Abstract
Purpose
Prostate specific antigen is used for prostate cancer screening but its specificity is limited. Specificity might be increased by considering genotype associated prostate specific antigen levels.
Materials and Methods
We examined associations between single nucleotide polymorphisms on chromosomes 10 and 19 (previously shown to be associated with prostate specific antigen) with prostate specific antigen and prostate cancer in 505 men from the Baltimore Longitudinal Study of Aging.
Results
In a model with age and date the risk ratio for prostate cancer was 1.18 (95% Cl 1.13–1.23) per unit increase in prostate specific antigen. Including the interaction between alleles and prostate specific antigen significantly altered the risk ratio for prostate cancer (Cox proportional hazards p <0.001). Specifically prostate cancer risk per unit increase in prostate specific antigen was significantly different in carriers than in noncarriers of a minor allele (1.28 vs 1.10, respectively, Cox proportional hazards p <0.001), whereas men with a minor allele had a significantly higher risk of prostate cancer at prostate specific antigen levels greater than 6 ng/ml.
Conclusions
Our data suggest that genotype influences the risk of prostate cancer per unit increase in prostate specific antigen. Prostate cancer risk stratification using prostate specific antigen and genotype could improve prostate specific antigen test performance.
Keywords: prostate-specific antigen; prostatic neoplasms; polymorphism, single nucleotide; mass screening; genetics
Recently genome wide association studies have identified sequence variants (single nucleotide polymorphisms) in numerous chromosomal regions that are significantly associated with prostate cancer risk.1 – 4 These associations could be causal or indirect as a result of linkage disequilibrium.
Eeles et al reported that SNPs on chromosomes 10 and 19 were associated with serum PSA concentration and CaP risk. 5 Strong associations were found for rs2735839 and rs2659056 (chromosome 19) as well as rs10993994 (chromosome 10). Of note, rs2735839 is in close proximity to KLK3, the gene that encodes PSA, whereas rs10993994 is near the transcription start site of the microseminoprotein beta gene that encodes PSP94, a prostatic secretory protein whose expression is decreased in the androgen independent state and may suppress tumor growth. 6
The notion of a relationship between genotype and PSA is intriguing since the majority of CaP is currently detected through PSA based screening and there is no PSA threshold below which CaP can be excluded with certainty.7 Thus, methods for improving the specificity of PSA are needed. We examined the association of genotype, serum PSA and prostate cancer risk in a longitudinal aging study.
MATERIALS AND METHODS
Our study population consisted of participants in the Baltimore Longitudinal Study of Aging, a previously described prospective cohort study initiated in 1958.8 All subjects provided written informed consent and the study protocol was approved by the institutional review board. At each evaluation participants underwent a complete medical examination, including CaP screening with PSA and digital rectal examination beginning in 1991. PSA measurements of participants enrolled before 1991 were obtained retrospectively using frozen serum samples when available. After 1991 prostate biopsy was recommended for a PSA greater than 4.0 ng/ml or abnormal digital rectal examination.
From 1,806 male Baltimore Longitudinal Study of Aging participants we excluded those who had no PSA measurements (605), men with CaP who had no PSA data before diagnosis (38) or before prostate surgery for prostatic enlargement (80), men who took finasteride (Proscar®) at any time (47), men with an unknown cause of death (54), men with a single outlier PSA value suspected to be laboratory error (2) and men with no genetic information (475). Thus, the final study population included 505 men with PSA measurements and a DNA sample, including 61 with CaP and 444 with no known CaP diagnosis.
All 505 men underwent genome-wide genotyping using the Illumina Infinium HumanHap 550K platform. Several quality control criteria were used to screen the SNPs including minor allele frequency 1% or greater, genotyping completeness 99% or greater and Hardy-Weinberg equilibrium (p <0.0001). We specifically evaluated SNPs previously associated with PSA such as rs10993994 (chromosome 10), rs2659056 (chromosome 19) and rs2735839 (chromosome 19).5 The outcome of interest in this study was CaP diagnosis (event).
Study group characteristics including age, PSA and followup time were compared using t tests (assuming unequal variance) with a Welch modification to the df. Because of the longitudinal nature of the study with 1 or more PSA measurements for each man during a period up to more than 40 years, mixed effects models were used to examine the relationship of SNPs, age and log (PSA + 1) with random effects for intercept and time. A likelihood ratio test was used to evaluate 3 models for each SNP in men without prostate disease. The base model included age at first evaluation, time from first evaluation (time), time squared, date, and random effects for time and subject. The second model then added the SNP, and the third model then added the interaction of SNP, time and time squared. A second PSA analysis included men with prostate cancer, and considered a baseline model including cancer status and interactions of cancer status, time and time squared. A likelihood ratio test was used to compare this baseline model to a second model which added the SNP, and a third model that subsequently included interactions between SNP, time and time squared.
To address whether genotype was associated with CaP risk, time dependent Cox proportional hazards models were examined individually for each genotype by PSA with and without adjustment for age and date. Furthermore, the sum of minor alleles was calculated for each subject. Time dependent proportional hazard models were examined for the total count, as well as comparing the presence of 1 or more minor allele(s) with participants who had no minor alleles. The time dependent models were based on the Anderson-Gill formulation as a counting process using survival functions developed by Therneau,9 and were evaluated using a likelihood ratio test. Statistical tests were considered significant for p <0.05 and all tests were 2-sided.
RESULTS
In the overall cohort the racial distribution was 75% white, 19% black and 6% other. The mean age (±SD) at initial evaluation was 48.9 (±15.1) and 52.2 (± 12.8) years for men with and without CaP, respectively (p = 0.06). The mean (±SD) initial PSA was 1.09 (±1.65) ng/ml for men without CaP and 2.07 (±2.70) ng/ml for those in whom CaP developed (p = 0.007). The mean (±SD) followup time was similar for men with and without CaP (16.92 ± 10.17 vs 18.11 ± 10.47, p = 0.39). The raw genotype data are shown in table 1, stratified by race.
Table 1.
Allele frequency by race in the study population
| No. rs10993994 |
No. rs2735839 |
No. rs2659056 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CC | TC | TT | GG | AG | AA | AA | AG | GG | |
| White | 144 | 178 | 56 | 285 | 81 | 12 | 211 | 149 | 18 |
| Black | 21 | 39 | 36 | 43 | 42 | 11 | 84 | 12 | 0 |
| Other | 7 |
19 |
5 |
17 |
13 |
1 |
11 |
18 |
2 |
| Totals | 172 | 236 | 97 | 345 | 136 | 24 | 306 | 179 | 20 |
In men without CaP the increment in PSA with age was independent of genotype (rs2735839, p = 0.22; rs10993994, p = 0.15; rs2659056, p = 0.28; likelihood ratio test). However, when comparing PSA increments including men with CaP, an interaction was found between genotype and time (p <0.001). After adjustment for age and date of evaluation there were associations between the genotype by time interaction and the likelihood of CaP (p = 0.01).
CaP frequency, PSA and age were similar when comparing men with and those without a minor allele (table 2). Compared to men without a minor allele those with a minor allele had a lower risk of CaP at a PSA of 0 ng/ml (risk ratio 0.39, 95% CI 0.20–0.78).
Table 2.
Risk of prostate cancer in the baseline Cox proportional hazards model with PSA, age and date as well as the interaction model including genotype
| Interaction Model |
|||
|---|---|---|---|
| Baseline | Minor Allele | No Minor Allele | |
| No. controls | 444 | 388 | 56 |
| No. cases (%) | 61 (12.1) | 49 (11.2) | 12 (17.6) |
| Total PSA evaluations | 2,637 | 2,253 | 384 |
| Mean age controls (SD) | 57.9 (14.8) | 58.1 (14.7) | 56.7 (15.9) |
| Mean age cases (SD) | 60.5 (11.2) | 61.1 (10.6) | 58.2 (12.8) |
| Mean ng/ml PSA controls (SD) | 1.34 (1.57) | 1.37 (1.61) | 1.17 (1.26) |
| Mean ng/ml PSA cases (SD) | 3.72 (4.40) | 3.64 (4.19) | 3.99 (5.14) |
| Risk ratio at PSA 0 (95% CI) | 1 | 0.39 (0.20–0.78) | 1 |
| Risk ratio/ng/ml PSA (95% CI) | 1.18 (1.13–1.23) | 1.28 (1.20–1.38) | 1.10 (1.06–1.15) |
Cox proportional hazards models were examined with the baseline model including PSA, age and date, with data updated at each evaluation. An interaction model was then examined including PSA, age and date of evaluation, plus a term for whether a subject had a minor allele as well as the interaction term between the presence of a minor allele and PSA.
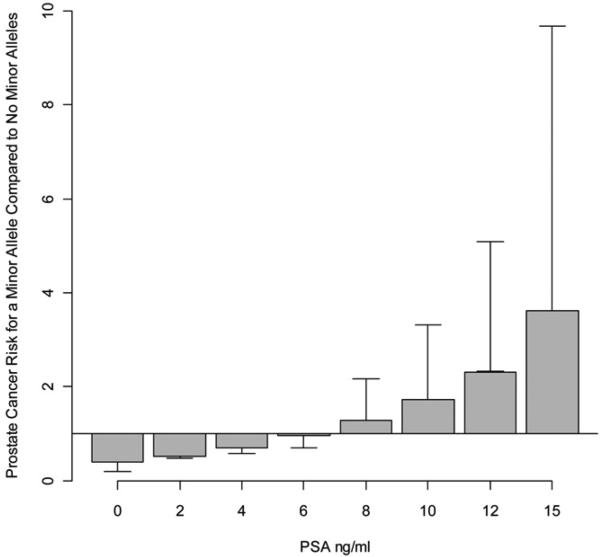
In the base model with age and date the risk ratio for CaP was 1.18 per unit increase in PSA (95% CI 1.13–1.23). In a model that also included genotype and the interaction between genotype and PSA, there was a significant difference in the risk ratio per unit increase in PSA compared to the base model (Cox proportional hazards model p <0.001). Moreover, in the interaction model the risk of CaP per unit increase in PSA was significantly different for men with (risk ratio 1.28, 95% CI 1.20–1.38) and without (risk ratio 1.10, 95% CI 1.06–1.15) a minor allele (Cox proportional hazards model p <0.001). Compared to men without a minor allele those with a minor allele had a risk of CaP that varied by PSA, with a lower risk at a PSA less than 6.0 ng/ml vs a higher risk at a PSA greater than 6.0 ng/ml (see figure).
Prostate cancer risk ratio between carriers and noncarriers of minor allele derived from interactional proportional hazards model. At PSA greater than 6 ng/ml carriers of minor allele had significantly higher risk of prostate cancer whereas at PSA less than 6 ng/ml carriers of minor allele had significantly lower risk of prostate cancer.
Because the frequency of these genetic variants differs by race we performed a subset analysis exclusively in white participants. In a model including genotype and the interaction between genotype and PSA, there was a significant difference in the risk ratio for prostate cancer per unit increase in PSA compared to the base model with age and date (Cox proportional hazards model p <0.001).
Finally we performed a subset analysis including only rs10993994 and rs2659056 in the model due to their similar effect on PSA in the study by Eeles et al. 5 Based on these 2 alleles we similarly found a significant difference in the risk ratio for prostate cancer per unit increase in PSA with the interaction model compared to the base model (Cox proportional hazards model p <0.001).
DISCUSSION
A significant limitation of PSA based prostate cancer screening is that PSA does not have both high sensitivity and high specificity at any given cut point.7 Accordingly it is difficult to choose a threshold at which men should undergo prostate biopsy to rule out cancer.
In light of prior reports suggesting an association between germline sequence variants and serum PSA, we hypothesized that the risk of prostate cancer at a given PSA might be dependent on genotype. Interestingly unlike Eeles et al we did not find a significant association between genotype and PSA by age in controls.5 While this discrepancy may be due to our smaller sample size and/or population differences, Savblorn et al similarly reported that polymorphisms at rs266882 and rs925013 (chromosome 19) within the PSA gene promoter did not influence serum PSA in men without CaP.9
Nevertheless, we did find a significant difference in the risk ratio for prostate cancer at a given PSA based on genotype. Specifically the presence of a minor allele was associated with a high PSA in the presence of CaP. Thus, a low PSA in a man with a minor allele is less likely to represent CaP compared to a man without a minor allele.
These data suggest that CaP risk stratification may be enhanced through the incorporation of PSA and genotype. This in turn might help inform a decision about the need for prostate biopsy and potentially reduce the proportion of unnecessary biopsies and CaP over detection.10 For example, sequence variants associated with PSA and CaP could drive the production of more PSA in the presence of CaP. Thus, men with this variant may have a low risk of harboring CaP at PSAs that are frequently considered to be abnormal and trigger prostate biopsies.
It is noteworthy that in the study by Eeles et al the minor allele for rsl0993994 and rs2659056 was associated with a higher PSA in controls whereas the rs2735839 minor allele was associated with a lower PSA.5 Although we did not find a significant association between genotype and PSA by age among the controls in our cohort, nonetheless we performed a separate Cox proportional hazards model including only the minor alleles for rsl0993994 and rs2659056 with similar results.
Several limitations of our study deserve mention. Data on pathological tumor features were not uniformly available, precluding an analysis of the relationship among genotype, PSA and CaP aggressiveness. In addition, the sample size was relatively small, since many men enrolled before 1991 did not undergo PSA testing or have frozen serum samples available for analysis. Moreover, a limited number of Baltimore Longitudinal Study of Aging participants had genotype data. When comparing the men included in this study to those excluded, included men were significantly younger at the first PSA measurement (mean age 49 vs 57 years, t test p = 0.001) and had a lower mean PSA (1.2 vs 1.7 ng/ml, t test p <0.001). However, the racial distribution of excluded men was similar (81% white, 18% black and 1% unknown).
Finally our cohort was primarily white and there are considerable differences in allele frequencies among racial groups. Nevertheless, excluding black men from the analysis did not change the results of the Cox proportional hazards model.
Despite these limitations our ability to assess the risk of CaP per unit increase in PSA by genotype is unique for this study with multiple PSA measures over time. With the rapidly evolving field of personal genomic testing, data on how to incorporate this new information into screening protocols will become increasingly important. Indeed, our results suggest that the presence of specific alleles influences the risk of prostate cancer at a given PSA. Nevertheless, further study will be necessary to determine whether improvements in risk stratification using genetic information will translate into better clinical outcomes.
CONCLUSIONS
After controlling for age and date of evaluation we found that the presence of a specific genetic variant was significantly associated with the risk of prostate cancer per unit increase in PSA. Further study is warranted to help evaluate the potential role of genetic markers in the clinical interpretation of PSA for prostate cancer risk assessment.
Acknowledgments
Supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Abbreviations and Acronyms
- CaP
prostate cancer
- PSA
prostate specific antigen
- SNP
single nucleotide polymorphism
REFERENCES
- 1.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Suuriniemi M, Agalliu I, Schaid OJ, Johanneson B, McDonnell SK, Iwasaki L. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 4.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 5.Eeles RA, Kote-Jarai Z, Giles GG, Dlama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 6.Beke L, Nuytten M, Van Eynde A, Beullens M, Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene. 2007;23:4590. doi: 10.1038/sj.onc.1210248. [DOI] [PubMed] [Google Scholar]
- 7.Thompson IM, Ankerst OP, Chi C, Lucia MS, Goodman PJ, Crowley JJ, et al. Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/ml or lower. JAMA. 2005;294:66. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 8.Carter HB, Pearson JD, Metter EJ, Brant LJ, Chan OW, Andres R, et al. Longitudinal evaluation of prostate-specific antigen levels in men with and without prostate disease. JAMA. 1992;267:2215. [PMC free article] [PubMed] [Google Scholar]
- 9.Savblom C, Giwercman A, Maim J, Hallden C, Lundin K, Lilja H, et al. Association between polymorphisms in the prostate-specific antigen (PSA) promoter and release of PSA. Int J Androl. 2008 doi: 10.1111/j.1365-2605.2008.00882.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Dall'Era M, Carroll PR. Prostate cancer-more information and more questions. J Urol. 2007;177:1607. doi: 10.1016/j.juro.2007.02.008. [DOI] [PubMed] [Google Scholar]