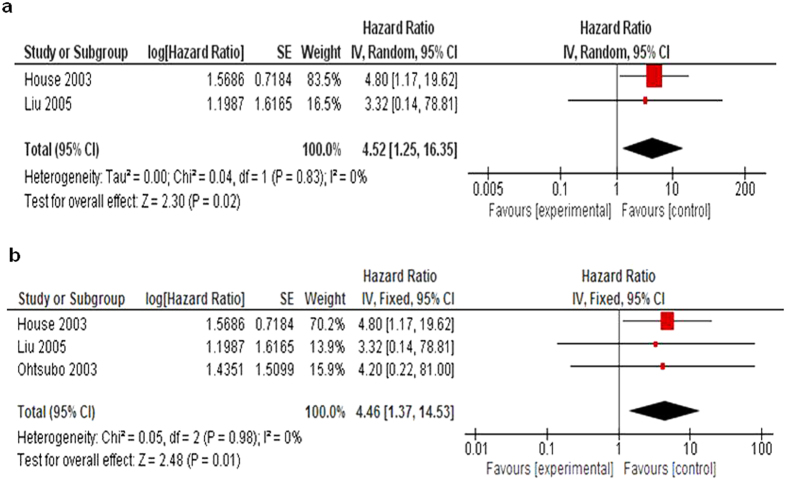
Only two included studies18,22 estimated the relationship between overall survival (OS) in PENs and CDKN2A methylation, the pooled results (Fig. 6a) showed the presence of prognostic impact of CDKN2A gene methylation on PENs patients (OR = 4.52, 95% CI = 1.25–16.35, Z = 2.30, P = 0.02). The hazard ratio is shown numberically in the fifth column, the confidence interval of the summary of hazard ratio does not include 1.0 (it is 1.25–16.35) suggesting that the association is statistically significant. Ohtsubo et al.24 reported that the survival period was significantly shorter in patients with pancreatic carcinoma with CDKN2A hypermethylation than those with a normal CDKN2A gene expression. Combined survival data from all three studies (Fig. 6b) showed OS tended to be shorter in pancreatic cancer patients with epigenetic abnormalities of CDKN2A than in PCs with normal expression of CDKN2A gene (OR = 4.46, 95% CI = 1.37–14.53, Z = 2.48, P = 0.01). In detail, the hazard ratio is shown numberically in the fifth column, the confidence interval of the summary of hazard ratio does not include 1.0 (it is 1.37–14.53) suggesting that the association is statistically significant. Another study30 was excluded from the analysis because only the narrative description can be found, and we were unable to calculate the pooled HR for OS.
Figure 6. All three included studies estimated the relationship between OS and CDKN2A methylation.
(a).The pooled HR for OS showed that CDKN2A hypermethylation was associated with worse survival in PENs, HR = 4.52, 95% CI = 1.25–16.35, P = 0.02. (b). The pooled HR for OS showed that CDKN2A hypermethylation was associated with worse survival in pancreatic cancer, HR = 4.46, 95% CI = 1.37–14.53, P = 0.01.
Association between smoking and CDKN2A methylation status
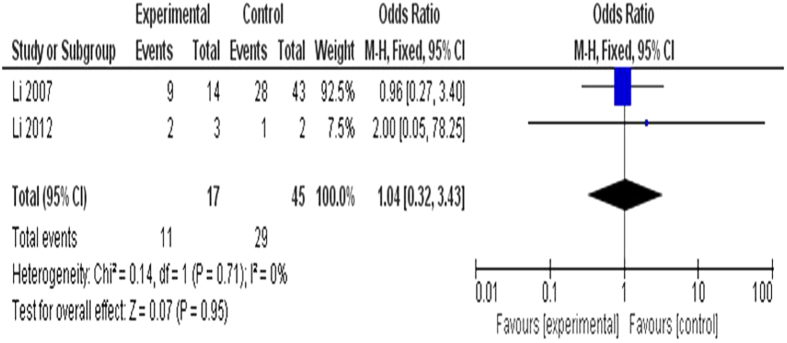
Cigarette smoking has been considered to increase the risk of pancreatic cancer;31 therefore, we evaluated the relationship between methylation of CDKN2A and smoking status, in another word, the changes in frequency of CDKN2A alterations by smoking status. The pooled OR from two studies including 17 pancreatic cancer patients and 45 controls was 1.04 (95% CI = 0.32–3.43, Z = 0.07, P = 0.95), indicating no correlation between smoking and hypermethylation status of CDKN2A gene (Fig. 7).
Figure 7. Association between smoking and CDKN2A methylation status.
The pooled OR from two studies including 17 pancreatic cancer patients and 45 controls is 1.04 (95% CI = 0.32–3.43, Z = 0.07, P = 0.95) which indicated no correlation exists between smoking and hypermethylation status of CDKN2A gene.
Sensitivity analyses and publication bias
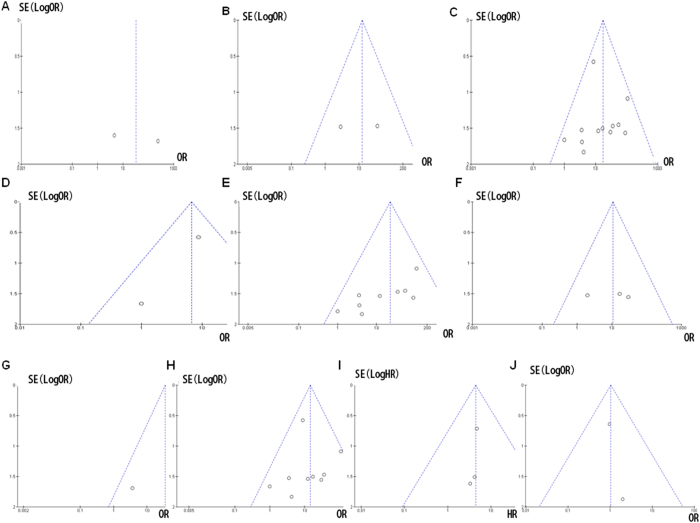
A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled ORs were not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric suggesting there were no publication biases in the meta-analysis of CDKN2A gene methylation/expression and clinicopathological features (Fig. 8).
Figure 8. Funnel plot of publication bias in the meta-analysis of CDKN2A hypermethylation and clinicalpahological features.
CDKN2A methylation in CP (A), PanINs (B), PC (C), blood (D), pancreatic tissue (E), pancreatic juice (F), PENs (G), PDA (H), overall survival (I), and smoking (J).
Discussion
Located in the retroperitoneum of individuals who show non-specific symptoms, pancreatic carcinoma is unlikely to be detected until it has reached an advanced stage in most of patients32. It has remained one of the most devastating and difficult tumors to diagnose and treat. Due to the absence of disease-specific manifestations, there is an urgent need for reliable biomarkers and new therapeutic target(s) in pancreatic carcinoma. Although the tumor suppressor genes K-ras, p53, CDKN2A and SMAD4 have found to be the central molecular genetic pathways in pancreatic cancer33,34,35,36, the gained survival advantage targeting aforementioned pathways remain limited. Other molecular events such as epigenetic changes have recently been identified to contribute to the initiation and progression of pancreatic cancer37. In the current study, we concluded that (1) CDKN2A inactivation through methylation plays an important role in the pathogenesis of pancreatic cancer (both in PDA and PENs), and could be one of the determinants for its malignancy as supported by higher CDKN2A methylation frequency in premalignant lesion, PanINs than in normal controls, (2) all type of samples such as blood, pancreatic tissue and juice have a potential role for CDKN2A methylation analysis in diagnosing pancreatic cancer, (3) overall survival tend to be shorter in pancreatic patients with epigenetic abnormalities of CDKN2A than in PCs with normal expression of CDKN2A gene, and (4) no correlation exists between smoking and hypermethylation status of CDKN2A gene.
As the differential diagnosis between pre-malignant/malignant diseases and normal/benign lesions, the detection of specific tumor markers in blood and pancreatic tissues is a convenient and attractive diagnostic tool. In addition to qualitative analysis, Li et al. further quantitatively evaluated methylation levels using the SIRPH (SNuPE combined with ion pair reverse phase HPLC)38,39 protocol and compared the DNA methylation levels in peripheral blood and cancer tissue for a panel of genes in pancreatic cancer. They found three different groups of methylation patterns. The first group of genes presented higher methylation levels in cancer tissues than in blood DNA (CDKN2A, APC, and DAPK1). In the second group, methylation levels were approximately equal (BCL2, CD44 and TNFRSF10), while in the third group, the DNA methylation levels of ACIN1 were lower in pancreatic tissues than in blood. Overall, methylation alterations in blood could provide a promising approach for early detection of pancreatic cancer12. The sensitivity, specificity and other parameters based on Table 2 demonstrated that sensitivity is 41.0% (a/a + c = 0.41/0.41 + 0.59), which indicated that the probability of being test positive when disease present; specificity is 73.0% (d/d + b = 0.73/0.27 + 0.73), which indicated the probability of being test negative when disease absent; positive predictive value (PPV) is 57.6% (a/a + b = 0.41/0.41 + 0.27), which indicated the probability of patient having disease when test is positive; and negative predictive value (NPV) is 57.6% (0.73/0.59 + 0.73), which indicated the probability of patient not having disease when test is negative. More interestingly, the methylation rates detected in pancreatic secretions endoscopically retrieved from the pancreatic duct proved to be somewhat higher than those detected in pancreatic tissues21. In parallel, some studies have investigated the alterations of molecular biomarkers other than methylation patterns in pancreatic secretions from pancreatic cancer patients. For instance, K-ras mutations in pancreatic juice of pancreatic cancer patients have been considered as a potential diagnosis tool for pancreatic cancer with acceptable specificity and sensitivity28.
Chronic pancreatitis is an important predisposing condition resulting in pancreatic malignancy40. The analysis obtained by us displayed that CDKN2A hypermethylaion in chronic pancreatitis is higher than those of normal pancreatogram and lower than those of pancreatic carcinoma21, suggesting a specific role of CDKN4A in the development of malignant pancreactic diseases, although the difference of CDKN2A methylation frequency between chronic pancreatitis patients and controls in the present study did not reach statistical significance. In another word, CDKN2A changes, especially promoter hypermethylation might imply high-risk precursors in chronic pancreatitis that might develop to cancer26. However, larger studies are needed to be carried out to explore the true situation. A study published by Moore et al.41 has addressed that distinct molecular pathways may be involved in exocrine and endocrine tumorigenesis of the pancreas. In this context, exclusively exocrine pancreatic adenocarcinomas of ductal origin (PDA) and endocrine origin (PENs) have been included, de novo CDKN2A promoter hypermethylation was detected and shown to contribute to the tumorigenesis in both types.
In support of our conclusion that overall survival tends to be shorter in pancreatic patients with epigenetic abnormalities of CDKN2A than in PCs with normal expression of CDKN2A gene, Gerdes et al.30 found reduced survival in patients with CDK2A alterations indicating CDKN2A is a prognostic marker in resected ductal pancreatic cancer. This article was excluded from the study because of no matched control.
It needs to be emphasized that the epigenetic alterations other than CDKN2A promoter hypermethylation also contribute to the development of pancreatic cancer. The other well known epigenetic mechanisms are histone modifications [histone deacetylation by histone deacetylases (HDACs) and histone methylation by histone methyltransferases (HMT)] and microRNAs (miRNAs). TGFBR242 and CDH143 are examples which can be regulated by HDACs while EZH244 and SUV39H137,45 are regulated by HMT in pancreatic cancers. An increasing number of miRNAs have been shown to be associated with pancreatic tumors46,47. The most well-known one is miR-21, which is upregulated in pancreatic cancer and targets many apoptosis related genes including PTEN and PDCD4, resulting in inhibited apoptosis and consequently, increased tumorigenicity.
No single technique of DNA methylation detection is appropriate for every application. Some limitations exist in this current analysis. First, DNA methylation at specific loci are dependent on modification of DNA by sodium bisulfate. Although the studies used quantitative gene-specific methylation analysis to link DNA methylation to functional outcomes, bisulfate sequencing is the ideal standard for mapping allele-specific methylation across CpG locations. Without allele-specific measurement, the difference between the status of mosaic methylation of individual alleles or complete methylation of a subpopulation of alleles cannot be distinguished48. Second, the current protocol is unable to differentiate 5-methylcytosin (5mc) from 5-hydroxy-cytosine (5hmc)49,50. Third, similar to gene expression microarrays improving the study of transcriptional regulation, locus-specific DNA methylation on a genome-wide scale will revolutionize and facilitate the DNA methylation analysis50. It would be possible to map-out genome-wide DNA methylation patterns from distinct subtype of pancreatic cancer patients. This analysis would result in new insight into the pathogenesis of cancer and open up new avenues of drug discovery and targeted therapies for all kinds of cancer patients.
Taken all together, due to the poor prognosis of pancreatic cancer, understanding the molecular events such as epigenetic changes in pancreatic cancer that drive this aggressive disease is the core for development of early diagnostic tools and more effective therapeutic strategies.
Conclusion
The results of our study strongly suggest that CDKN2A methylation is correlated with an increased risk of pancreatic cancer. CDKN2A methylation plays a critical role in pancreatic carcinogenesis and may serve as a prognostic marker.
Methods
Search strategy
Medline, Pubmed, Web of Science, Scopus and Embase were searched in August 2014 using the search terms: ‘p16’, ‘p16INK4a’, “CDKN2A”, ‘methylation’, ‘pancreatic cancer’, “pancreatic carcinoma” and ‘clinical studies’. Investigations identified through the search approach as described above were screened by titles first, then by abstracts of the publications. After exclusion of non-relevant publications and identifications of duplicates from the different databases, the remaining papers were evaluated in the full text version for in- and exclusion criteria and for relevant articles in the reference lists. All clinical studies except case reports were chosen, for instance, randomized controlled trials (RCTs), cohort studies, case-controls studies and case series. The language of publication was restricted to English. All searched data were retrieved. Authors’ bibliographies and references of selected studies were also searched for other relevant studies. The most complete study was chosen to avoid duplication if same patient populations were reported in several publications.
Selection criteria
We collected all eligible articles about the relationship between CDKN2A methylation and/or expression and clinicopathological features and clinical outcomes in pancreatic cancer patients for this meta-analysis. Studies meeting the following inclusion criteria were included: (1) CDKN2A methylation and/or expression which were evaluated in the circulation, pancreatic juice, and/or pancreatic tissues, (2) researches which revealed the relationship between CDKN2A methylation and/or expression and pancreatic cancer clinicopathological parameters and prognosis, (3) CDKN2A methylation and/or expression which were examined by methylation specific polymerase chain reaction (MSP), (4) articles which were published as a full papers in English, (5) articles which provided sufficient information to estimate hazard ratio (HR) about overall survival (OS) and 95% confidence interval (CI) and probabilities for overall survival (OS) where applicable. The exclusion criteria included the following: (1) letters, reviews, case reports, conference abstracts, editorials, expert opinion, and non-English language papers; (2) articles having no information on OS or those that could not calculate the HR about OS from the given information; and (3) all publications regarding in vitro/ex vivo studies, cell lines and human xenografts were also excluded.
Data extraction
Two investigators independently extracted data from eligible studies. Disagreements were resolved by discussion till consensus were achieved. Two investigators reviewed all of the articles that fit inclusion and exclusion criteria. The following information was recorded for each study: the name of first author, year of publication, sample source, number of cases, clinicopathological parameters, stage, CDKN2A methylation and/or expression, and patient survival. Data for study characteristics and clinical response were summarized and the data were turned into table format. Heterogeneity of investigation was evaluated to determine whether or not the data of the various studies could be analyzed in a meta-analysis.
Statistical analysis
Analysis was conducted using the Stata 12.0 (Stata Corporation,TX, USA) and Review Manager 5.2 (Cochrane Collaboration, Oxford, UK). Comparisons of dichotomous measures were done by pooled estimates of odds ratios (ORs) as well as their 95% CIs. P value of <0.05 was considered to be statistically significant. Heterogeneity was examined by a chi-square test with significance being set at P < 0.10; the total variation among studies was estimated by I square. We used I square statistic to assess heterogeneity. The I square value is an estimate of variance due to between-study heterogeneity rather than chance (the Cochran Q statistics). Substantial heterogeneity exists when I square exceeding 50%. If there was heterogeneity among studies, we used a random effect model to pool the ORs; otherwise, a fixed effect model was selected.
The database search generated 70 articles from Medline, Pubmed, the Web of Science, Scopus and Embase. After initial screening of all titles, abstracts and eligibility, 14 full-text studies were retrieved for a more detailed assessment. The search of the article references did not produce additional publications. Eventually, 14 publications met the inclusion criteria for qualitative study and meta-analysis. The article search and study selection are depicted in Fig. 1.
Additional Information
How to cite this article: Tang, B. et al. Clinicopathological Significance of CDKN2A Promoter Hypermethylation Frequency with Pancreatic Cancer. Sci. Rep. 5, 13563; doi: 10.1038/srep13563 (2015).
Acknowledgments
This research was supported in part by The National Natural Science Foundation of China (No. 81360367 and No. 81160066 and No. 30870719), Science and technology university research project in Guangxi (2013ZD046). Guangxi Health Department of Pharmaceutical Technology special projects (GZPT13-45). Guangxi repair liver damage and Molecular Medicine Laboratory Open Fund Project (QT2013025). Liver damage in Guangxi Key Laboratory of Molecular Medicine and repair projects (SYS2013009). Guangxi Distinguished Experts Special Fund. The Guangxi culture of new century academic and technical leader of special funds.
Footnotes
Author Contributions B.T., Y.L. and S.H. participated in the design of the study and identify related studies, as well as drafted the manuscript. G.Q., S.G.Y. and Z.W. and S.P.Y. reviewed and extracted data from eligible studies. Y.L., G.Q., B.L. and S.H. participated in the search the study and performed the statistical analysis.
References
- Neureiter D., Jager T., Ocker M. & Kiesslich T. Epigenetics and pancreatic cancer: pathophysiology and novel treatment aspects. World J Gastroenterol 20, 7830–7848, 10.3748/wjg.v20.i24.7830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2012. CA Cancer J Clin 62, 10–29, 10.3322/caac.20138 (2012). [DOI] [PubMed] [Google Scholar]
- Schnelldorfer T. et al. Long-term survival after pancreatoduodenectomy for pancreatic adenocarcinoma: is cure possible? Annals of surgery 247, 456–462, 10.1097/SLA.0b013e3181613142 (2008). [DOI] [PubMed] [Google Scholar]
- Rossi M. L., Rehman A. A. & Gondi C. S. Therapeutic options for the management of pancreatic cancer. World J Gastroenterol 20, 11142–11159, 10.3748/wjg.v20.i32.11142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B. & Kinzler K. W. Cancer genes and the pathways they control. Nat Med 10, 789–799, 10.1038/nm1087 (2004). [DOI] [PubMed] [Google Scholar]
- Almoguera C. et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53, 549–554, 0092-8674(88)90571-5 (1988). [DOI] [PubMed] [Google Scholar]
- Koh Y. X., Chok A. Y., Zheng H. L., Tan C. S. & Goh B. K. A systematic review and meta-analysis of the clinicopathologic characteristics of cystic versus solid pancreatic neuroendocrine neoplasms. Surgery 156, 83–96 e82, S0039-6060(14)00119-6 (2014). [DOI] [PubMed] [Google Scholar]
- Singhi A. D. et al. Cystic pancreatic neuroendocrine tumors: a clinicopathologic study. Am J Surg Pathol 36, 1666–1673, 10.1097/PAS.0b013e31826a0048 (2012). [DOI] [PubMed] [Google Scholar]
- Boninsegna L. et al. Pancreatic cystic endocrine tumors: a different morphological entity associated with a less aggressive behavior. Neuroendocrinology 92, 246–251, 000318771 (2010). [DOI] [PubMed] [Google Scholar]
- Ahrendt S. A., Komorowski R. A., Demeure M. J., Wilson S. D. & Pitt H. A. Cystic pancreatic neuroendocrine tumors: is preoperative diagnosis possible? J Gastrointest Surg 6, 66–74, S1091255×01000208 (2002). [DOI] [PubMed] [Google Scholar]
- Simon B. & Lubomierski N. Implication of the INK4a/ARF locus in gastroenteropancreatic neuroendocrine tumorigenesis. Ann N Y Acad Sci 1014, 284–299 (2004). [DOI] [PubMed] [Google Scholar]
- Dauksa A. et al. Whole blood DNA aberrant methylation in pancreatic adenocarcinoma shows association with the course of the disease: a pilot study. PLoS One 7, e37509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Ji Y., Liu C., Li J. & Zhou Y. Reduced levels of p15INK4b, p16INK4a, p21cip1 and p27kip1 in pancreatic carcinoma. Mol Med Rep 5, 1106–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. H. et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep 18, 1225–1230 (2007). [PubMed] [Google Scholar]
- Jiao L. et al. K-ras mutation and p16 and preproenkephalin promoter hypermethylation in plasma DNA of pancreatic cancer patients: in relation to cigarette smoking. Pancreas 34, 55–62 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi H. et al. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer Res 66, 1208–1217 (2006). [DOI] [PubMed] [Google Scholar]
- Peng D. F. et al. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis 27, 1160–1168 (2006). [DOI] [PubMed] [Google Scholar]
- Liu L. et al. Epigenetic alterations in neuroendocrine tumors: methylation of RAS-association domain family 1, isoform A and p16 genes are associated with metastasis. Mod Pathol 18, 1632–1640 (2005). [DOI] [PubMed] [Google Scholar]
- Chan A. O. et al. CpG island methylation in carcinoid and pancreatic endocrine tumors. Oncogene 22, 924–934 (2003). [DOI] [PubMed] [Google Scholar]
- Yan L. et al. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology 128, 2124–2130 (2005). [DOI] [PubMed] [Google Scholar]
- Klump B. et al. Methylation status of p14ARF and p16INK4a as detected in pancreatic secretions. Br J Cancer 88, 217–222 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- House M. G. et al. Aberrant hypermethylation of tumor suppressor genes in pancreatic endocrine neoplasms. Annals of surgery 238, 423–431 discussion 431-422 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima N. et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol 160, 1573–1581 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K. et al. Abnormalities of tumor suppressor gene p16 in pancreatic carcinoma: immunohistochemical and genetic findings compared with clinicopathological parameters. J Gastroenterol 38, 663–671, 10.1007/s00535-003-1119-6 (2003). [DOI] [PubMed] [Google Scholar]
- Fukushima N. et al. Diagnosing pancreatic cancer using methylation specific PCR analysis of pancreatic juice. Cancer Biol Ther 2, 78–83, 183 (2003). [DOI] [PubMed] [Google Scholar]
- Gerdes B. et al. p16(INK4a) alterations in chronic pancreatitis-indicator for high-risk lesions for pancreatic cancer. Surgery 129, 490–497 (2001). [DOI] [PubMed] [Google Scholar]
- Zamboni G., Hirabayashi K., Castelli P. & Lennon A. M. Precancerous lesions of the pancreas. Best practice & research. Clinical gastroenterology 27, 299–322, 10.1016/j.bpg.2013.04.001 (2013). [DOI] [PubMed] [Google Scholar]
- Liu S. L., Chen G., Zhao Y. P., Wu W. M. & Zhang T. P. Diagnostic accuracy of K-ras mutation for pancreatic carcinoma: a meta-analysis. Hepatobiliary Pancreat Dis Int 12, 458–464 (2013). [DOI] [PubMed] [Google Scholar]
- Parikh R., Mathai A., Parikh S., Chandra Sekhar G. & Thomas R. Understanding and using sensitivity, specificity and predictive values. Indian J Ophthalmol 56, 45–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes B. et al. p16INK4a is a prognostic marker in resected ductal pancreatic cancer: an analysis of p16INK4a, p53, MDM2, an Rb. Annals of surgery 235, 51–59 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford A. et al. Genetic mutations associated with cigarette smoking in pancreatic cancer. Cancer Res 69, 3681–3688, 0008-5472.CAN-09-0015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathis A. & Moore M. J. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol 7, 163–172, nrclinonc.2009.236 (2010). [DOI] [PubMed] [Google Scholar]
- Jones S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806, 1164368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C. A. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut 61, 1085–1094, gut.2010.236026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes B. et al. Tumor-suppressing pathways in cystic pancreatic tumors. Pancreas 26, 42–48, 00006676-200301000-00008 (2003). [DOI] [PubMed] [Google Scholar]
- Gerdes B. et al. Multiple primary tumors as an indicator for p16INK4a germline mutations in pancreatic cancer patients? Pancreas 21, 369–375 (2000). [DOI] [PubMed] [Google Scholar]
- van Kampen J. G. et al. Epigenetic targeting in pancreatic cancer. Cancer Treat Rev 40, 656–664, S0305-7372(13)00265-X (2014). [DOI] [PubMed] [Google Scholar]
- El-Maarri O., Herbiniaux U., Walter J. & Oldenburg J. A rapid, quantitative, non-radioactive bisulfite-SNuPE- IP RP HPLC assay for methylation analysis at specific CpG sites. Nucleic Acids Res 30, e25 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O. SIRPH analysis: SNuPE with IP-RP-HPLC for quantitative measurements of DNA methylation at specific CpG sites. Methods Mol Biol 287, 195–205, 1-59259-828-5:195 (2004). [DOI] [PubMed] [Google Scholar]
- Teich N. & Mossner J. Molecular analysis of pancreatic juice: a helpful tool to differentiate benign and malignant pancreatic tumors? Dig Dis 22, 235–238, 82794 (2004). [DOI] [PubMed] [Google Scholar]
- Moore P. S. et al. Pancreatic tumours: molecular pathways implicated in ductal cancer are involved in ampullary but not in exocrine nonductal or endocrine tumorigenesis. Br J Cancer 84, 253–262, 10.1054/bjoc.2000.1567 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Venkatasubbarao K., Li S. & Freeman J. W. Requirement of a specific Sp1 site for histone deacetylase-mediated repression of transforming growth factor beta Type II receptor expression in human pancreatic cancer cells. Cancer Res 63, 2624–2630 (2003). [PubMed] [Google Scholar]
- von Burstin J. et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 137, 361–371, 371 e361-365, S0016-5085(09)00545-9 (2009). [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B. & Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745, S0092-8674(07)00190-0 (2007). [DOI] [PubMed] [Google Scholar]
- Baumgart S. et al. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology 142, 388–398 e381-387, S0016-5085(11)01514-9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T. et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther 8, 1067–1074, 1535-7163.MCT-08-0592 (2009). [DOI] [PubMed] [Google Scholar]
- Dillhoff M., Liu J., Frankel W., Croce C. & Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg 12, 2171–2176, 10.1007/s11605-008-0584-x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanlan Shen R. A. W. Methods of DNA methylation analysis. Curr Opin Nutr Metab Care, 10, 576–581 10.1097/MCO.0b013e3282bf6f43 (2007). [DOI] [PubMed] [Google Scholar]
- Tahiliani M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935, 1170116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888, 10.1371/journal.pone.0008888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]