Abstract
Peptide splicing, in which two distant parts of a protein are excised and then ligated to form a novel peptide, can generate unique MHC class I–restricted responses. Because these peptides are not genetically encoded and the rules behind proteasomal splicing are unknown, it is difficult to predict these spliced Ags. In the current study, small libraries of short peptides were used to identify amino acid sequences that affect the efficiency of this transpeptidation process. We observed that splicing does not occur at random, neither in terms of the amino acid sequences nor through random splicing of peptides from different sources. In contrast, splicing followed distinct rules that we deduced and validated both in vitro and in cells. Peptide ligation was quantified using a model peptide and demonstrated to occur with up to 30% ligation efficiency in vitro, provided that optimal structural requirements for ligation were met by both ligating partners. In addition, many splicing products could be formed from a single protein. Our splicing rules will facilitate prediction and detection of new spliced Ags to expand the peptidome presented by MHC class I Ags.
Introduction
Following malignant transformation or viral infection a broad repertoire of antigenic peptides is presented on the cell surface by MHC class I complexes, enabling CD8+ T cells to sense alterations in intracellular protein content including new Ags that then instruct the T cells for eliminating the APC (1). The 26S proteasome, a large threonine protease complex, is largely responsible for the generation of these antigenic peptides in vivo (2, 3). The 26S proteasome consists of a 20S core, in which the catalytic activity resides, and 19S regulatory caps, which regulate unfolding and entry of substrates into the 20S core. The 20S core is formed by four stacked rings consisting of seven subunits each, with an overall architecture of α (1–7)β (1–7)β (1–7)α (1–7). Catalytic activity is provided by three β subunits, termed β1, β2, and β5, which are responsible for the caspase-like, tryptic, and chymotryptic activities of the proteasome, respectively. In lymphoid tissues or under the influence of the cytokine IFN-γ, these subunits are replaced by their immunoproteasomal counterparts, termed β1i, β2i, and β5i, to form the immunoproteasome (4).
The protease trypsin has long been known to also reverse cleavage resulting in ligation of two peptide sequences by transpeptidation under the correct conditions. This reversed cleavage has also been shown for other proteases including the proteasome that then would yield new spliced Ags that are not genetically coded yet can be presented to the immune system (5–9). To date, five spliced Ags that are immunogenic in vivo have been described: [RTK][QLYPEW] (5), [NTYAS][PRFK] (6), [SLPRGT][STPK] (7), [IYMDGT][ADFSF] (8), and [RSYVPLAH][R] (9), all of which are produced by the proteasome through a transpeptidation mechanism (5, 7, 10, 11). During normal proteasomal peptide hydrolysis, the N-terminal threonine residue of a catalytically active subunit reacts with the scissile peptide bond to form an O-acyl enzyme intermediate, resulting in the release of the C-terminal part of the peptide. In a second step, water reacts with the intermediate ester, resulting in the release of the N-terminal part of the peptide. During a proteasomal transpeptidation reaction, in contrast, the amino group of a second peptide competes with water for reaction with the intermediate ester, resulting in the formation of a new peptide bond and thus a spliced peptide. Splicing of ligation partners in the proteasome has been described to occur both in-line [i.e., in the order in which they occurred in the parent protein sequence (5, 6)] and in the reverse order [i.e., a posterior sequence is spliced to the anterior end of its neighboring sequence (7–9)]. In addition, peptide splicing by the proteasome can be combined with other posttranslational processes, such as asparagine deamidation (8), and can be followed by further trimming to produce peptides of the right HLA binding properties (9).
Proteasomal splicing thus results in a genuine posttranslational modification, which can increase the diversity of the antigenic peptides presented but which may also serve other yet unknown functions. A greater diversity of epitopes increases the chance that one or more of these epitopes are recognized by CD8+ T cells, which ultimately may result in more efficient elimination of infected or malignant cells (12–14). Because spliced Ags may mediate T cell responses to cancer, transplants, and viral infection, predicting their generation would be important for the development of vaccines and immunotherapies or to pharmacologically suppress T cell responses (12). Although proteasomal peptide ligation can readily be detected in vitro (11, 15), identifying spliced Ags in vivo has proven to be difficult with only five immunomodulatory spliced epitopes identified (5–9). Classically, novel epitopes are identified by cell surface elution and mass spectroscopy (MS) analysis, followed by matching against protein databases (16, 17). Alternatively, epitopes may be predicted from protein sequences (18, 19) and tested for TCR recognition, for example, by incorporation into fluorescent MHC class I tetramers, which can subsequently be used to stain T cells in (FACS) assays (20, 21). However, because spliced Ags are not contiguous, they neither match existing databases nor can they be predicted from contiguous protein sequences. Knowledge of the rules—if any—that govern the production of noncontiguous Ags would facilitate their prediction and identification, but it is currently not known whether proteasome splicing is determined by explicit rules or whether it is a random event (5, 10, 11).
In this study, we use short peptides to identify amino acid sequences that are likely to promote transpeptidation reactions, enabling us for the first time, to our knowledge, to deduce splicing rules that we validate in vitro and in cells. We quantify peptide ligation of splicing-prone ligation partners to demonstrate that splicing can occur efficiently in vitro and in cells. The elucidation of these splicing rules will facilitate the prediction and detection of spliced Ags to study their roles in immunity.
Materials and Methods
Peptide building blocks were purchased from Novabiochem and appropriately functionalized resins from Applied Biosystems. 15N-Glycine-N-Fmoc was purchased from Cambridge Isotope Laboratories. All solvents were purchased from Biosolve at the highest grade available. All other chemicals were purchased from Aldrich at the highest available purity. All solvents and chemicals were used as received. HPLC purifications and analyses were performed on a Shimadzu LC-20AT prominence liquid chromatography system, coupled to a Shimadzu SPD-20A prominence UV/Vis detector and a Shimadzu CTO-20A prominence column oven. Liquid chromatography (LC)/MS analyses of in vitro ligation assays were carried out on a Waters LCT mass spectrometer in line with a Waters 2795 HPLC system and a Waters 2996 photodiode array detector, using an XBridge BEH300 C18 column (3.5 μm; 2.1 × 100 mm; Waters) with a linear gradient (5–50% acetonitrile in H2O containing 0.1% formic acid).
Peptide synthesis
Peptides were synthesized using standard Fmoc-based solid-phase peptide synthesis protocols and appropriately functionalized polyethylene glycol–polystyrene Wang resins. Functionalized resins were subjected to coupling cycles, in which deprotection of the Fmoc group with piperidine/NMP (1/4 v/v) was followed by coupling with four equivalents each of Fmoc protected amino acid, di-isopropyl ethylamine and (benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate (PyBop). Reactions were carried out in NMP at a volume of 1 ml/0.1 g of resin. After the final coupling step, the Fmoc group was removed, and peptides were released from the resin and fully deprotected by treating the resin with trifluoroacetic acid (TFA)/H2O/triisopropylsilane (93/5/2 v/v) for 2.5 h. Peptides were precipitated with cold diethylether/pentane (3/1 v/v) and lyophilized from H2O/acetonitrile/acetic acid (65/25/10 v/v). Peptides were purified by HPLC over an Atlantis Preparative T3 column (5 μm; 20 × 150 mm; Waters) to >99.5% purity using a linear gradient (5–50% ACN in H2O containing 0.05% TFA). Peptide-containing fractions were pooled and lyophilized.
Cell culture
FLYRD18 is a human fibrosarcoma retroviral packaging cell line (European Cell Culture Collection number 95091902). JY cell line is an EBV-immortalized, HLA-A2,-B7 homozygous positive B cell lymphoblastoid line. Both cell lines were cultured in IMDM (Invitrogen Life Technologies), 5% FBS (Greiner Bio-one), and penicillin/streptomycin (Roche Diagnostics). The human melanoma MelJuso-A2 cell line (22) was cultured in DMEM, 10% FBS, and penicillin/streptomycin.
Proteasome purification
Proteasome was purified from bovine liver as described previously (23). After each step, proteasome purity was monitored by incubating fractions with a fluorescent proteasome activity probe, followed by SDS-PAGE and scanning of the resulting gel for fluorescence emission as described previously (24, 25). Briefly, bovine liver was homogenized in PBS, followed by precipitation using 40% saturated ammonium sulfate as a first precipitation step. The proteasome was subsequently precipitated by increasing the concentration of saturated ammonium sulfate to 60%. Following dialysis, the proteasome was further purified using a 10–40% sucrose gradient and anion exchange column chromatography, using DEAE Sephadex A25 resin. Proteasome containing fractions were pooled and concentrated, and protein concentrations were determined using the Bradford assay (Bio-Rad). The quality of the isolation was further confirmed by SDS-PAGE and Coomassie staining.
In vitro ligation assays
All ligation reactions were performed in ligation buffer (50 mM Tris-HCl [pH 8.5]) at 37°C. YLDWSY (0.67 mM) was incubated with a 5-fold molar excess of XLPSV, KXPSV, or KLXSV (where X is any proteogenic amino acid) and proteasome (0.067 mg/ml) for 16 h; control samples contained YLDWSY and proteasome only. YLGXSY and YLXLSY (0.67 mM) were incubated with an equimolar amount of RLPSV and proteasome (0.067 mg/ml) for various time periods up to 24 h. YLGDSYRLPSV (0.67, 1.3, or 2.7 mM was incubated with proteasome (0.067, 0.13, and 0.27 mg/ml, respectively) for various time periods up to 8 h. YLGDSYRLGSV (0.67 mM) and YL15NGDSYRL15NGSV (0.67 mM or a 1:1 mixture of both peptides (each at 0.33 mM) were incubated with 0.067 mg/ml proteasome for 2 h. Various amounts of synthetic GSSFTIMTDGPSD-GLASYKIFKIEKGKV (N1.1; 100–400 nmol, 1.3–5.3 mM), 2 mM 15NGSSFTIMTDGPSD15NGLASY-KIFKIEKGKV (15N-N1.1), or a 1:1 mixture of both peptides (both at 1 mM) were incubated with proteasome for various time periods up to 24 h. For fluorescence polarization (FP) analysis, ligation mixtures were snap-frozen in liquid N2, lyophilized, dissolved in 50 μl DMSO, and the volume was adjusted to 1 ml with H2O. Samples were purified over Strata-X 33 μm polymeric reversed-phase disposable columns (Phenomenex). To ensure equal amounts of non–HLA-A2–binding peptides in all samples XLPSV, KXPSV, or KLXSV was added to control samples prior to purification. Ligation products were eluted with ACN/H2O (2/3 v/v), lyophilized, and dissolved in FP buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5 mg/ml bovine gammaglobulins). For LC-MS and HPLC analysis, ligation mixtures were snap-frozen in liquid N2, lyophilized, and dissolved in ACN/H2O (1/1 v/v). Samples were filtered over Strata-X 33-μm polymeric reversed-phase disposable columns (Phenomenex) to remove proteasomal proteins and eluted directly.
FP assays
FP HLA-A*0201 binding assays with UV-mediated peptide exchange were performed as described previously (26) with minor changes. Assays were performed in 384-well low-volume black nonbinding surface assay plates (Corning 3820). Wells were loaded with 30 μl total volume containing 0.5 μM HLA-A*0201-peptide complex, 1 nM tracer peptide, and competitor sample/peptide in FP buffer. The peptide KILGFVFJ1V (in which J1 is UV-cleavable 3-amino-3-(2-nitrophenyl)propionic acid) was used as a conditional ligand and fluorescently labeled FLPSDC(TMR)FPSV (in which TMR is maleimido-linked tetramethylrhodamine) was used as a tracer peptide. To screen ligation preferences of the primed sites, in vitro ligation mixtures were used as competitor samples, and binding curves of serial 2-fold dilutions of the expected ligation products were determined as a reference. To determine the affinities of N1.1 spliced peptides for HLA-A*0201, 3-fold serial dilutions of synthetic products were used as competitor peptide. The plate was spun for 1 min at 1000 × g at room temperature to ensure proper mixing of all components. To start UV-mediated cleavage of the conditional ligand and peptide exchange the plate was placed under a 365-nm UV lamp at 10 cm distance (366-nm UV lamp, 2 × 15 W blacklight blue tubes, l × w × h = 505 × 140 × 117 mm; Uvitec) located in a cold room (4°C). After 30 min irradiation, the plate was sealed with thermowell sealing tape (Corning) and incubated at room temperature for 4 or 24 h, allowing cleaved peptide fragments to dissociate and to be exchanged for competitor and/or tracer peptide. Subsequently, FP measurements were performed as described previoiusly (27). Data were analyzed using GraphPad Prism software (GraphPad). The relative ligation efficiency of ligation mixtures was calculated by comparing each sample to the corresponding control sample and ligation efficiencies were normalized to the efficiency of [YLDW][KLPSV] formation, which was included in all experimental runs. The HLA binding score of each N1-1 spliced product was defined as the percentage inhibition of tracer peptide binding and the binding affinity (IC50) as an HLA binding score of 50%.
Splicing assays in cells
Peptide sequences YLGDSYRLPSV and YLWGRPLSV were translated into codon-optimized minigenes using Gene Designer 2.0 (DNA2.0) to obtain minigenes, which were cloned into the pMX retroviral vector to obtain a pMX-minigene-IRES-GFP vector which has been used previously (28). FLYRD18 packaging cells were plated in 10-cm plates at 1.2 × 106 cells/plate. One day after seeding, cells were transfected with 10 μg retroviral vector DNA using FuGENE TM6 (Roche Diagnostics). Cell medium was refreshed 24 h later. Retroviral supernatant was collected 48 h after transfection. JY cells were resuspended in retroviral supernatant, transferred to 24-well plates coated with RetroNectin (Takara) at 0.1 × 106 cells/ml, and centrifuged for 90 min at 880× g. Transduced cells were transferred back into culture medium 24 h after transduction. Cells were expanded and 1 × 108 cells per sample were frozen for peptide elutions. In case of a frequency lower than 90% of GFP+ cells within the living cells, flow cytometry sorting was performed prior to freezing. mYFP Ubiquitin YLGNSYRLPSV was generated by PCR on a ubiquitin template with the following primers 5′-CCCATGATCAATGCAGATCTTCGTGAAGAC-3′ (forward) and 5′-cccaGAATTCTCACACGCTGGGCAGTCTGTAGCTGTTGCCCAGGTACCCACCTCTGAGACGGAGCA-3′ (reverse). The amplified product was digested with BclI and EcoRI and cloned into the BglII and EcoRI sites of mYFP-C1 vector (Neefjes Lab). The construct was sequence verified. MelJuso-A2-GFP cells were transfected using calcium phosphate, expanded, and FACS sorted for cells coexpressing GFP and YFP. Data were acquired and analyzed on a FACSCalibur (BD Biosciences) with FlowJo software (Tree Star). Cell sorts were carried out using FACSAria (BD Biosciences) or Moflow Highspeed Sorter (Beckmann Coulter). Peptides were eluted from transfected JY and MelJuso cells as described previously (29). In short, cells were lysed in lysis buffer (50 mM Tris·HCl [pH 8], 0.3 M NaCl, 5 mM Na2EDTA·2 H2O, 5 mM MgCl2·6 H2O, and 1% CHAPS) containing protease inhibitors (20 mM iodoacetamide, 1 μg/μl pepstatin A, 20 μM MG132, inhibitor mixture (Roche) by shaking for 4 h at 4°C. Lysates were centrifuged for 10 min at 1,000 × g, followed by 1 h at 12,000 × g and the supernatant was filtered over a 0.45-μm filter. Supernatants were precleared with CNBr Sepharose 4B capped beads and CNBr Sepharose 4B 12CA5 (anti-hemagglutinin) beads, before being incubated with Sepharose W6/32 (anti-MHC class I beads to immunoprecipitate MHC class I–peptide complexes. The Sepharose W6/32 beads were washed thoroughly and the complexes were eluted and dissociated with 10% acetic acid, before being incubated for 15 min at 70°C. Samples were freeze-dried prior to further LC-MS/MS analysis.
LC-MS/MS analysis of splicing reactions in cells
Samples were dried and reconstituted in water + 0.1% (v/v) TFA, also containing two test peptides, each at a concentration level of 5 pmol/μl and injected onto the strong cation exchange (SCX) system. In this system, peptides were initially trapped onto a Hypercarb trapping column (5 mm × 200 μm) for 4 min, which was subsequently switched in-line with the analytical SCX column (PolySULFOETHYL Aspartamide, 13 cm × 200 μm). Peptides were transferred from the trapping column onto the SCX column with a 10 μl plug of water/ACN (1:1, v/v) + 0.5% acetic acid and eluted using a KCl salt/ACN gradient. Fractions were collected based on the retention behavior of the two test peptides and synthetic standards of the expected ligation products. The collected fractions were dried and reconstituted in water + 5% (v/v) DMSO + 0.1% (v/v) formic acid and also containing the test peptide coded S081-04 (amino acid sequence SGYSGIFSVEGK) at a concentration level of 1 fmol/μl. An aliquot of each fraction was subjected to nanoscale LC-MS analysis (30), comprising a home-built nanoscale LC system coupled to an LTQ Orbitrap XL system (Thermo Scientific). The nanoscale LC system contained a Reprosil-Pur C18-AQ 5 μm 120 Å trapping column (20 mm length × 50 μm I.D.) and a Reprosil-Pur C18-AQ 3 μm 120 Å analytical column (25.4 cm length × 25 μm I.D.). Samples were separated using a linear gradient (7.5–47.5% ACN in H2O + 0.1% [v/v] formic acid at 2%/min) at a flow rate of 30 nL/min. Analytes were introduced into the mass spectrometer using a nanoelectrospray interface comprising a gold-carbon–coated fused silica tip (25 μm I.D.), manually tapered to a tip I.D. of 3.5 μm. An electrospray voltage of 1.8 kV was applied to the spray tip. The mass spectrometric detection of the peptides was performed with a LTQ Orbitrap XL instrument (Thermo Scientific). Mass spectra were acquired at a mass resolution of 60,000 full width at half maximum with internal lock mass calibration at m/z 391.28428 Da. The acquisition of CID–fragmentation spectra was performed in the ion trap and only triggered on the presence of the doubly charged ions of the expected ligation products and the test peptide S081-04 within a mass accuracy of ± 3 ppm.
Results
Distinct peptide motifs either promote or abolish Ag splicing
In principle, two types of fragments participate in proteasomal ligation reactions: N-terminal precursors that fit the nonprimed proteasomal substrate binding pockets, called S1, S2, S3…Sn depending on their proximity to the scissile peptide bond, and C-terminal ligation partners that may fit the primed proteasomal pockets S1′, S2′, S3′…Sn′, but have also been postulated to bind to a distinct proteasomal splicing product binding site (11). Amino acid residues in both ligation partners that interact with these proteasomal substrate binding pockets are here referred to as P1, P2, P3…Pn and P1′, P2′, P3′…Pn′, accordingly (Fig. 1A). For both types of ligation partners, rules predicting proteasomal peptide ligation were deduced by comparing small libraries of potential ligation partners in ligation assays.
FIGURE 1.
Splicing rules predict splicing-prone peptide sequences that can participate in highly efficient ligation reactions. (A) Summary of the splicing rules determined for the C-terminal fragment (gray circles, P1′, P2′, and P3′ residues) and the N-terminal precursor (black circles, P1, P2 residues). Sx′/Sx = proteasomal substrate binding pockets. Φ = hydrophobic residue. (B) Relative ligation efficiencies of different C-terminal ligation fragments measured in fluorescence polarization assays, indicating splicing preferences for the P1′, P2′, and P3′ positions. Efficiencies were normalized to the efficiency of [YLDW][KLPSV] formation, which was included in all experiments. Values are averaged over at least two independent experiments; error bars represent SD. (C and D) Relative maximum ligation efficiencies of different N-terminal ligation precursors measured in LC-MS assays, indicating splicing preferences for the P1 and P2 positions. Maximum efficiencies are averages of the two highest efficiencies as measured in time-course experiments and normalized to the maximum efficiency of [YLGL][RLPSV] formation, which was included in all experiments. Error bars represent SD. (E) Ligation efficiencies of different ligation products resulting from digestion of the optimized splicing precursor YLGD-SY-RLPSV, measured at various time points and precursor peptide concentration.
To quantitatively determine which C-terminal amino acid sequences are prone to ligation, we developed an FP-based ligation assay. For this FP assay, precursor peptides were designed such that they each contained one anchor residue needed for binding to HLA-A2. These peptides themselves display no or very low HLA-A2 affinity. However, the combination of both precursors through a splicing reaction may form a product that then displays high affinity for MHC class I. Using this FP assay (26, 27), the HLA-A*0201 binding affinities of reaction mixtures containing various C-terminal ligation partners were measured to determine the relative ligation efficiency of each precursor. A series of C-terminal precursors XLPSV or KXPSV or KLXSV (in which X is any proteogenic amino acid) were each incubated with purified 20S proteasome in the presence of the N-terminal precursor YLDW-SY to allow formation of the transpeptidation products [YLDW][XXXSV]. Ligation efficiencies of [YLDW][XXXSV] formation were normalized to the efficiency of [YLDW][KLPSV] formation, which was included as a standard in all experiments (Fig. 1B).
A limitation of the FP assay described above is that it requires an equal rate of N-terminal precursor processing by the proteasome to allow for quantitative comparison of transpeptidation efficiencies between reaction mixtures. However, changing the sequence of the N-terminal precursors changes its rate of hydrolysis (Supplemental Fig. 1B). We therefore switched to LC-MS–based assays to enable the analysis of both hydrolysis and ligation products for different N-terminal precursors. To determine which N-terminal precursors were likely to promote transpeptidation, a screen was performed in which YLGX-SY was incubated with proteasome in the presence of C-terminal ligation partner RLPSV, and the resulting reaction mixtures were analyzed by LC-MS. For five of the YLGX-SY precursors, in which X is C, H, K, P, or R, the formation of ligation product was not observed (Fig. 1C, top panel). YLGP-SY was not cleaved by the proteasome. Processing of YLGX-SY precursors in which X was a basic amino acid (H, K, and R) resulted in no or trace amounts of ligation products. YLGC-SY suffered from poor detection with LC-MS, which hampered further analysis. These five precursors were therefore not further studied in detail.
Next, series of N-terminal precursors YLGX-SY, YLXL-SY, or YLXX-SY were incubated with proteasome in the presence of C-terminal ligation partner RLPSV and reaction mixtures were analyzed by LC-MS at multiple time points. Ligation efficiency was defined as the percentage of YLXX-SY cleavages that resulted in the formation of the ligation product [YLXX][RLPSV] and was determined for each precursor at each time point. Because the absolute ligation efficiencies and hydrolysis rates showed variation between experiments, YLGL-SY was included as a standard in parallel in all experiments. Maximum ligation efficiency was defined as the average efficiency of the top two most efficient time points per precursor. All maximum ligation efficiencies were subsequently normalized to the maximum ligation efficiency of benchmark peptide YLGL-SY determined simultaneously in a corresponding, parallel experiment, which was set to 1 (Fig. 1C). The hydrolysis and transpeptidation products in these different reaction mixtures differ by a single amino acid only. Mass spectra of mixtures of all 20 synthetic YLGXRLPSV transpeptidation products showed that many of these peptides, including benchmark peptide YLGLRLPSV, displayed close to average peak intensities when compared side-by-side by LC-MS (Supplemental Fig. 1A). However, for some peptides, individual peak intensities deviated up to ∼20% from the average peak intensity. This deviation impedes a formal quantitative comparison of normalized ligation efficiencies. Instead, we considered the analysis of these LC-MS–based assay data to be semiquantitative, allowing us to screen for sequences that are either prone or refractory to splicing.
Taken together, these data revealed distinct peptide motifs that are likely to either promote or abolish Ag splicing for both the N- and C-terminal ligation partner, which is summarized in Fig. 1A. C-terminal ligation partners that promoted ligation mostly contained a basic residue (K/R) at P1′ combined with hydrophobic residue at the P2′ position. An additional hydrophobic residue at P3′ improved ligation further. In contrast, peptides containing a negatively charged residue at P2′ or P3′ did not serve as C-terminal ligation partners. The binding curves of synthetic ligation products (Supplemental Fig. 1C) of both ligation-prone (red) and ligation-poor (blue) precursors are similar, indicating that the differences observed in this FP assay are not caused by differences in binding affinity between ligation products. The one exception are peptides with two positively charged residues, which bind to HLA-A*0201 with reduced affinity. Thus, although peptides containing a positively charged residue at P2′ or P3′ did not seem to serve as C-terminal ligation partners, this might, at least in part, be caused by the reduced binding affinity of these specific ligation products.
N-terminal precursors that contained aspartate or a polar uncharged residue at P1 showed the highest ligation efficiencies, as well as the highest absolute amounts of ligation product (Supplemental Fig. 1B). For these peptides, degradation rates were typically slow (Supplemental Fig. 1B), as compared with the rate of YLGL-SY hydrolysis in the corresponding parallel experiment (Supplemental Fig. 1B, dotted lines). Ligation also occurred with precursors that contained a hydrophobic residue at P1, especially when combined with a small, polar or negatively charged residue at P2 (Fig. 1C). Although this second type of precursor showed lower splicing efficiencies, processing rates were high, resulting in substantial amounts of ligation products (Supplemental Fig. 1B). To investigate whether the requirement for a small or polar residue at P2 was also correct for precursors with nonhydrophobic residues at P1, we repeated the assay while varying both P1 and P2 positions within the most splicing-prone sequences (Fig. 1D). For all residues tested at P1, ligation efficiencies were higher with a small residue (G) compared with a hydrophobic residue (L). However, ligation did also occur with a hydrophobic residue at P2, especially when combined with serine at P1. As can be seen in Supplemental Fig. 1A, many ligation products YLGXRLPSV, in which X represents a negatively charged or polar amino acid (D, E, T, and Q), showed slightly lower peak intensities when compared side-by-side with products containing a hydrophobic residue (I, W, and F). This suggests that the differences in normalized maximum ligation efficiency between these two groups of precursors may have been underestimated to some extent in Fig. 1C.
Splicing-prone peptide sequences participate in highly efficient ligation reactions
To validate that peptides following these rules are indeed prone to proteasomal splicing, we designed model peptide YLGD-SY-RLPSV, which combined the optimized N-terminal precursor YLGD with the optimized C-terminal ligation fragment RLPSV, separated by a two amino acid spacer sequence. This peptide was incubated with proteasome at various concentrations and the resulting reaction mixtures were analyzed at various time points by LC-MS and by monitoring tyrosine absorption at 280 nm by HPLC to quantify the amounts of hydrolysis and ligation products (Fig. 1E, Supplemental Fig. 2A). HPLC and LC-MS spectra revealed two main cleavage sites in this optimized precursor peptide, resulting in the formation of various hydrolysis products including YLGD, YLGDSY, and SYRLPSV. In addition, four ligation products were clearly detectable at 280 nm, which could be identified as [YLGD][RLPSV], [YLGD][YLGD], [YLGD][YLGDSY], and [YLGD][YLGDSYRLPSV] (Supplemental Fig. 2A), which indicated that any splicing-prone sequence with a free N terminus, be it an unprocessed precursor or a ligation/hydrolysis product, can potentially serve as a ligation partner.
Over time, the amount of precursor peptide rapidly decreased, whereas the amount of YLGD rapidly increased. The amounts of YLGDSY and SYRLPSV remained relatively constant (Supplemental Fig. 2B), suggesting that both these peptides are subjected to secondary processing (to YLGD and RLPSV, respectively), especially at low precursor peptide concentrations. Ligation efficiency, defined as the percentage D-S cleavages that resulted in a ligation event reached 14% after 2 h (Fig. 1E) and gradually decreased over time. As also the ratio of different ligation and hydrolysis products shifted to shorter peptides over time (Fig. 1E, Supplemental Fig. 2B), this decrease in ligation efficiency is probably caused by the degradation of formed ligation products. Increasing the concentration of precursor peptides by a factor four doubled the ligation efficiency at 2 h from 15 to 30% (Fig. 1E), without changing the cleavage specificity of the proteasome (Supplemental Fig. 2B). This suggests that under in vitro conditions as many as one of three cleavages of splicing-prone peptide sequences can result in a ligation event. At higher concentrations of YLGDSYRLPSV, an increased formation of especially [YLGD][YLGDSYRLPSV] was observed (Fig. 1E), indicating that not only the sequence but also the concentration of C-terminal ligation partner in the proteasome is an important determinant for ligation with again particular preferences for the product. Remarkably, although the amounts of trans-ligated products (i.e., ligation of two different peptides, e.g., [YLGD][YLGD]) decreased over time, the amounts of [YLGD][RLPSV], which can also be formed by cis-splicing (i.e., ligation of two fragments of the same peptide) increased over time. As ultimately similar amounts of YLGD and RLPSV are formed from the precursor peptide, this suggests that the efficiency of cis-splicing events is less controlled by the amount of the C-terminal ligation partners but rather depends on the nature of both the N- and C-terminal peptide sequences. Taken together, these data show that splicing can be a highly efficient process that is governed both by the sequence and the concentration of peptides involved and confirm that cleavage after the optimal N-terminal GD motif, as predicted, results in high ligation efficiencies.
Splicing rules facilitate the prediction of potential spliced Ags.
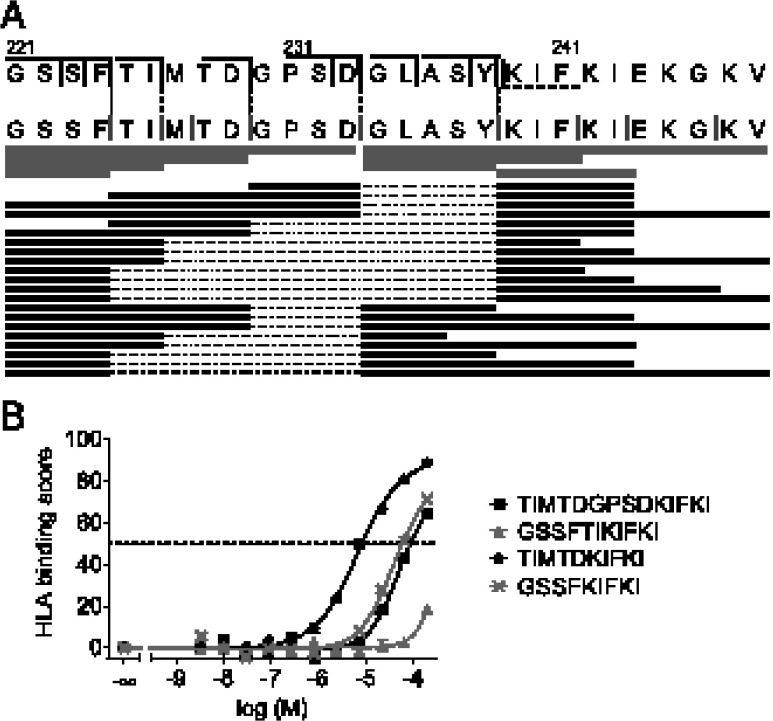
To test and refine our splicing rules, we set out to validate these rules using a longer polypeptide from a known immunogenic protein. Because of the need to perform in vitro digestions, the length of such a polypeptide was constrained to ∼30 aa. Therefore, we predicted ligation sites in a neuraminidase 1 (N1) protein (from Influenza A virus A/Puerto Rico/8/34) and we selected the 28-mer peptide 221–248, which contained relatively many potential N- and C-terminal ligation partners. Finally, we replaced the cysteine residue on position 223 by a serine residue to facilitate LC-MS analysis, resulting in peptide 221–248/C223S (N1.1; GSSFTIMTDGPSDGLASYKIFKIEKGKV), which we used in further studies. The various N-terminal precursor sequences in N1.1 as well as its C-terminal splicing motif are indicated in Fig. 2A (vertical black solid and dotted lines, respectively). Peptide N1.1 was synthesized and incubated with proteasome and the resulting digestion mixtures were analyzed by LC-MS. Nine proteasomal cleavage sites were identified in N1.1, indicated by the gray vertical lines (Fig. 2A). Processing of N1.1 resulted in the accumulation of two longer fragments (GSSFTIMTDGPSD and GLASYKIFKIEKGKV), both of which can be generated through a single cleavage event at D233. In addition, a variety of shorter cleavage products was formed (Fig. 2A, gray horizontal bars).
FIGURE 2.
Splicing rules facilitate the prediction of spliced epitopes from a long polypeptide. (A) Top panel, Peptide 221-248/C223S (N1.1) derived from the neuraminidase 1 protein of Influenza A virus A/Puerto Rico/8/34, showing several predicted N-terminal precursor sequences (black solid vertical lines) and one predicted C-terminal ligation motif (black dotted vertical lines). Middle panel, Actual cleavage sites in peptide 221-248 (gray vertical lines) and detected hydrolysis products (gray horizontal bars), measured by LC-MS. Bottom panel, Overview of ligation products measured in digestion mixtures of N1.1 by LC-MS, indicated by black horizontal bars, where the excised fragment is indicated by a dotted line. (B) HLA-A*0201 binding curves of selected splicing products originating from N1.1.
A total of 21 ligation products could be identified in digestion mixtures at various concentrations of peptide and proteasome, indicated by the black horizontal bars in Fig. 2A. Table I shows the measured peak intensities of all splicing products and the corresponding hydrolysis products. It should be noted, however, that these intensities cannot formally be compared in a quantitative way because different peptides display different ionization efficiencies. The identity of ligation products was confirmed by comparing retention times and MS spectra with those of the corresponding synthetic peptides, as exemplified in Supplemental Fig. 3. Importantly, ligation occurred onto four of the five predicted N-terminal precursor sequences that were actually created by the proteasome (F224, I226, D229, and D233). In contrast, no ligation was observed onto those cleavage sites that did not match our splicing rules (M230, F241, I243, and G246). Peptides starting with either amino acids KIF or GLA served as C-terminal ligation partners, whereas only amino acid sequence KIF was predicted by the model.
Table I. MS peak areas of ligation products and the corresponding hydrolysis products identified in digestion mixtures of peptide N1.1.
| Incubation Time (nmol Peptide/10 μg Proteasome) | MS Peak Area/10 μg Proteasome |
IC50 | |||
|---|---|---|---|---|---|
| 20 h |
8 h |
||||
| 100 | 200 | 400 | 150 | ||
| GSSF | 1,644 | 1,972 | 2,722 | 1,960 | |
| GSSF-GLASYKIFKIEKGKV | 176 | 398 | 578 | 400 | ND |
| GSSF-GLASYKIFKI | 88 | 114 | 136 | 104 | ND |
| GSSF-GLASY | 334 | 416 | 434 | 477 | >1 mM |
| GSSF-KIFKIEKGKV | 126 | 152 | 236 | 193 | ND |
| GSSF-KIFKIEKG | 82 | 86 | 94 | 79 | ND |
| GSSF-KIFKI | 776 | 888 | 1,106 | 1,304 | 39 μM |
| GSSFKIF | 134 | 158 | 270 | 319 | ND |
| GSSFTI | 3,630 | 3,764 | 3,962 | 2,923 | |
| GSSFTI-GLASYKIFKI | 18 | 40 | 64 | 38 | ND |
| GSSFTI-GLA | 44 | 50 | 108 | 112 | ND |
| GSSFTI-KIFKIEKGKV | 58 | 80 | 116 | 102 | ND |
| GSSFTI-KIFKI | 186 | 242 | 452 | 334 | ND |
| GSSFTI-KIF | 236 | 256 | 344 | 333 | ND |
| GSSFTIMTD | 5,446 | 5,898 | 5,892 | 4,572 | |
| GSSFTIMTD-GLASYKIFKIEKGKV | 42 | 56 | 94 | 124 | ND |
| GSSFTIMTD-GLASYKIFKI | 0 | 14 | 18 | 23 | ND |
| GSSFTIMTD-GLASY | 0 | 0 | 0 | 40 | 49 μM |
| GSSFTIMTD-KIFKI | 30 | 56 | 106 | 109 | ND |
| TIMTD-KIFKI | 28 | 42 | 0 | 64 | 6.8 μM |
| GSSFTIMTDGPSD | 13,732 | 16,356 | 17,014 | 12,156 | |
| GSSFTIMTDGPSD-KIFKIEKGKV | 54 | 118 | 136 | 164 | ND |
| GSSFTIMTDGPSD-KIFKI | 74 | 202 | 292 | 328 | ND |
| GPSD-KIFKI | 70 | 98 | 122 | 183 | ND |
Taken together, these data suggest that the N-terminal ligation partner is the dominant factor determining ligation efficiency because ligation will only take place if the N-terminal ligation precursor has a splicing-prone sequence. On the basis of the limited number of C-terminal ligation sequences that were identified in this peptide, ligation appears to depend both on the availability of peptides and on the sequence of the C-terminal ligation fragment. Length was not a determining factor because both short and long peptides were equally suited to serve as C-terminal ligation partners. However, C-terminal fragments shorter than 3 aa were not found to participate in ligation reactions, in line with results from previous studies (9).
To investigate the affinity of spliced N1.1 products for HLA-A2.1, binding curves of all products, which consisted of 14 residues or fewer and contained at least one anchor residue (L/M/I at P2 and/or V/L/I as C-terminal residue), were measured using an FP assay (26, 31) (Fig. 2B). In this assay, peptides have to compete with a tracer peptide for binding, and their HLA binding score is defined as the percent inhibition of tracer peptide binding. IC50s (Table I) are defined as the concentration of peptide that shows a half-maximal binding score. Although N1.1 is a small protein fragment containing only 28 residues, N1.1 processing resulted in the splicing of four peptides that displayed affinity for HLA-A*0201. The peptide [TIMTD][KIFKI], which contains both anchor residues and has the proposed optimal length for binding to HLA-A*0201, showed the highest affinity for HLA-A*0201 (Fig. 2B) with an IC50 in the low micromolar range (Table I). The splicing products [GSSF][KIFKI] and [TIMTDGPSD][KIFKI], which displayed suboptimal anchor residues and length, respectively, displayed intermediate affinity and showed IC50s in the high micromolar range, whereas [GSSFTI][KIFKI] had only low affinity for HLA-A*0201. Considering the many types of HLA molecules, each with their own anchor residue preference, and considering the fact that spliced peptides can be trimmed to the correct length by the proteasome and peptidases in the cytosol (1), these data suggest that many potential HLA-A binding epitopes may be formed by splicing from an average protein.
Splicing of longer peptides predominantly occurs via cis-splicing
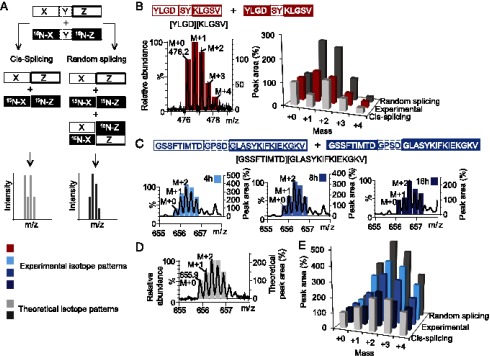
Splicing of short peptides has been described to occur both between fragments of the same peptide (cis-splicing) and between fragments derived from different peptides (trans-splicing) (10, 11, 15). For spliced peptides to play a role in immunity, one would assume that splicing occurs only with defined peptide sequences from the same protein that are ligated together multiple times. Random recombination of fragments from different proteins is likely to either result in amounts of spliced peptides that are too low to evoke an immune response or in autoimmunity because of a lack of consistent negative selection in the thymus. We hypothesize that cis-splicing is likely driven by the simultaneous generation of both splicing partners in the proteasome in situ. Trans-splicing, however, is unlikely to be driven by any mechanism, but rather the result of a random recombination that would generate equal amounts of both cis- and trans-spliced products (Fig. 3A). To study this aspect of the splicing mechanism in more detail, for our splicing-prone short peptide, a precursor, which contained a 15N-labeled glycine residue in both the N- and C-terminal fragments, was synthesized, mixed with its unlabeled counterpart in a 1:1 ratio, and subjected to proteasomal processing, followed by LC-MS analysis. As a control, both peptides were digested and analyzed separately. In this experimental setup, cis-splicing should yield equal amounts of two splicing products with masses M+0 and M+2. Random recombination on the other hand should result in the formation of four different peptides with masses M+0, M+1, and M+2 in a 1:2:1 ratio. Cis- and random splicing, therefore, result in distinct isotope distribution patterns, which can be distinguished by LC-MS (Fig. 3A). In the single-isotope digestions of 15N-Gly–labeled and unlabeled YLGD-SY-KLGSV, peaks at m/z 476.2 (M+0) and 477.2 (M+2) corresponding to the splicing products [[YLGD][KLGSV]+2H]2+ and [[YL15NGD][KL15NGSV]+2H]2+, respectively, could readily be detected, each with a similar experimental isotope pattern (Supplemental Fig. 4A). These patterns were used to predict the theoretical isotope patterns corresponding to cis-splicing and random splicing of a 1:1 mixture of the unlabeled and 15N-Gly–labeled peptide (Supplemental Fig. 4A). We next mixed 15N-Gly–labeled and unlabeled YLGD-SY-KLGSV in a 1:1 ratio and analyzed the isotope pattern of the resulting [YLGD][KLGSV] peptide (Fig. 3B, black trace). A comparison of this experimentally determined isotope pattern (Fig. 3B, red bars) with the theoretical patterns (Fig. 3B, light and dark gray bars) showed the experimental pattern to be an intermediate between cis- and random splicing, confirming that splicing of a short peptide indeed occurs through a combination of both.
FIGURE 3.
Splicing of long peptides predominantly occurs via cis-splicing. (A) Splicing of a mixture of 15N-Gly–labeled (indicated in bold) and unlabeled precursor peptides through cis-splicing or through random splicing results in differently labeled spliced products and therefore in different isotope patterns that can be distinguished by LC-MS. (B) Left panel, Mass spectrum (black trace) and the corresponding experimental isotope pattern (red bars) of [YLGD][KLGSV] resulting from the digestion of a mixture of 15N-labeled and unlabeled precursor YLGD-SY-KLGSV. Right panel, Comparison of the experimental isotope pattern with theoretical isotope patterns for cis- and random splicing. (C) Mass spectra (black traces) and the corresponding experimental isotope pattern (blue bars) of [GSSFTIMTD][GLASYKIFKIEKGKV], resulting from the digestion of a mixture of 15N-Gly–labeled (indicated in bold) and unlabeled precursor GSSFTIMTD-GPSD-GLASYKIFKIEKGKV for different time periods. (D) Mass spectrum (black trace) of a 1:1 mixture of single-isotope incubation samples, in which the labeled and 15N2-labeled splicing products are present in equal amounts, compared with the theoretical isotope pattern for cis-splicing (gray bars). (E) Comparison of the experimental isotope patterns (blue bars) determined in (C) with theoretical isotope patterns for cis- and random splicing.
A high concentration of relatively short peptides within the proteasomal catalytic chamber likely increases the chance that two polypeptide strands are degraded simultaneously, favoring random recombination. We therefore hypothesized that cis-splicing would be favored for longer precursor peptides, especially at decreasing precursor peptide concentrations. To test this hypothesis, 15N-labeled N1.1, in which G221 and G234 were replaced with 15N-glycine, and unlabeled N1.1 were first incubated separately with proteasome and followed by LC-MS analysis. The formation of [(15N)GSSFTIMTD][(15N)GLASYKIFKIEKGKV] by the proteasome was studied by LC-MS. In the single-isotope digestion mixtures of N1.1 and 15N-N1.1, peaks for both the unlabeled and the 15N-labeled [GSSFTIMTD][GLASYKIFKIEKGKV] could be observed for the quadruple charged peptides at m/z 655.9 (M+0) and 656.3 (M+2), as expected (Supplemental Fig. 4B). Mixing these samples postdigestion resulted in the isotope pattern shown in Fig. 3D (black trace), which closely resembled the calculated isotope pattern for cis-splicing (Fig. 3D, 3E, light gray bars), as expected. Next, a 1:1 mixture of N1.1 and 15N-N1.1 was incubated with proteasome for time periods up to 16 h. Over time, the isotope pattern of the splicing product in this incubation mixture should now shift from random to cis-splicing because of the decreasing precursor peptide concentration within the proteasomal catalytic chamber. Indeed, when the isotope patterns at different time points were compared, a clear decrease in M+1/M+0 ratio was observed over time, indicative of a shift toward cis-splicing (Fig. 3C, 3E). After a 16-h incubation period, the resulting MS spectrum closely matched the theoretical spectrum for complete cis-splicing (Fig. 3E). Collectively, these data indicate that splicing of long protein fragments occurs predominantly via cis-splicing, especially at lower (more physiological) precursor peptide concentrations.
Splicing rules predict splicing events in cells
Finally, we set out to confirm that the splicing rules determined by us can also predict splicing of Ags in cells. To this end, we transiently transfected the HLA-A2 epitope splicing precursor YLGN-SY-RLPSV, N-terminally fused to mYFP-ubiquitin, into MelJuSo cells stably overexpressing HLA-A2.1-GFP. We also stably transduced JY cells (that naturally express HLA-A*0201) with a minigene encoding the HLA-A2 epitope splicing precursor YLGD-SY-RLPSV or the HLA-A2 epitope YLWGRPLSV in a pMX-minigene-IRES-GFP vector. In these transfected JY and MelJuSo/HLA-A2-GFP cells, splicing of the precursor sequence should result in the formation of [YLGD][RLPSV] and [YLGN][RLPSV], respectively. Both these splicing products have high affinity for HLA-A2.1 and should be presented at the cell surface, if formed. The HLA-A*0201 epitope YLWGRPLSV, in contrast, should be presented on the cell surface directly without involvement of proteasomal processing.
To test the formation of the predicted splicing products, cells were sorted by flow cytometry to select for epitope (precursor) expression and all MHC class I binding peptides were eluted from the cell surface. Eluted peptides were mixed with a small amount of reference peptides and fractionated by SCX chromatography. Fractions were collected based on the SCX retention behavior of synthetic standards of all expected peptides relative to the reference peptides and analyzed with nanoscale LC-MS/MS. Fig. 4A shows the ion traces of [YLGN][RLPSV] and [YLGD][RLPSV] and YLWGRPLSV as eluted from the cell surface, as well as that of the test peptide, constructed using stringent mass windows of 2 ppm around the theoretical m/z values of the doubly charged target peptides to avoid false-positive hits. Retention times relative to the test peptide matched the expected values. In addition, MS/MS data confirmed the amino acid sequence of these peptides, as exemplified for [YLGD][RLPSV] in Fig. 4B. These data confirm that the splicing products are formed by the proteasome in cells as predicted and are presented on the cell surface by MHC class I. Because this method is semiquantitative, the amounts of eluted peptide in each sample could be calculated based on the measured ion intensities. Approximately 1–5 fmol of the spliced products [YLGN][RLPSV] and [YLGD][RLPSV] was eluted from cells expressing the splicing precursor peptide, compared with 150 fmol YLWGRPLSV, which is presented without any proteasomal processing (Fig. 4A). From these results, we conclude that the rules determined in this study predict splicing events in cells.
FIGURE 4.
Splicing rules predict efficient splicing reactions in cells. (A) Ion traces and estimated amounts of the test peptide SGYSGIFSVEGK (10 fmol on column), the spliced peptides [YLGN][RLPSV] and [YLGD][RLPSV], and the HLA-A*0201 binding peptide YLWGRPLSV, as eluted from the cell surface of JY or MelJuso-A2 cells. (B) MS/MS spectrum of peptide [YLGD][RLPSV] as eluted from the cell surface of JY cells.
Discussion
In the past decade, it has become apparent that noncontiguous and thus not genetically encoded Ags that consist of two posttranslationally fused peptides are formed in cells and are presented to the immune system by MHC class I (5–9). In the current study, we aimed at deciphering splicing rules that govern the production of noncontiguous Ags to facilitate their prediction and identification. Spliced epitopes are produced by the proteasome via a transpeptidation mechanism (5, 7–11), but whether this process occurs randomly or follows specific rules has been a matter of debate. In contrast, the transpeptidation model could implicate that proteasomal splicing is not determined by a particular sequence motif but can occur at any major cleavage site (5, 10). This hypothesis is also in line with the observation that the concentration of precursor fragments is one of the driving factors of the splicing reaction (11). In contrast, Mishto et al. (11) show that peptides involved in proteasomal splicing are often derived from minor proteasome cleavage sides, suggesting that the splicing process is dependent on unknown sequence specificities. Our results clearly indicate that splicing rules exist. We have deciphered these rules for the first time, to our knowledge, and they are summarized in Table II. For both N-terminal precursors and C-terminal ligation fragments, the anchor residues (L at position 2 and V on position 9) were not varied. It cannot be excluded that these residues influence the relative ligation efficiencies determined in this study, and therefore, the rules we deduced are formally restricted to the HLA-A*02 haplotype. However, we consider it likely that these rules can—at least in part—be extended to other haplotypes, especially to those with similar anchor residue requirements.
Table II. Summary of the splicing rules identified in this study.
| The N-terminal ligation precursor |
| Primarily determines the ligation efficiency |
| Is most efficient when slowly hydrolyzed by the proteasome and therefore possesses a longer half-life |
| Is most efficient when there is a negatively charged or polar residue at P1 combined with a small or polar residue at P2 |
| Can participate in ligation if a hydrophobic residues at P1 is combined with a basic, small, or—to a lesser extent—polar or negatively charged residue at P2 |
| The C-terminal ligation partner |
| Has less stringent structural requirements, although not all fragments are suitable ligation partners |
| Influences the ligation efficiency more through its mere presence and concentration than by its precise sequence. |
| Cis-splicing or random recombination? |
| Splicing from short peptides occurs both through cis-splicing and via random recombination |
| Splicing from longer precursor peptides is non-random and occurs predominantly via cis-splicing, especially at lower precursor peptide concentrations |
Previous studies have so far identified five in vivo and 39 in vitro splicing products. Strikingly, many of the described in vitro splicing products contain an N-terminal precursor with a motif that is in accord with our splicing rules: either a hydrophobic residue at P1 combined with a small, polar, or negatively charged amino acid at P2 or a polar or negatively charged residue at the P1 position. This also holds true for three of the five in vivo N-terminal precursors (NTYAS, SLPRGT, and IYMDGT), which all contain a polar residue at P1 combined with a small amino acid at P2. Although such combinations are abundant in the human proteome, their prevalence in spliced peptides does confirm that these motifs facilitate the proteasomal splicing reaction and suggests that our splicing rules are an important first step toward predicting the occurrence of spliced epitopes. Two previously described in vivo N-terminal precursors contained a K or H on P1 (5, 9). In our hands, however, peptides with a basic residue on P1 failed to serve as N-terminal ligation precursors. The presence of many hydrophobic residues in the ligation precursors investigated in this study may have biased the results toward splicing in the β5 subunit, whereas peptides with a basic residue on P1 are predominantly cleaved (and therefore likely spliced) by the β2 subunit. Further research is needed to elucidate the contribution of different subunits to the splicing process.
Splicing has been hypothesized to be a rare event: the in vivo ligation efficiencies of splicing reactions for two reported spliced epitopes, RTK-QLYPEW (5) and NTYAS-PRFK (6), have been estimated to be 0.01 and 0.0002%, respectively (5, 10). Notably, these low splicing efficiencies were sufficient to produce immunogenic epitopes that were presented on the cell surface by MHC class I and evoked an immune response. In contrast, our data indicate that splicing-prone precursors can be ligated with up to 30% efficiency in vitro. One to 5 fmol of spliced products could be eluted from the surface of cells expressing a precursor peptide, compared with 150 fmol of a peptide that was presented without any proteasomal processing. Although cell surface quantities of a processed spliced peptide cannot be directly compared with those of an unprocessed peptide, these numbers suggest that splicing and cell surface presentation of optimized splicing fragments occurs with substantial efficiency to allow for detection by MS. That they are not often observed in the various analyses of the MHC-associated peptidome by MS may be the consequence of the mode of analysis that only considers peptides that are genetically encoded. Spliced peptides are then automatically ignored. Taken together, these data suggest that spliced epitopes may be presented on the surface of cells to a much larger extent than previously assumed.
Although ligation has been described to occur between fragments of different peptides in vitro (trans-splicing) (10, 11, 15), Dalet et al. (10) have shown that cis-splicing is the predominant event in vivo. Whereas two cis-splicing precursors are always generated together in situ, the chance that two trans-splicing precursors are repeatedly generated simultaneously inside one proteasome is statistically negligible. Thus, random splicing would reduce actual peptide levels to very low numbers, and the cytosolic peptidase activities may then be expected to clear the random spliced peptides before they can be presented (32). Indeed, our data indicate that cis-splicing of long peptides is strongly preferred over random splicing, especially at low peptide concentration, which can be considered a model for protein splicing in vivo. Thus, our data support the view that protein ligation occurs via cis-splicing in vivo, whereas the random splicing of peptides can be observed and is possible under in vitro conditions.
In the proteasomal active sites, activated and preoriented water molecules, which form a tight hydrogen-bonding network, are responsible for the hydrolysis of the O-acyl enzyme intermediate (33). Ligation can thus only take place if the C-terminal ligation partner is able to compete with these water molecules that are constantly present in the active site (12, 14). Therefore, splicing is most likely to occur onto N-terminal precursors that form intermediate esters with relatively long half-lives. It has been suggested that this relatively long half-life could result from a high affinity of the N-terminal precursor for the nonprimed sites, suggesting that splicing mainly occurs with tight-binding substrates (14, 34). In striking contrast, we find that N-terminal ligation precursors seemed more likely to be involved in ligation if the P1 and P2 residue side chains bind suboptimally to the substrate binding pockets, assuming that these precursors are mainly cleaved by the β5 subunit, which preferably cleaves after hydrophobic amino acids. In line with this, we indeed find that most efficient N-terminal ligation precursors are hydrolyzed at a relatively slow rate by the proteasome. We therefore propose a model in which a perfect fit of the substrate to the active site pockets may orientate the O-acyl enzyme intermediate such that the attack of one of the preoriented water molecules on the intermediate ester is facilitated. A suboptimal fit of charged and polar residues in the N-terminal precursor, in contrast, may orient the O-acyl enzyme intermediate away from these water molecules and toward the C-terminal ligation partner. Alternatively, such polar or charged residues may interfere with the hydrogen-bonding network that is required to activate water molecules for hydrolysis. This would result in an intermediate complex that cannot be hydrolyzed swiftly by water and is therefore more prone to ligation.
In conclusion, we show that splicing is not a random event, and we have determined, to our knowledge for the first time, rules that dominate the splicing process, as summarized in Table II. In addition, we show that splicing-prone sequences are ligated with high efficiency, that many spliced products may be formed from a single protein, and that splicing of long peptides predominantly occurs via cis-splicing, all suggesting that protein splicing may play a much more significant role in immunity than previously assumed. In an accompanying article, we show that the proteasome can form a novel type of Ag containing an isopeptide linkage, further extending the spliced Ag repertoire (35). Ag splicing would ensure that a more diverse peptide repertoire reaches the cell surface, which increases the chance of recognition by CD8+ T cells and ultimately of elimination of the malignant or infected cell by the immune system (3, 12–14). The rules predicting splicing, which are described in this paper, will facilitate the detection of such spliced Ags to elucidate their role in immunity.
Supplementary Material
Acknowledgments
We thank Henk Hilkmann for peptide synthesis.
C.R.B., J.J.N., T.N.M.S., and H.O. are members of the Institute for Chemical Immunology.
This work was supported by Netherlands Organization for Scientific Research Grant 819.02.003 and Dutch Cancer Society Grant NKI 2005-3368 (to H.O.).
The online version of this article contains supplemental material.
- FP
- fluorescence polarization
- LC
- liquid chromatography
- MS
- mass spectroscopy, MS/MS, tandem MS
- SCX
- strong cation exchange
- TFA
- trifluoroacetic acid.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Neefjes J., Jongsma M. L., Paul P., Bakke O. 2011. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11: 823–836. [DOI] [PubMed] [Google Scholar]
- 2.Sijts E. J., Kloetzel P. M. 2011. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell. Mol. Life Sci. 68: 1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vigneron N., Van den Eynde B. J. 2012. Proteasome subtypes and the processing of tumor antigens: increasing antigenic diversity. Curr. Opin. Immunol. 24: 84–91. [DOI] [PubMed] [Google Scholar]
- 4.Basler M., Kirk C. J., Groettrup M. 2013. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 25: 74–80. [DOI] [PubMed] [Google Scholar]
- 5.Vigneron N., Stroobant V., Chapiro J., Ooms A., Degiovanni G., Morel S., van der Bruggen P., Boon T., Van den Eynde B. J. 2004. An antigenic peptide produced by peptide splicing in the proteasome. Science 304: 587–590. [DOI] [PubMed] [Google Scholar]
- 6.Hanada K., Yewdell J. W., Yang J. C. 2004. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427: 252–256. [DOI] [PubMed] [Google Scholar]
- 7.Warren E. H., Vigneron N. J., Gavin M. A., Coulie P. G., Stroobant V., Dalet A., Tykodi S. S., Xuereb S. M., Mito J. K., Riddell S. R., Van den Eynde B. J. 2006. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 313: 1444–1447. [DOI] [PubMed] [Google Scholar]
- 8.Dalet A., Robbins P. F., Stroobant V., Vigneron N., Li Y. F., El-Gamil M., Hanada K., Yang J. C., Rosenberg S. A., Van den Eynde B. J. 2011. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. USA 108: E323–E331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaux A., Larrieu P., Stroobant V., Fonteneau J. F., Jotereau F., Van den Eynde B. J., Moreau-Aubry A., Vigneron N. 2014. A spliced antigenic peptide comprising a single spliced amino acid is produced in the proteasome by reverse splicing of a longer peptide fragment followed by trimming. J. Immunol. 192: 1962–1971. [DOI] [PubMed] [Google Scholar]
- 10.Dalet A., Vigneron N., Stroobant V., Hanada K., Van den Eynde B. J. 2010. Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor-5. J. Immunol. 184: 3016–3024. [DOI] [PubMed] [Google Scholar]
- 11.Mishto M., Goede A., Taube K. T., Keller C., Janek K., Henklein P., Niewienda A., Kloss A., Gohlke S., Dahlmann B., et al. 2012. Driving forces of proteasome-catalyzed peptide splicing in yeast and humans. Mol. Cell. Proteomics 11: 1008–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkers C. R., de Jong A., Ovaa H., Rodenko B. 2009. Transpeptidation and reverse proteolysis and their consequences for immunity. Int. J. Biochem. Cell Biol. 41: 66–71. [DOI] [PubMed] [Google Scholar]
- 13.Shastri N. 2006. Cell biology: peptides, scrambled and stitched. Science 313: 1398–1399. [DOI] [PubMed] [Google Scholar]
- 14.Borissenko L., Groll M. 2007. Diversity of proteasomal missions: fine tuning of the immune response. Biol. Chem. 388: 947–955. [DOI] [PubMed] [Google Scholar]
- 15.Liepe J., Mishto M., Textoris-Taube K., Janek K., Keller C., Henklein P., Kloetzel P. M., Zaikin A. 2010. The 20S proteasome splicing activity discovered by SpliceMet. PLOS Comput. Biol. 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson R. A., Michel H., Sakaguchi K., Shabanowitz J., Appella E., Hunt D. F., Engelhard V. H. 1992. HLA-A2.1‑associated peptides from a mutant cell line: a second pathway of antigen presentation. Science 255: 1264–1266. [DOI] [PubMed] [Google Scholar]
- 17.Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A. L., Appella E., Engelhard V. H. 1992. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255: 1261–1263. [DOI] [PubMed] [Google Scholar]
- 18.Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanović S. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50: 213–219. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen M., Lundegaard C., Worning P., Lauemøller S. L., Lamberth K., Buus S., Brunak S., Lund O. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 12: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toebes M., Coccoris M., Bins A., Rodenko B., Gomez R., Nieuwkoop N. J., van de Kasteele W., Rimmelzwaan G. F., Haanen J. B., Ovaa H., Schumacher T. N. 2006. Design and use of conditional MHC class I ligands. Nat. Med. 12: 246–251. [DOI] [PubMed] [Google Scholar]
- 21.Altman J. D., Moss P. A., Goulder P. J., Barouch D. H., McHeyzer-Williams M. G., Bell J. I., McMichael A. J., Davis M. M. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274: 94–96. [DOI] [PubMed] [Google Scholar]
- 22.Grommé M., Uytdehaag F. G., Janssen H., Calafat J., van Binnendijk R. S., Kenter M. J., Tulp A., Verwoerd D., Neefjes J. 1999. Recycling MHC class I molecules and endosomal peptide loading. Proc. Natl. Acad. Sci. USA 96: 10326–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. 2008. Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berkers C. R., van Leeuwen F. W., Groothuis T. A., Peperzak V., van Tilburg E. W., Borst J., Neefjes J. J., Ovaa H. 2007. Profiling proteasome activity in tissue with fluorescent probes. Mol. Pharm. 4: 739–748. [DOI] [PubMed] [Google Scholar]
- 25.de Jong A., Schuurman K. G., Rodenko B., Ovaa H., Berkers C. R. 2012. Fluorescence-based proteasome activity profiling. Methods Mol. Biol. 803: 183–204. [DOI] [PubMed] [Google Scholar]
- 26.Rodenko B., Toebes M., Celie P. H., Perrakis A., Schumacher T. N., Ovaa H. 2009. Class I major histocompatibility complexes loaded by a periodate trigger. J. Am. Chem. Soc. 131: 12305–12313. [DOI] [PubMed] [Google Scholar]
- 27.Bakker A. H., Hoppes R., Linnemann C., Toebes M., Rodenko B., Berkers C. R., Hadrup S. R., van Esch W. J., Heemskerk M. H., Ovaa H., Schumacher T. N. 2008. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl. Acad. Sci. USA 105: 3825–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendle G. M., Linnemann C., Hooijkaas A. I., Bies L., de Witte M. A., Jorritsma A., Kaiser A. D., Pouw N., Debets R., Kieback E., et al. 2010. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 16: 565–570. [DOI] [PubMed] [Google Scholar]
- 29.Meiring, H. D., E. C. Soethout, A. P. de Jong, and C. A. van Els. 2007. Targeted identification of infection-related HLA class I-presented epitopes by stable isotope tagging of epitopes (SITE). Curr. Protoc. Immunol. Chapter 16: Unit 16.13. [DOI] [PubMed]
- 30.Meiring H., van der Heeft E., ten Hove G., de Jong A. 2002. Nanoscale LC-MS(n): technical design and applications to peptide and protein analysis. J. Sep. Sci. 25: 557–568. [Google Scholar]
- 31.Hoppes R., Oostvogels R., Luimstra J. J., Wals K., Toebes M., Bies L., Ekkebus R., Rijal P., Celie P. H., Huang J. H., et al. 2014. Altered peptide ligands revisited: vaccine design through chemically modified HLA-A2‑restricted T cell epitopes. J. Immunol. 193: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reits E., Griekspoor A., Neijssen J., Groothuis T., Jalink K., van Veelen P., Janssen H., Calafat J., Drijfhout J. W., Neefjes J. 2003. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity 18: 97–108. [DOI] [PubMed] [Google Scholar]
- 33.Groll M., Huber R., Potts B. C. 2006. Crystal structures of Salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of β-lactone ring opening and a mechanism for irreversible binding. J. Am. Chem. Soc. 128: 5136–5141. [DOI] [PubMed] [Google Scholar]
- 34.Groll M., Götz M., Kaiser M., Weyher E., Moroder L. 2006. TMC-95‑based inhibitor design provides evidence for the catalytic versatility of the proteasome. Chem. Biol. 13: 607–614. [DOI] [PubMed] [Google Scholar]
- 35.Berkers C. R., de Jong A., Schuurman K. G., Linnemann C., Geenevasen J. A. J., Schumacher T. N. M., Rodenko B., Ovaa H. 2015. Peptide splicing in the proteasome creates a novel type of antigen with an isopeptide linkage. J. Immunol. 195: 4075–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.