Abstract
The proteasome is able to create spliced Ags, in which two distant parts of a protein are excised and ligated together to form a novel peptide, for presentation by MHC class I molecules. These noncontiguous epitopes are generated via a transpeptidation reaction catalyzed by the proteasomal active sites. Transpeptidation reactions in the proteasome follow explicit rules and occur particularly efficiently when the C-terminal ligation partner contains a lysine or arginine residue at the site of ligation. Lysine contains two amino groups that theoretically may both participate in ligation reactions, implying that potentially not only peptide but also isopeptide linkages could be formed. Using nuclear magnetic resonance spectroscopy, we demonstrate in the present study that the proteasome can use the ε-amino group of an N-terminal lysine residue in transpeptidation reactions to create a novel type of posttranslationally modified epitopes. We show that the overall efficiency of ε ligation is only 10-fold lower as compared with α ligation, suggesting that the proteasome can produce sufficient isopeptide Ag to evoke a T cell response. Additionally, we show that isopeptides are more stable toward further proteasomal processing than are normal peptides, and we demonstrate that isopeptides can bind to HLA-A2.1 and HLA-A3 with high affinity. These properties likely increase the fraction of ε-ligated peptides presented on the cell surface for CD8+ T cell surveillance. Finally, we show that isopeptide Ags are immunogenic in vivo. We postulate that ε ligation is a genuine posttranslational modification, suggesting that the proteasome can create a novel type of Ag that is likely to play a role in immunity.
Introduction
The eukaryotic 26S proteasome is responsible for the degradation of redundant and misfolded proteins and for the turnover of regulatory proteins involved in a wide range of cellular processes, including cell proliferation and survival, cell cycle control, and cellular stress responses (1). Additionally, the proteasome is responsible for the generation of peptides that are presented on the cell surface by MHC class I proteins (2, 3). The proteasome generates peptides both from self and foreign proteins, which are subsequently transported into the endoplasmic reticulum, loaded onto MHC class I, and transported to the cell surface. CD8+ T cells continuously scan the MHC class I–peptide complexes on the cell surface, thereby receiving a blueprint of the intracellular protein content. This enables CD8+ T cells to sense viral infection or malignant transformation, which ultimately results in killing of the APC.
The 26S proteasome consists of a barrel-shaped 20S core, complexed at one or both ends with 19S regulatory particles. The 19S caps deubiquitinate and unfold protein substrates and regulate their entry into the 20S core, where the proteolytic activity resides. The 20S catalytic chamber consists of four stacked heptameric rings and has an overall architecture of α(1–7)β(1–7)β(1–7)α(1–7). The two outer α rings regulate entry into the complex and provide anchor points for the attachment of 19S regulatory caps, whereas the actual catalytic activity resides within the inner β rings. Once entered into the 20S core, a protein is degraded via the action of three catalytically active subunits, termed β1, β2, and β5, which all have different cleavage specificities and which are responsible for the proteasomal caspase-like, tryptic, and chymotryptic activities, respectively. In lymphoid tissues, these subunits are largely replaced by their immunoproteasome counterparts, termed β1i, β2i, and β5i, to form the immunoproteasome, which has been hypothesized to favor the production of antigenic peptides (4).
Interestingly, the proteasome does not only produce contiguous peptides for presentation on MHC class I. Noncontiguous Ags, in which two distant parts of a protein are excised and ligated together to form a novel peptide, are presented on the cell surface and can evoke immune responses (5–9). Proteasomal splicing thus results in a genuine posttranslational modification and has been hypothesized to increase diversity of the antigenic peptides presented, ultimately resulting in a more efficient recognition and elimination of infected or malignant cells by CD8+ T cells (3, 10–12). Noncontiguous Ags are produced by the proteasome via a transpeptidation mechanism (5, 7, 13, 14). During the transpeptidation event, an active site N-terminal threonine hydroxyl group attacks the scissile peptide bond, resulting in the formation of an O-acyl enzyme intermediate and the release of the C-terminal part of the peptide. In a second step, an aminolysis reaction takes place, in which this intermediate ester is captured by an amino group of a second peptide (the C-terminal ligation fragment), leading to the formation of a novel peptide bond and a spliced peptide in which two separate peptide fragments are combined. Transpeptidation thus competes with normal hydrolysis, in which the intermediate ester is hydrolyzed by water molecules present in excess, resulting in the release of the N-terminal part of the peptide (12).
All peptides produced in the proteasomal catalytic core have a free N terminus and can therefore serve as a C-terminal ligation partner in ligation reactions. Indeed, peptide splicing involving a large variety of C-terminal fragments can be readily observed in vitro (14, 15). Although the concentration of precursor fragments is one of the driving factors of the splicing reaction (14), we show in an accompanying article (16) that splicing is also governed by explicit rules, as revealed by preferred sequence motifs within both splicing partners (16). For the C-terminal ligation partner, ligation occurs particularly efficiently when a basic amino acid residue (lysine or arginine) is present at the N terminus (16). Lysine has a second free amino group on the Cε position (the ε-amine) in addition to its free N terminus (the α-amine) and, theoretically, both these amino groups may be able to participate in ligation reactions. Whereas ligation with the α-amine results in the formation of a normal peptide bond, ligation with the ε-amine results in the formation of an isopeptide. Such isopeptide linkage-containing epitopes may have unique properties and may form a novel class of posttranslationally modified peptides that can be presented on MHC class I. Isopeptide bonds are notoriously difficult to identify and have not been observed in epitopes so far. In this study we show that the proteasome can use both the α- and ε-amino group of lysine in ligation reactions and can form both spliced peptides and spliced isopeptides. Additionally, we demonstrate that these isopeptides are immunogenic in vivo and we show how their unique properties may contribute to their efficient presentation by MHC class I molecules. Collectively, these data strongly suggest that ε ligation may contribute to the pool of Ags that can be scanned by the T cell repertoire. Development of sensitive methods that can be used to detect such isopeptide linked Ags will be of value to further probe their potential role.
Materials and Methods
Peptide building blocks were purchased from Novabiochem and appropriately functionalized resins were from Applied Biosystems. 15N-ε-l-lysine HCl was purchased from C/D/N Isotopes (Pointe-Claire, QC, Canada). All solvents were purchased from Biosolve at the highest grade available. All other chemicals were purchased from Sigma-Aldrich at the highest available purity. All solvents and chemicals were used as received. Liquid chromatography–mass spectrometry (LC-MS) analyses were carried out on a Waters LCT mass spectrometer in line with a Waters 2795 HPLC system and a Waters 2996 photodiode array detector, using an XBridge BEH300 C18 column (3.5 μm; 2.1 × 100 mm; Waters) with a linear gradient (5–50% acetonitrile [ACN] in H2O containing 0.1% formic acid). All 1H and 1H-15N heteronuclear single quantum coherence (HSQC) experiments were carried out in DMSO-d6 using a Bruker Avance 300 (1H, 300 MHz; 15N, 30.4 MHz) spectrometer. Heteronuclear multiple quantum coherence (HMQC) experiments were performed on a Bruker ARX 400 (1H, 400 MHz; 15N, 40.5 MHz) spectrometer. Nuclear magnetic resonance (NMR) data were processed and analyzed using Bruker v2.1 Topspin software.
Peptide synthesis
Peptides were synthesized using standard Fmoc-based solid-phase peptide synthesis protocols and appropriately functionalized polyethylene glycol–polysterene Wang resins. Functionalized resins were subjected to coupling cycles, in which deprotection of the Fmoc group with piperidine/NMP (1:4 [v/v]) was followed by coupling with four equivalents each of Fmoc-protected amino acid, di-isopropylethylamine, and (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate. Reactions were carried out in NMP at a volume of 1 ml/0.1 g resin. After the final coupling step, the Fmoc group was removed and peptides were directly fully deprotected and released from the resin directly by treating the resin with trifluoroacetic acid (TFA)/H2O/triisopropylsilane (93:5:2 [v/v]) for 2.5 h. Alternatively, peptides were treated with four equivalents each of acetic anhydride and di-isopropylethylamine for 40 min to acetylate the N terminus prior to deprotection and release from the resin. Peptides containing Nε-linked lysine residues were synthesized using Boc-l-lysine(Fmoc)-OH. Incorporation of this building blocked followed by removal of the Fmoc group ensures that peptide synthesis continues at the lysine ε-amine group, resulting in isopeptide linkage containing peptides. Peptides were precipitated with cold diethyl ether/pentane (3:1 [v/v]) and lyophilized from H2O/ACN/acetic acid (65:25:10 [v/v]).
Synthesis of 15N-ε-Fmoc-Lys(Boc)-OH
Nα-(9-fluorenyl)methoxycarbonyl-15Nε-tert-butyloxycarbonyl-l-lysine was synthesized as described (17). 1H NMR (400 MHz, d6-DMSO) δ = 12.53 (bs, COOH), 7.91 (d, J = 7.5 Hz, 2H, HAr), 7.73 (d, J = 7.5 Hz, 2H, HAr), 7.62 (d, J = 7.6 Hz, 1H, NH), 7.42 (t, J = 7.5 Hz, 2H, HAr), 7.33 (t, J = 7.5 Hz, 2H, HAr), 6.79 (dt, JNH = Hz, JHH = 6.8 Hz, 1H, 15NεH), 4.29–4.26 (m, 2H, OCH2), 4.24–4.20 (m, 1H, OCH2CH), 3.93–3.88 (m, 1H, Hα), 2.91–2.78 (m, 2H, CH2), 1.70–1.52 (m, 2H, CH2), 1.36 [s, 9H, C(CH3)3], 1.33–1.27 (m, 4H, 2× CH2). 13C NMR APT (300 MHz, CDCl3) δ = 175.51 (COOH), 156.29 (2× CO), 143.74 (2× Cq-Ar), 141.30 (2× Cq-Ar), 127.69 (2× CHAr), 127.08 (2× CHAr), 125.14 (2× CHAr), 119.95 (2× CHAr), 79.57 (CCH3), 67.04 (CH2O), 53.71 (CαH) 47.16 (CHCH2O), 40.15 (CεH2, 1JCN = 11 Hz), 31.76 (CβH2), 29.54 (CδH2), 28.41 [(CH3)3], 22.31 (CγH2). 15N1H-HMQC (6.79 ppm, 83.7 ppm). MS (ESI): [M + H]+calc = 470.2, [M + H]+found = 470.1.
Proteasome purification
Proteasome was purified from bovine liver as described (18). After each step, proteasome purity was monitored by incubating fractions with a fluorescent proteasome activity probe, followed by SDS-PAGE and scanning of the resulting gel for fluorescence emission as described (19, 20). Briefly, bovine liver was homogenized in PBS, followed by precipitation using 40% saturated ammonium sulfate as a first precipitation step. The proteasome was subsequently precipitated by increasing the concentration of saturated ammonium sulfate to 60%. Following dialysis, the proteasome was further purified using a 10–40% sucrose gradient and anion exchange column chromatography, using DEAE Sephadex A25 resin. Proteasome-containing fractions were pooled, concentrated, and protein concentrations were determined using the Bradford assay (Bio-Rad). The quality of the isolation was further confirmed by SDS-PAGE and Coomassie staining.
In vitro ligation assays
All ligation reactions were performed in ligation buffer (50 mM Tris-HCl [pH 8.5]) at 37°C unless indicated otherwise. YLGDSY (0.67 mM) was incubated with a 5-fold molar excess (acetylated) KLI or KLISV and proteasome (0.067 mg/ml) for 16 h. Then, 0.67 mM YLDWα/εKLISV, GILGFα/εKTL GILGFVα/εKL, GILGα/εKFTL, GILα/εKVFTL, or GIα/εKFVFTL was incubated with proteasome (0.067 mg/ml) for different time periods up to 26 h. Incubation mixtures were snap frozen, lyophilized, dissolved in DMSO/H2O/ACN (1:2:1 [v/v]), and filtered over Strata-X 33-μm polymeric reversed-phase disposable columns (Phenomenex). Eluted fractions were analyzed by LC-MS. For MALDI analysis, 1.6 μM C*SLPRGTASSR (where * is maleimido-linked tetramethylrhodamine) was incubated with proteasome (0.02 mg/ml) for 16 h at 45°C in the presence or absence of 100 μM MG132, 100 nM bortezomib, 0.5 M lysine, and 0.5 M Nα-acetylated lysine or 0.5 M Nε-acetylated lysine. For NMR experiments, 16 μmol ε(15N)KLI was incubated with 80 μmol YLGDSY and 2.8 mg proteasome in 23.2 ml ligation buffer for 24 h. The reaction mixture was snap frozen, lyophilized, dissolved in 12 ml ACN/H2O (1:1), and filtered over a Strata-X 33-μm polymeric reversed-phase disposable column (Phenomenex) to remove proteasomal proteins.
MALDI
Reaction mixtures were freeze-dried and dissolved in ACN/H2O (5:95 [v/v]) containing 0.1% TFA prior to purification and desalting using reversed-phase ZipTipC18 tips (Millipore, C18, spherical silica, 15 μm, 200 Å pore size). Peptides were eluted from the ZipTip pipette with ACN and eluates were analyzed by MALDI mass spectrometry. MALDI-TOF experiments were carried out on an Autoflex, linear MALDI-TOF mass spectrometer (Bruker Daltonik, Bremen, Germany). Desalted samples were mixed 1:1 with 10 mg/ml 2,5-dihydroxybenzoic acid (Bruker Daltonik) matrix solution in 0.1% TFA and spotted onto a MALDI target plate. Peptide calibration standard (molecular mass 1046–3147 Da, Bruker Daltonik) was used for calibration. Spectra were analyzed with Bruker Daltonics FlexAnalysis software. Ligation efficiencies were calculated using the following formula: efficiency = [IL/(IL + IH)] × 100%, where IL and IH are the peak intensities of the ligation and hydrolysis products, respectively.
Nuclear magnetic resonance
For NMR analysis, the filtered reaction mixture (see In vitro ligation assays above) was lyophilized again, dissolved in 7.5 ml DMSO/H2O (1:4) and diluted, and purified by HPLC over an Atlantis Preparative T3 column (5 μm; 20 × 150 mm; Waters) using a linear gradient (5–50% ACN in H2O containing 0.05% TFA). HPLC purifications were performed on a Shimadzu LC-20AT prominence liquid chromatography system, coupled to a Shimadzu SPD-20A prominence UV/Vis detector and a Shimadzu CTO-20A prominence column oven. The YLGD-ε(15N)KLI–containing fraction was lyophilized and dissolved in 0.4 ml DMSO-d6 for NMR spectroscopy analysis. Reference spectra of synthetic YLGDαKLI, YLGDεKLI, and YLGDYLGD were recorded using a 50 mM peptide solution in DMSO-d6. HSQC experiments were performed using a Bruker preprogrammed gradient enhanced echo/antiecho HSQC pulse program (21, 22) (hsqcetgp, avance version, 2D H-1/X correlation via double inept transfer, phase sensitive using echo/antiecho-TPPI gradient selection with decoupling during acquisition, using trim pulses in inept transfer) optimized for the detection of 15N signals, and 15N signals and were compared with a 15NH3 reference signal. The relative percentage of ε ligation versus α ligation was calculated using the following formula: efficiency of ε ligation = I1/(0.44 × IN) × 0.37/98 × 100%, where I1 is the integral of the 15Nε-enriched peak 1, IN is the integral of peak N, resulting from a naturally abundant amide resonance, 0.44 is the fraction [YLGD][α/εKLI] in the ligation sample as compared with total peptide content, 0.37 is the percentage of naturally abundant 15N, and 98 is the percentage of 15N enrichment of 15Nε-enriched peak 1. The efficiency of ε ligation was calculated for peaks three to five and averaged.
Fluorescence polarization HLA-A2.1 and HLA-A3 binding assay
Fluorescence polarization (FP) HLA-A*0201 and HLA-A*0301 binding assays with UV-mediated peptide exchange were performed as described previously (23, 24) with minor changes. For the HLA-A*0201 binding assay, KILGFVFJ1V (in which J1 is UV-cleavable 3-amino-3-(2-nitrophenyl)propionic acid was used as conditional ligand and fluorescently labeled FLPSDCTMRFPSV (in which TMR is maleimido-linked tetramethylrhodamine) was used as tracer peptide. For the HLA-A*0301 binding assay, RIYRJ1GATR was used as conditional ligand and KVPCTMRALINK was used as tracer peptide. Serial 2- or 3-fold dilutions of different (iso)peptides were used as competitor peptides. Assays were performed in 384-well low-volume black nonbinding surface assay plates (Corning 3820). Wells were loaded with 15 or 30 μl total volume containing 0.5 μM HLA-A–peptide complex, 1 nM tracer peptide, and serial dilutions of competitor peptide in PBS containing 0.5 mg/ml bovine γ-globulin. The plate was spun for 1 min at 1000 × g at room temperature to ensure proper mixing of all components. To start UV-mediated cleavage of the conditional ligand and peptide exchange, the plate was placed under a 365-nm UV lamp at 10 cm distance (366 nm ultraviolet lamp, 2 × 15 W blacklight blue tubes, 505 × 140 × 117 mm [length × width × height], UVItec, Cambridge, U.K.) located in a cold room (4°C). After 30 min irradiation, the plate was sealed with thermowell sealing tape (Corning) and incubated at room temperature for 4 or 24 h, allowing cleaved peptide fragments to dissociate and to be exchanged for competitor and/or tracer peptide. Subsequently, fluorescence polarization measurements were performed as described (24). The binding affinity (IC50 value) of each competitor peptide was defined as the concentration that inhibited 50% of tracer peptide binding. Data were analyzed using GraphPad Prism software (GraphPad Software).
Mouse immunizations and T cell staining
HHD mice that are transgenic for a chimeric MHC molecule consisting of the HLA-A*0201 α1 and α2 domain and the H-2Dd α3 domain (The Jackson Laboratory, Bar Harbor, ME) (n = 4/group) were injected at both sides of the tail base with 150 μg isopeptide and 50 μg CpG oligonucleotide 1826 (InvivoGen), emulsified in IFA (Sigma-Aldrich). One-hundred microliters blood was collected from the tail tip at the indicated time points, subjected to erylysis, and then used for MHC multimer staining. To create MHC multimers, isopeptides in DMSO were added to biotinylated MHC monomers (25 μg/ml in PBS) to a final concentration of 50 μM and UV irradiated for 30 min. Samples were left at room temperature for another 30 min. Subsequently, plates were centrifuged for 5 min at 3300 × g to remove any aggregated MHC molecules (25). Streptavidin–R-PE or streptavidin-allophycocyanin (Life Technologies) was added to the exchanged MHC monomers to a final concentration of 13.5 μg/ml, and 2 μl PE-labeled MHC multimers and 4 μl allophycocyanin-labeled MHC multimers were used for dual staining of blood samples (26). Following staining, samples were taken up in FACS buffer (1× PBS, 0.5% BSA, 0.02% sodium azide) containing 1% propidium iodide to distinguish between live and dead T cells by flow cytometry. Samples were analyzed on either a Beckman Coulter CyAn ADP analyzer or a BD FACSCalibur machine, and data were analyzed using FlowJo 7.6.1 (Tree Star).
Results
The proteasome can form both normal and isopeptide bonds during ligation
In contrast to other amino acids, a C-terminal lysine residue that is ligated onto an N-terminal splicing precursor has two available amino groups that may perform the aminolysis reaction: the α-amino and the ε-amino group that can form a peptide and an isopeptide bond, respectively (Supplemental Fig. 1A). To investigate whether the ε-amino group of lysine could react with the O-acyl enzyme intermediate during ligation reactions, unprotected lysine, Nε-acetylated lysine (Kε(Ac)), and Nα-acetylated lysine (Kα(Ac)) were compared in ligation experiments. Because only the unprotected amino groups of Kε(Ac) and Kα(Ac) can participate in ligation reactions (Supplemental Fig. 1B), the use of monoacetylated lysine makes it possible to discriminate α from ε ligation. As N-terminal precursor we used the peptide C*SLPRGTASSR, an N-terminally extended and fluorescently labeled version of the known splicing precursor peptide SLPRGTASSR (7). This precursor was subjected to proteasomal degradation in the presence of proteasome inhibitors or (acetylated) lysine, followed by MALDI analysis. As both hydrolysis and ligation products that were formed during these digestions contained a positively charged TMR label (indicated with the asterisk), which functioned as an ionization enhancer and acted as the main determinant of signal intensities, peak heights in MALDI spectra could be semiquantitatively compared. The 20S proteasome preparation used throughout this study was purified from bovine liver and almost exclusively contained constitutive proteasome, as indicated both by mass spectrometry experiments and by using a fluorescent proteasome activity probe (18).
Upon incubation with purified 20S proteasome (∼27 nM), C*SLPRGTASSR at m/z 1616 disappeared, whereas two peaks appeared at m/z 1372 and 1214, which could be assigned to the hydrolysis products C*SLPRGTAS and C*SLPRGT, respectively (Fig. 1A). Hydrolysis was completely abolished by the addition of the pan-proteasome inhibitor MG132, whereas the addition of 100 nM proteasome inhibitor bortezomib only hampered the formation of C*SLPRGTAS, but not C*SLPRGT (Fig. 1A). Bortezomib has been described to inhibit the chymotryptic and caspase-like activities, but not the tryptic activity of 20S proteasome purified from (immature) RBCs, as measured by fluorogenic substrates (27, 28). This indicates that bortezomib is a β1/β5 inhibitor in vitro, a finding that could also be confirmed in cell lysates (29). Based on these data, we consider it likely that C*SLPRGTAS is formed by the β5 subunit, whereas C*SLPRGT is probably a β2 hydrolysis product. When excess lysine was added to the reaction mixture, direct ligation of lysine onto the precursor peptide occurred, as evidenced by the appearance of additional peaks at m/z 1500 (Fig. 1B) and m/z 1342 (data not shown), which could be assigned to the β5 ligation product [C*SLPRGTAS][K] and the β2 ligation product [C*SLPRGT][K], respectively. When either Kα(Ac) or Kε(Ac) was added to the digestion mixture, both MALDI spectra showed an additional peak at m/z 1542 (Fig. 1B), indicative of the formation of the β5 ligation product [C*SLPRGTAS][KAc] with both types of acetylated lysine.
FIGURE 1.
The proteasome accepts α-acetylated amino acids and peptides as C-terminal ligation partner during splicing reactions. (A) MALDI spectra of C*SLPRGTASSR and of proteasomal digestion mixtures of C*SLPRGTASSR, C*SLPRGTASSR and MG132, and C*SLPRGTASSR and bortezomib. (B) MALDI spectra of proteasomal digestion mixtures of C*SLPRGTASSR, C*SLPRGTASSR and lysine, C*SLPRGTASSR and Nε-acetylated lysine C*SLPRGTASSR, and Nα-acetylated lysine. (C) Ligation efficiencies for the formation of the indicated β2 and β5 ligation products by lysine and acetylated lysine, calculated from MALDI experiments. (D) Ligation efficiencies for the formation of [YLGD][KLISV] and [YLGD][KLI] using nonacetylated or Nα-acetylated C-terminal ligation partners, calculated from LC-MS experiments. Ligation efficiency was defined as the percentage of cleavage that resulted in ligation as opposed to hydrolysis. An asterisk indicates maleimido-linked TMR.
Using nonacetylated lysine and based on the obtained MALDI spectra, ligation efficiencies (defined as the percentage of cleavages that resulted in ligation as opposed to hydrolysis) were 26 and 13% for the β5 and β2 active sites, respectively (Fig. 1C). Peptide bond formation by Kε(Ac) in the β5 site occurred with an estimated ligation efficiency of 29%. Strikingly, Kα(Ac) was able to form isopeptide bonds with a similar ligation efficiency of 33% (Fig. 1C). The α- and ε-amino groups of lysine are thus equally capable of participating in proteasomal ligation reactions in the β5 active site. In contrast, formation of the β2 ligation product C*SLPRGT][KAc] was only observed upon addition of Kε(Ac) and occurred at a lower ligation efficiency of 7% (Fig. 1C), indicating that the β2 active site only accepts α-amino groups. Taken together, these data suggest that both peptide and isopeptide bonds can be formed by the single amino acid K during proteasomal splicing reactions.
It is very well possible that a single lysine residue is capable of fitting into the specificity pockets of the proteasomal active sites in multiple orientations, which would allow for both α and ε ligation to occur. However, for a longer peptide substrate, orientation of the amino group may potentially only be right for α ligation, as this is also the orientation prior to cleavage. To investigate whether peptides containing an acetylated N-terminal lysine residue could also serve as C-terminal ligation partners, the β5 N-terminal splicing precursor YLGD-SY (16) was incubated with proteasome and either unprotected or acetylated KLI and KLISV. The formation of the hydrolysis product YLGD and the ligation products [YLGD][KLI], [YLGD][KAcLI], [YLGD][KLISV], and [YLGD][KAcLISV] was monitored by LC-MS. As expected, ligation of Nε-acetylated peptides proceeded with similar efficiency as in reactions using nonacetylated peptides (data not shown). Surprisingly, ligation of Nα-acetylated peptides also readily occurred, albeit with a lower efficiency compared with nonacetylated peptides (Fig. 1D). Similar to what is observed for α-linked peptides (16), the efficiency of isopeptide bond formation with Nα-acetylated peptides was independent of the length of the C-terminal ligation fragments.
ε Ligation can compete with α ligation in proteasomal ligation events
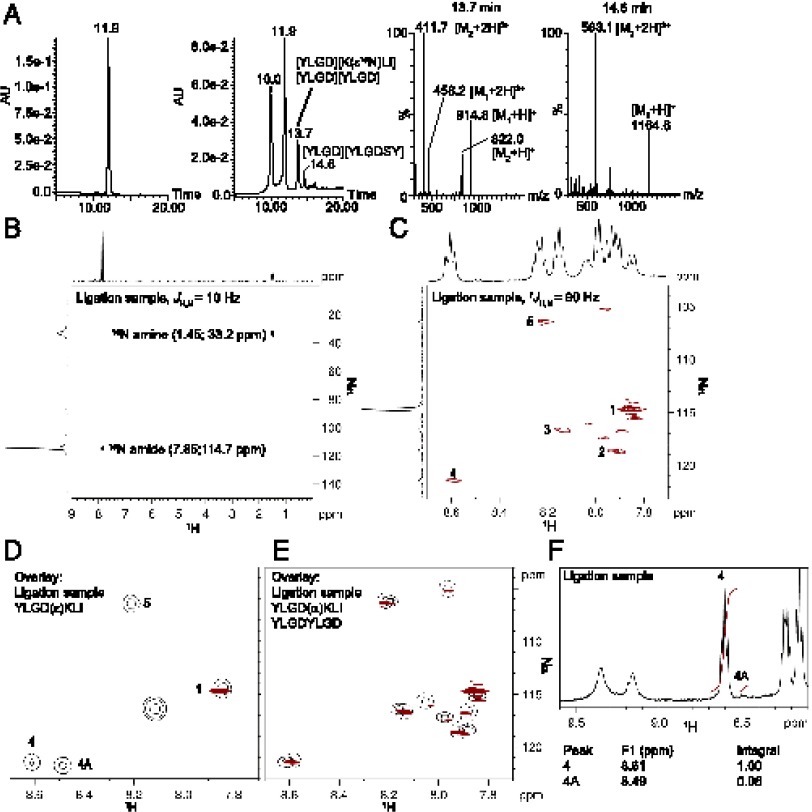
From the data above we conclude that the proteasome can form isopeptide bonds between two peptide fragments in a situation in which the α-amino group is unavailable. We next wanted to determine whether the ε-amino group of an N-terminal lysine residue could compete with the α-amino group for ligation in the physiological situation in which both amines are unprotected. To this end, a ligation experiment was performed using KLI in which the ε-amine was 15N enriched (Kε15NLI, Supplemental Fig. 1C). In this case, ε ligation will result in deformation of a 15N-enriched amide group, whereas α ligation will lead to a ligation product containing a 15N-enriched amino group, which are distinguishable by NMR spectroscopy. 15N-ε-Fmoc-Lys(Boc)-OH was synthesized and incorporated into Kε15NLI by standard solid-phase peptide synthesis. Subsequently, Kε15NLI was incubated with proteasome and YLGD-SY and the resulting digestion mixture was analyzed by LC-MS (Fig. 2A, left and second panels, respectively). After incubation, the precursor YLGD-SY (eluting at 11.9 min), the hydrolysis product YLGD (eluting at 10.0 min), and different ligation products could be detected in the digestion mixture. MS analysis of the peak eluting at 13.7 min showed that this peak contained two peptides with masses of 821 Da (M1) and 913.8 Da (M2) (Fig. 2A, third panel), corresponding to the ligation products [YLGD][Kε15NLI] and [YLGD][YLGD]. The peak eluting at 14.6 min contained a third ligation product with a mass of 1163.6 Da (M3) (Fig. 2A, right panel), corresponding to [YLGD][YLGDSY].
FIGURE 2.
ε Ligation can compete with α ligation during proteasomal splicing reactions. (A) HPLC profile (UV detection at 280 nm) of ligation mixture A containing YLGDSY, Kε15NLI, and proteasome before and after incubation (left and second panels, respectively). In-line MS spectra of the peaks eluting at 13.7 min (third panel) and 14.6 min (right panel). M1, [YLGD][KLI]; M2, [YLGD][YLGD]; M3, [YLG][YLGDSY]. (B) 1H-15N HMQC spectrum of purified ligation mixture A, recorded using JH,N = 10 Hz. One-dimensional projections of the 1H and 15N signals are shown on top and left axes, respectively. (C) 1H-15N HSQC spectrum of purified ligation mixture A, recorded using 1JH,N = 90 Hz to visualize all amide resonances. One-dimensional projections of the 1H and 15N signals are shown on top and left axes, respectively. (D) Overlay of the 1H-15N HSQC spectra of 15Nε-enriched purified ligation mixture A (red) and synthetic, nonenriched YLGDεKLI (black). (E) Overlay of the 1H-15N HSQC spectra of 15Nε-enriched purified ligation mixture A (red) and synthetic, nonenriched YLGDαKLI (black, solid lines) and synthetic nonenriched YLGDYLGD (black, dotted lines). (F) 1H spectrum of purified ligation mixture A and integrals of peak 4, visible in the 1H spectra of all products, and peak 4A, visible in the 1H spectrum of YLGDεKLI only (Supplemental Fig. 2C, 2D). In all NMR spectra, peaks were assigned as follows: 1, εK5; 2, not assigned; 3, L2 or D4; 4, L2 or D4; 4A, not assigned; 5, G3 (see Supplemental Fig. 2B).
Next, the ligation mixture was purified using HPLC and the fraction containing the coeluting products [YLGD][Kε15NLI] and [YLGD][YLGD] was analyzed by NMR. If both α-linked and ε-linked [YLGD][Kε15NLI] are formed during the ligation reaction, a 15N NMR spectrum should show two distinct peaks, one corresponding to the 15N amine and the other to the 15N amide group (Supplemental Fig. 1C). As direct detection of 15N atoms suffers from low sensitivity and long relaxation times (30), 15N signals are usually inverse detected via the proton signal in a 1H-15N HSQC experiment using a fixed JH,N coupling constant. Whereas amide groups can be readily detected in an HSQC experiment via the 1JH,N coupling, NMR characterization of lysine amine groups via the 1JH,N coupling is challenging, due to rapid hydrogen exchange with water (Supplemental Fig. 2A) (31). In accordance, in HSQC experiments of the purified ligation mixture we could not detect the 15N amine signal using amine JH,N coupling constant ranging from 55 to 90 Hz (data not shown), although these covered the whole range of reported amine 1JH,N coupling constants (55–80 Hz) (30). Alternatively, when 15N signals are hand inverse detected via long-range 2JH,N and 3JH,N couplings (JH,N = 0.3–16 Hz) (30), both amide and amine resonances can be measured simultaneously in a single experiment (Supplemental Fig. 2A). Therefore, we performed a 1H-15N HMQC measurement using a JH,N coupling constant of 10 Hz. The resulting HMQC spectrum showed two cross peaks, with shifts of 7.85 (114.7 ppm) and 1.45 (33.2 ppm) (Fig. 2B). Because amines and amides display 15N chemical shifts of 0–70 and 80–170 ppm compared with a 15NH3 reference signal, respectively (30), we conclude that during proteasomal digestion of YLGD-SY in the presence of Kε15NLI, ligation products containing 15N-enriched amines and 15N-enriched amides were both formed, strongly indicating that ε ligation can compete with α ligation in proteasomal splicing reactions.
ε Ligation occurs in 1 of 10 ligation events
Next, we set out to quantify the relative efficiency of ε ligation. It is, however, not possible to directly compare the 15N amide and 15N amine signals in a single HSQC measurement. 1H-15N correlations of the lysine amine group are often difficult to observe in HSQC experiments optimized for backbone amides, even when the water exchange rate is slow enough to permit their detection by 1H NMR (31), for several reasons. First, the 15N transverse relaxation of lysine amine groups has been shown to be highly affected by water exchange, resulting in broadening of 15N line shapes and a decrease in resolution and sensitivity (31), as visible in Fig. 2B. Second, the 15N chemical shift of lysine amine groups lies ∼90 ppm upfield from the backbone amide signals (Fig. 2B). As the radio frequency field strength that can be used for 15N pulses is limited, application of a 15N 180° pulse at 115 ppm (close to the amide resonance frequency) leads to an imperfect pulse at 30 ppm (close to the lysine amine resonance frequency) and therefore to loss of signal intensity, and vice versa (31). With standard HSQC pulse programs it is therefore not possible to obtain maximal signal intensities for 15N amide and 15N amine groups simultaneously.
All amide signals in a single HSQC spectrum are, however, directly proportional to each other. Therefore, the relative amounts of ε and α ligation products were estimated by quantifying the signal intensity of the newly formed 15N-enriched amide bond using amide backbone resonances due to naturally abundant 15N within the peptide as an internal reference (Supplemental Fig. 2B). To ensure maximum signal intensities of amide resonances, the [YLGD][Kε15NLI]-containing fraction was analyzed again in an HSQC experiment using a 1JH,N coupling constant of 90 Hz (32). In the resulting spectrum, one intense cross peak was visible at 7.86 (114.7 ppm), originating from the 15N-enriched isopeptide amide in [YLGD][Kε15NLI] (Fig. 2C, Supplemental Fig. 2B, peak 1). Additionally, several weaker signals (peaks 2–5) from naturally abundant 15N amide signals of different ligation products could be detected. As LC-MS analysis indicated that the ligation sample contained three ligation products, [YLGD][αKLI], [YLGD][εKLI], and [YLGD][YLGD] (Fig. 2A), we synthesized these three peptides and recorded 1H and 1H-15N HSQC spectra as a reference to assign 1H-15N cross peaks to one or more peptides (Supplemental Fig. 2C). An overlay of the 1H-15N HSQC spectra of the 15N-enriched ligation sample and synthetic YLGDεKLI confirmed that peak 1 resulted from the 15N-enriched εK5 amide resonance (Fig. 2D). Additionally, the overlay of HSQC spectra of the ligation sample, synthetic YLGDαKLI, and synthetic YLGDYLGD showed that all weaker signals could be attributed to naturally abundant 15N amide resonances in one or more ligation products (Fig. 2E). For quantification purposes, we focused on cross peaks 3–5 that were shared by all three peptides. [YLGD][YLGD] and [YLGD][α/εKLI] were present in the ligation sample at an estimated ratio of 56:44 based on the 1H spectrum (Supplemental Fig. 2D), suggesting that 56% of the signal of overlapping cross peaks originated from [YLGD][YLGD], whereas 44% originated from [YLGD][α/εKLI]. We integrated the signals of the εK5 cross peak 1 and the overlapping cross peaks 3–5 and calculated the signal intensities originating from [YLGD][YLGD] only (Supplemental Table I). Taking the abundance of 15N signals into account, the 15N-enriched εK5 signal intensity of 2 × 109 should be accompanied by naturally abundant signal intensities from other amide groups within the ε ligation product [YLGD][εKLI] of ∼8 × 105 and the remaining signal of cross peaks 3–5 therefore originates from amide groups in the α ligation product [YLGD][αKLI]. The relative frequency of ε versus α ligation could thus be estimated at 10.8 ± 1.7% (average ± SD) (Supplemental Table I).
The relative frequency of ε ligation could also be derived from the 1H spectra of the ligation sample (Fig. 2F). In this spectrum, the triplet at 8.61 ppm originates from naturally abundant peak 4, which is present in all peptides, whereas the doublet at 8.49 ppm originates from naturally abundant peak 4A, which is only present in [YLGD][εKLI] (Supplemental Fig. 2C). From the integral of these resonances, we derived that [YLGD][εKLI] constituted 6% of total ligation products ([YLGD][εKLI], [YLGD][αKLI], and [YLGD][YLGD]) and estimated the relative frequency of ε ligation versus α ligation to be 14%. Both these frequencies correlate well with the LC-MS experiments described above, in which relative ε ligation efficiencies of 10% were observed. We therefore conclude that ε ligation occurs in ∼10% of all ligation events involving an N-terminal lysine residue at the site of ligation.
Isopeptides have enhanced stability toward proteasomal degradation
Isopeptide Ags formed by the proteasome may have unique properties compared with normal peptide Ags. In particular, such epitopes may be less susceptible to processing by proteases, as intramolecular isopeptide bonds have been described to provide proteolytic stability to pili on the surface of Gram-positive bacteria (33). To investigate whether isopeptides showed enhanced stability to proteasomal processing, we first studied two peptides, YLDWα/εKLISV and GILGFα/εKTL, in which cleavage of the (iso)peptide bond is the only proteasomal cleavage occurring. All peptides were incubated with proteasome for time periods up to 16 h, the degradation mixtures were analyzed by LC-MS, and the amounts of input peptide were plotted as the percentage of total ion count versus time (Fig. 3A). Both α-linked peptides (gray dotted lines) were cleaved readily with approximate half-lives of 2 and 6 h, respectively. Strikingly, both isopeptides (black solid lines) were more resistant toward proteasomal degradation compared with their peptide counterparts, with YLDWεKLISV being particularly stable and showing <5% cleavage within 16 h.
FIGURE 3.
Isopeptides show enhanced stability toward proteasomal degradation. (A) Degradation of GILGFα/εKTL and YLDWα/εKLISV at the α/ε bond (indicated by a dash) by the proteasome as a function of time. The amounts of input peptides and degradation products were quantified using LC-MS. (B) Proteasomal degradation patterns of the indicated (iso)peptides. The amounts of input peptides and degradation products after 16 h incubation with proteasome were quantified using LC-MS. Cleavage sites are indicated with upward arrows, α/ε bonds are indicated with downward arrows.
To study the stability of isopeptides that were cleaved at positions other than the isopeptide bond, GILGFVα/εKL, GILGα/εKFTL, GILα/εKVFTL, and GIα/εKFVFTL were incubated with proteasome for 16 h and the resulting digestion mixtures were analyzed with LC-MS to assess the extent as well as the site of peptide cleavage. The amounts of input peptides and cleavage products were plotted as the percentage of total ion count as a measure of total peptide content (Fig. 3B). All isopeptides that were cleaved C-terminally of the isopeptide linkage showed differences in terms of both the rate and the site of hydrolysis compared their α-linked counterparts. These differences between peptide and isopeptide processing were most apparent when the isopeptide linkage was located close to the main cleavage site. For example, the peptide GILGαKFTL was processed almost completely during the digestion and was cleaved predominantly at the KF bond, located next to the αK bond. In the same time frame, only 20% of GILGεKFTL was processed, and its main cleavage site was the FT bond located two residues away from the isopeptide linkage (Fig. 3B). Similar patterns were found for GILα/εKVFTL and GIα/εKFVFTL. Both α-linked peptides were completely processed within 16 h, whereas 50 and 10% of GILεKVFTL and GIεKFVFTL, respectively, remained intact. Both isopeptides were also preferentially cleaved between residues that were located farther away from the lysine residue compared with their normal counterparts (Fig. 3B). No differences were found between digestion mixtures of GILGFVαKL and GILGFVεKL that were cleaved N-terminally of the α/εK linkage (data not shown). Taken together, these data suggest that peptides containing an isopeptide linkage are less susceptible to proteasomal degradation. This holds true both when the isopeptide linkage is the preferred site of cleavage and when the isopeptide linkage is located close to the preferred cleavage site. Additionally, when proteasomal processing of isopeptides did occur, the actual cleavage site was located farther away from the εK residue compared with an αK residue.
Isopeptide Ags can bind to HLA class I molecules and are immunogenic in vivo
Finally, we set out to investigate whether isopeptide Ags are able to bind to HLA class I molecules and can be recognized by TCRs. To address the first issue, the affinity of a set of isopeptide bond–containing peptides for HLA-A*0201 and HLA-A*0301 was measured. Isopeptides were based on three known epitopes, GILGFVFTL (influenza A matrix protein 158–66) (34), NLVPMVATV (human CMV pp65495–503) (35), which both bind to HLA-A*0201, and SLPRGTSTPK (spliced together from SP110 nuclear body protein 296–301 and 286–289) (7), a HLA-A*0301 ligand. As the length of an Nε-linked lysine residue in a peptide backbone is twice that of an Nα-linked lysine residue (see for example Supplemental Fig. 2B), we replaced two adjacent amino acid residues at different positions in the epitope by one Nε-linked lysine. To ensure binding was not altered because of modification of the major HLA class I anchor residues, amino acids P2 and P9 (P10 in case of SLPRGTSTPK) were not modified. As a comparison, the affinities of GILG-based epitopes with identical sequences but a normal αK peptide bond were also determined. These peptides are 8 aa in length and are therefore expected to have low affinity for HLA-A*0201, which only binds peptides of 9–11 aa with high affinity.
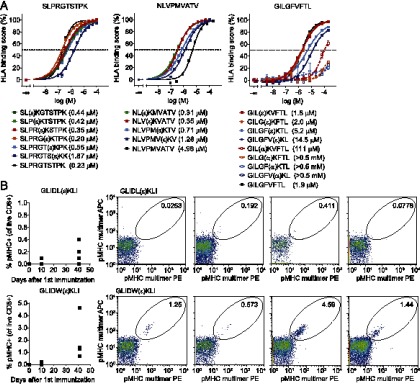
To study binding to HLA-A*0201 or HLA-A*0301, a UV-mediated MHC exchange FP assay was used (23, 24). In this assay HLA-A*0201 or HLA-A*0301 complexes loaded with a UV-cleavable peptide ligand are subjected to UV light, which cleaves the ligand, resulting in the generation of “empty” HLA-A*0201 or HLA-A*0301 (36). These empty HLA complexes quickly disintegrate, unless they are rescued by the addition of a rescue peptide. When rescue is performed by addition of both a fluorescent tracer peptide and increasing concentrations of a competitor peptide, the affinity of this competitor peptide can be measured. Binding curves of the NLVPMVATV-, SLPRGTSTPK-, and GILGFVFTL-based isopeptides were measured (Fig. 4A, solid lines), with the HLA binding score being defined as the percentage inhibition of tracer binding, and IC50 values for binding (i.e., an HLA binding score of 50) were determined after 4 h.
FIGURE 4.
Isopeptides bind HLA-A complexes and are immunogenic in vivo. (A) Binding curves of (iso)peptides based on the HLA-A*0301–specific epitope SLPRGTSTPK and the HLA-A*0201–restricted epitopes NLVPMVATV and GILGFVFTL. (α)K indicates a normally linked lysine residue; (ε)K indicates the location of the isopeptide linkage. IC50 values for binding are defined as an HLA binding score of 50% and are indicated between brackets. Error bars represent SD. (B) Plots showing the percentage of isopeptide–MHC multimer-reactive CD8+ T cells in the blood of HLA-A2 (HHD) transgenic mice (n = 4/group) that were vaccinated with the isopeptides GLIDLεKIL and GLIDWεKIL (day 0) and received a boost-vaccination at day 34. FACS plots show the percentage of isopeptide–MHC multimer reactive CD8+ T cells in individual mice at day 41.
The presence of an isopeptide linkage did not abrogate HLA-A*0201 or HLA-A*0301 binding; all peptides were able to bind to HLA-A*0201 or HLA-A*0301 complexes with similar affinities as the parent peptides (0.2–5 μM as measured in similar assays). The isopeptide linkage was well tolerated in the middle or toward the N terminus of the epitope without loss of HLA class I affinity (Fig. 4A, red/orange and green lines, respectively). Alternatively, when the isopeptide linkage was located toward the C-terminal side of the epitope, affinity decreased. As expected, the binding affinities of GILG-based peptides containing normal αK peptide bonds decreased by a factor 40–200 compared with their isopeptide counterparts (Fig. 4A, dotted lines). Taken together, these data suggest that isopeptides of various lengths can be presented at the cell surface by HLA class I alleles. Additionally, the presence of an isopeptide linkage enables 8-mer isopeptides to bind HLA complexes with a much increased affinity as compared with normal 8-mer peptides.
Having established that isopeptides can bind to HLA complexes with high affinity, we next investigated whether isopeptides are also immunogenic in vivo. To this end, HLA-A2 (HHD) transgenic mice (n = 4/group) that express a chimeric HLA-A*0201/H2-Dd MHC class I complex were vaccinated twice with the isopeptides GLIDLεKIL and GLIDWεKIL. Blood samples were taken before (boost) vaccination and at days 6–13 after vaccination, when peak T cell responses were expected, and stained with fluorescent MHC class I tetramers loaded with the isopeptide Ags of interest. As shown in Fig. 4B, GLIDWεKIL-reactive T cells could be observed in the blood of two of four vaccinated animals after the primary vaccination. Additionally, responses against both GLIDLεKIL (two of four) and GLIDWεKIL (four of four) could be measured after boost vaccination. HLA-A*0201 FP binding assays of GLIDLεKIL and GLIDWεKIL showed IC50 values of 14 ± 0.94 and 3.0 ± 0.23 μM (average ± SE), respectively, whereas α-linked GLIDLKIL and GLIDWKIL bound to HLA-A*0201 with much lower affinity (139 ± 9.6 and 81 ± 5.5 μM, respectively). Additionally, the binding affinity of GLIDLKIL and GLIDWKIL decreased almost 7-fold in 24 h, whereas binding of their isopeptide counterparts only decreased 2- to 3-fold (data not shown). Taken together, these data suggest that 8-mer α-linked peptides bind with very low affinity and high off-rates, indicating that stable peptide–MHC complexes cannot be formed between these 8-mers and HLA-A2*0201. Consequently, it is highly unlikely that T cells that are specific for peptides with an isopeptide bond can also be stimulated by a corresponding peptide containing α linkages only. From these data we conclude that isopeptides are immunogenic in vivo, and that isopeptide-specific T cells may exist in humans in vivo.
Discussion
Proteasomal splicing reactions in vivo can create a novel type of Ag, in which two peptide fragments are posttranslationally fused together via a transpeptidation mechanism (5–9, 13). During proteasomal transpeptidation reactions, a protein amide backbone linkage is attacked by one of the proteasomal catalytic N-terminal threonine residues, resulting in the formation of an O-acyl enzyme intermediate and the release of the C-terminal part of the protein. Subsequently, this intermediate ester reacts with an amino group, usually the N terminus, from a second peptide (the C-terminal ligation partner), resulting in the formation of a novel peptide linkage and a spliced product (5, 7). This transpeptidation reaction can apparently compete with normal hydrolysis, in which water reacts with the intermediate ester (12), in such a way that sufficiently new peptide can be formed to invoke an immune response against transpeptidation products. In an accompanying article in this issue by Berkers et al. (16), we show that transpeptidation reactions can efficiently compete with hydrolysis when specific structural requirements on the two ligating partners are met. Ligation occurs particularly efficiently when the C-terminal ligation partner has a basic amino acid residue (lysine or arginine) at its N terminus (16). As lysine has two amino groups that can theoretically both react with the O-acyl enzyme intermediate, this implies that the proteasome may be able to form isopeptide linkages.
In the current study, we show, to our knowledge for the first time, that the proteasome can use both the α- and the ε-amino group of a lysine residue in aminolysis reactions. This suggests that in addition to forming spliced peptides, the proteasome can also create spliced isopeptides to form a novel type of Ag, which contains a posttranslational modification that has not been described for MHC class I Ags before. When single lysine residues are ligated onto a precursor peptide, both amino groups are equally reactive and capable of participating in ligation reactions. Surprisingly, when ligation of lysine-containing C-terminal fragments was monitored by both LC-MS and NMR, ε ligation was also found to occur, albeit at a 10-fold lower frequency as compared with α ligation. Additionally, ε ligation only occurred in the β5 and not in the β2 active site. C-terminal fragments presumably bind to the specificity pockets of a distinct proteasomal splicing product binding site (14). Likely, a single lysine residue is capable of fitting into the specificity pockets of both the β2 and β5 active site in multiple orientations, thereby facilitating both α and ε ligation in both sites. A peptide, alternatively, will adopt an orientation in which multiple side chains fit multiple specificity pockets. This may favor orientation of the lysine residue in such a way that only the α-amino group is positioned optimally for a nucleophilic attack on the intermediate ester bond, resulting in normal peptide bond formation and disfavoring alternative ε ligation and isopeptide bond formation, especially in the β2 active site.
The data presented in the present study also show that isopeptides can bind different HLA alleles with high affinity and have unique properties that discern them from normal epitopes. The presence of an isopeptide linkage allows peptides that are comprised of only 8 aa to bind to HLA-A2 or HLA-A3, whereas normally only peptides of 9–11 residues in length bind to these HLA alleles with high affinity. In such isopeptide epitopes the isopeptide linkage is tolerated in the middle and toward the N terminus of different epitopes, but loss of HLA-A2 or HLA-A3 affinity is observed when the isopeptide linkage is located toward the C terminus of an epitope. Importantly, isopeptides are more stable toward further proteasomal processing than are normal peptides. Proteasomal splicing reactions can be followed by further proteasomal trimming (9). This may aid in the production of peptides with the right HLA-binding properties, but it can also destroy potential epitopes. Degradation of isopeptides, however, is slow compared with common peptides, when the isopeptide bond is located at or close to the preferred cleavage site. Additionally, the proteasome cleaves most isopeptides at a site that is located farther away from the lysine residue compared with normal peptides. This results in Ags that retain their isopeptide linkage at a location (toward the middle of the peptide) that does not affect HLA-A2 or HLA-A3 affinity. Based on the observation that isopeptides are more stable toward proteasomal degradation, we consider it likely that the isopeptide linkage also hampers hydrolysis by other (cytosolic) proteases. We therefore anticipate that there is an increased chance that an isopeptide Ag stays intact and can be efficiently loaded onto MHC, once it is formed, allowing it to reach the cell surface for CD8+ T cell surveillance.
The high diversity of the TCR repertoire that is created by V(D)J recombination is essential for protection from a constantly changing bacterial and viral repertoire (11). Similarly, proteasomal splicing may increase the diversity of the peptide repertoire that is available for presentation by MHC class I and recognition by CD8+ T cells, ultimately resulting in a more effective elimination of infected or malignant cells (3, 10–12). The data presented in the present study indicate that isopeptides are immunogenic in vivo, suggesting that proteasomal εK ligation reactions may further increase the antigenic space that can be seen by T cells. Splicing efficiencies in vivo have been estimated to range from 0.0002 to 0.01%, depending on the epitope (5, 13), suggesting that even very low splicing efficiencies lead to sufficient amounts of peptide for detection by CD8+ T cells. In an accompanying paper (16), we show that in vivo splicing and cell surface presentation of splicing-prone fragments can occur with up to 3% efficiency compared with a peptide that does not require any processing, suggesting that spliced epitopes may be presented on the surface of cells to a much larger extent than previously assumed. Thus, although the frequency of εK ligation reactions is 10-fold lower as compared with α ligation, ε ligation can be expected to produce sufficient amounts of peptide to evoke a T cell response, especially when taken into account that a large fraction of spliced isopeptide epitopes may be able to reach the cell surface as a result of their increased proteolytic stability.
In conclusion, although the ε linkage has not been observed before in MHC epitopes, which is easily explained by its difficult (and so far unanticipated) detection, we hypothesize that isopeptide epitopes exist based on our in vitro experiments. With evidence of proteolytic stability and therefore accumulation of this modification, we postulate K-ε ligation as a genuine posttranslational modification resulting from transpeptidation mechanisms. We consider it likely that our in vitro findings will be validated in vivo soon.
Supplementary Material
Acknowledgments
We thank Olaf van Tellingen and Dorothé Linders for help with MALDI measurements, Henk Hilkmann for peptide synthesis, and Jacques Neefjes for help with proteasome purification.
C.R.B., T.N.M.S., and H.O. are members of the Institute for Chemical Immunology.
This work was supported by Netherlands Organization for Scientific Research Grant 819.02.003 (to H.O.).
The online version of this article contains supplemental material.
- ACN
- acetonitrile
- FP
- fluorescence polarization
- HMQC
- heteronuclear multiple quantum coherence
- HSQC
- heteronuclear single quantum coherence
- LC-MS
- liquid chromatography–mass spectrometry
- Kα(Ac)
- Nα-acetylated lysine
- Kε(Ac)
- Nε-acetylated lysine
- NMR
- nuclear magnetic resonance
- TFA
- trifluoroacetic acid
- TMR
- tetramethylrhodamine.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Schmidt M., Finley D. 2014. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843: 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sijts E. J., Kloetzel P. M. 2011. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. 68: 1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigneron N., Van den Eynde B. J. 2012. Proteasome subtypes and the processing of tumor antigens: increasing antigenic diversity. Curr. Opin. Immunol. 24: 84–91. [DOI] [PubMed] [Google Scholar]
- 4.Basler M., Kirk C. J., Groettrup M. 2013. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 25: 74–80. [DOI] [PubMed] [Google Scholar]
- 5.Vigneron N., Stroobant V., Chapiro J., Ooms A., Degiovanni G., Morel S., van der Bruggen P., Boon T., Van den Eynde B. J. 2004. An antigenic peptide produced by peptide splicing in the proteasome. Science 304: 587–590. [DOI] [PubMed] [Google Scholar]
- 6.Hanada K., Yewdell J. W., Yang J. C. 2004. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427: 252–256. [DOI] [PubMed] [Google Scholar]
- 7.Warren E. H., Vigneron N. J., Gavin M. A., Coulie P. G., Stroobant V., Dalet A., Tykodi S. S., Xuereb S. M., Mito J. K., Riddell S. R., Van den Eynde B. J. 2006. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313: 1444–1447. [DOI] [PubMed] [Google Scholar]
- 8.Dalet A., Robbins P. F., Stroobant V., Vigneron N., Li Y. F., El-Gamil M., Hanada K., Yang J. C., Rosenberg S. A., Van den Eynde B. J. 2011. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. USA 108: E323–E331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaux A., Larrieu P., Stroobant V., Fonteneau J. F., Jotereau F., Van den Eynde B. J., Moreau-Aubry A., Vigneron N. 2014. A spliced antigenic peptide comprising a single spliced amino acid is produced in the proteasome by reverse splicing of a longer peptide fragment followed by trimming. J. Immunol. 192: 1962–1971. [DOI] [PubMed] [Google Scholar]
- 10.Borissenko L., Groll M. 2007. Diversity of proteasomal missions: fine tuning of the immune response. Biol. Chem. 388: 947–955. [DOI] [PubMed] [Google Scholar]
- 11.Shastri N. 2006. Cell biology. Peptides, scrambled and stitched. Science 313: 1398–1399. [DOI] [PubMed] [Google Scholar]
- 12.Berkers C. R., de Jong A., Ovaa H., Rodenko B. 2009. Transpeptidation and reverse proteolysis and their consequences for immunity. Int. J. Biochem. Cell Biol. 41: 66–71. [DOI] [PubMed] [Google Scholar]
- 13.Dalet A., Vigneron N., Stroobant V., Hanada K., Van den Eynde B. J. 2010. Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor-5. J. Immunol. 184: 3016–3024. [DOI] [PubMed] [Google Scholar]
- 14.Mishto M., Goede A., Taube K. T., Keller C., Janek K., Henklein P., Niewienda A., Kloss A., Gohlke S., Dahlmann B., et al. 2012. Driving forces of proteasome-catalyzed peptide splicing in yeast and humans. Mol. Cell. Proteomics 11: 1008–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liepe J., Mishto M., Textoris-Taube K., Janek K., Keller C., Henklein P., Kloetzel P. M., Zaikin A. 2010. The 20S proteasome splicing activity discovered by SpliceMet. PLOS Comput. Biol. 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkers C. R., de Jong A., Schuurman K. G., Linnemann C., Meiring H. D., Janssen L., Neefjes J. J., Schumacher T. N. M., Rodenko B., Ovaa H. 2015. Definition of proteasomal peptide splicing rules for high efficiency spliced peptide presentation by MHC class I molecules. J. Immunol. 195: 4085–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiejak S., Masiukiewicz E., Rzeszotarska B. 1999. A large scale synthesis of mono- and di-urethane derivatives of lysine. Chem. Pharm. Bull. (Tokyo) 47: 1489–1490. [DOI] [PubMed] [Google Scholar]
- 18.Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. 2008. Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkers C. R., van Leeuwen F. W., Groothuis T. A., Peperzak V., van Tilburg E. W., Borst J., Neefjes J. J., Ovaa H. 2007. Profiling proteasome activity in tissue with fluorescent probes. Mol. Pharm. 4: 739–748. [DOI] [PubMed] [Google Scholar]
- 20.de Jong A., Schuurman K. G., Rodenko B., Ovaa H., Berkers C. R. 2012. Fluorescence-based proteasome activity profiling. Methods Mol. Biol. 803: 183–204. [DOI] [PubMed] [Google Scholar]
- 21.Boyd J., Soffe N., John B., Plant D., Hurd R. 1992. The generation of phase-sensitive 2D N-15-H-1 spectra using gradient pulses for coherence-transfer-pathway selection. J. Magn. Reson. 98: 660–664. [Google Scholar]
- 22.Davis A., Keeler J., Laue E., Moskau D. 1992. Experiments for recording pure-absorption heteronuclear correlation spectra using pulsed field gradients. J. Magn. Reson. 98: 207–216. [Google Scholar]
- 23.Rodenko B., Toebes M., Celie P. H., Perrakis A., Schumacher T. N., Ovaa H. 2009. Class I major histocompatibility complexes loaded by a periodate trigger. J. Am. Chem. Soc. 131: 12305–12313. [DOI] [PubMed] [Google Scholar]
- 24.Bakker A. H., Hoppes R., Linnemann C., Toebes M., Rodenko B., Berkers C. R., Hadrup S. R., van Esch W. J., Heemskerk M. H., Ovaa H., Schumacher T. N. 2008. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl. Acad. Sci. USA 105: 3825–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toebes, M., B. Rodenko, H. Ovaa, and T. N. Schumacher. 2009. Generation of peptide MHC class I monomers and multimers through ligand exchange. Curr. Protoc. Immunol. Chapter 18: Unit 18.16. doi:10.1002/0471142735.im1816s87. [DOI] [PubMed] [Google Scholar]
- 26.Hadrup S. R., Bakker A. H., Shu C. J., Andersen R. S., van Veluw J., Hombrink P., Castermans E., Thor Straten P., Blank C., Haanen J. B., et al. 2009. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat. Methods 6: 520–526. [DOI] [PubMed] [Google Scholar]
- 27.Lightcap E. S., McCormack T. A., Pien C. S., Chau V., Adams J., Elliott P. J. 2000. Proteasome inhibition measurements: clinical application. Clin. Chem. 46: 673–683. [PubMed] [Google Scholar]
- 28.Chauhan D., Catley L., Li G., Podar K., Hideshima T., Velankar M., Mitsiades C., Mitsiades N., Yasui H., Letai A., et al. 2005. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from bortezomib. Cancer Cell 8: 407–419. [DOI] [PubMed] [Google Scholar]
- 29.Berkers C. R., Leestemaker Y., Schuurman K. G., Ruggeri B., Jones-Bolin S., Williams M., Ovaa H. 2012. Probing the specificity and activity profiles of the proteasome inhibitors bortezomib and delanzomib. Mol. Pharm. 9: 1126–1135. [DOI] [PubMed] [Google Scholar]
- 30.Marek R., Lycka A. 2002. 15N NMR spectroscopy in structural analysis. Curr. Org. Chem. 6: 35–66. [Google Scholar]
- 31.Iwahara J., Jung Y. S., Clore G. M. 2007. Heteronuclear NMR spectroscopy for lysine NH3 groups in proteins: unique effect of water exchange on 15N transverse relaxation. J. Am. Chem. Soc. 129: 2971–2980. [DOI] [PubMed] [Google Scholar]
- 32.Tjandra N., Grzesiek S., Bax A. 1996. Magnetic field dependence of nitrogen-proton J splittings in 15N-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 118: 6264–6272. [Google Scholar]
- 33.Kang H. J., Coulibaly F., Clow F., Proft T., Baker E. N. 2007. Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science 318: 1625–1628. [DOI] [PubMed] [Google Scholar]
- 34.Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. 1991. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351: 290–296. [DOI] [PubMed] [Google Scholar]
- 35.Solache A., Morgan C. L., Dodi A. I., Morte C., Scott I., Baboonian C., Zal B., Goldman J., Grundy J. E., Madrigal J. A. 1999. Identification of three HLA-A*0201-restricted cytotoxic T cell epitopes in the cytomegalovirus protein pp65 that are conserved between eight strains of the virus. J. Immunol. 163: 5512–5518. [PubMed] [Google Scholar]
- 36.Toebes M., Coccoris M., Bins A., Rodenko B., Gomez R., Nieuwkoop N. J., van de Kasteele W., Rimmelzwaan G. F., Haanen J. B., Ovaa H., Schumacher T. N. 2006. Design and use of conditional MHC class I ligands. Nat. Med. 12: 246–251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.