A long-held tenet in the field of diabetes is that the tipping point between insulin sensitivity and resistance resides at the level of insulin receptor/insulin receptor substrate–adaptor complexes. Here it is shown that activation of Gαi by GIV/Girdin is a decisive event within the metabolic insulin signaling cascade that reversibly orchestrates insulin sensitivity or resistance.
Abstract
Insulin resistance (IR) is a metabolic disorder characterized by impaired insulin signaling and cellular glucose uptake. The current paradigm for insulin signaling centers upon the insulin receptor (InsR) and its substrate IRS1; the latter is believed to be the sole conduit for postreceptor signaling. Here we challenge that paradigm and show that GIV/Girdin, a guanidine exchange factor (GEF) for the trimeric G protein Gαi, is another major hierarchical conduit for the metabolic insulin response. By virtue of its ability to directly bind InsR, IRS1, and phosphoinositide 3-kinase, GIV serves as a key hub in the immediate postreceptor level, which coordinately enhances the metabolic insulin response and glucose uptake in myotubes via its GEF function. Site-directed mutagenesis or phosphoinhibition of GIV-GEF by the fatty acid/protein kinase C-theta pathway triggers IR. Insulin sensitizers reverse phosphoinhibition of GIV and reinstate insulin sensitivity. We also provide evidence for such reversible regulation of GIV-GEF in skeletal muscles from patients with IR. Thus GIV is an essential upstream component that couples InsR to G-protein signaling to enhance the metabolic insulin response, and impairment of such coupling triggers IR. We also provide evidence that GIV-GEF serves as therapeutic target for exogenous manipulation of physiological insulin response and reversal of IR in skeletal muscles.
INTRODUCTION
Insulin resistance (IR) is a metabolic disorder in which adipocytes and muscle cells fail to take up and metabolize glucose in response to the hormone insulin. Although IR is a hallmark of type 2 diabetes mellitus (T2DM), IR alone in the absence of T2DM significantly increases the risk for stroke, heart failure, and atherosclerosis (Carter, 2005; Rundek et al., 2010).
Although multiple etiologic factors contribute to the pathogenesis of IR (Saltiel and Kahn, 2001), they all ultimately converge to suppress critical components of metabolic insulin signaling. Insulin binds its receptors (InsR, IGF1R), triggering receptor autophosphorylation and subsequent tyrosine phosphorylation of insulin receptor substrate 1 (IRS1), among others. This leads to the recruitment and activation of Src homology 2 (SH2) proteins such as p85α (PI3-kinase) and downstream activation of Akt (Taniguchi et al., 2006). Akt triggers the translocation of the 12-transmembrane glucose transporter 4 (GLUT4) to the plasma membrane (PM) by phosphoinhibiting the Rab GTPase-activating protein (GAP) AS160 (Miinea et al., 2005). Among the many adaptors that relay signals within the insulin cascade, IRS1 is widely believed to serve as the major node for orchestrating metabolic insulin signaling (Taniguchi et al., 2006).
Besides IRS1, metabolic insulin signaling also relies on the activation of heterotrimeric G proteins, another major hub in eukaryotic signal transduction. InsRs are functionally coupled to the pertussis toxin–sensitive Gαi/o proteins, for example, insulin can trigger their activation (Rothenberg and Kahn, 1988; Ciaraldi and Maisel, 1989), localization (Gohla et al., 2007), and phosphorylation (O’Brien et al., 1987; Krupinski et al., 1988). Activation of Gi augments insulin sensitivity (Chen et al., 1997; Song et al., 2001), enhances tyrosine phosphorylation of both InsR and IRS1 (Moxham and Malbon, 1996), and triggers efficient translocation of GLUT4 storage vesicles (GSVs) to the PM (Ciaraldi and Maisel, 1989; Kanoh et al., 2000; Song et al., 2001). Although multiple clues consistently point to a critical role of Gi activation in the insulin response, what couples and activates Gi downstream of InsR and how such activation may cross-talk with IRS1-dependent insulin signaling and trigger downstream metabolic events remain unknown. Additionally, little is known about how G-protein pathways are altered in IR.
With regard to the pathogenesis of IR, suppression of metabolic insulin signaling via the IRS1/PI3K pathway is an invariable hallmark (Kahn and Flier, 2000; Pessin and Saltiel, 2000; Le Roith and Zick, 2001). Such suppression occurs via common mechanisms that involve cellular accumulation of lipid metabolites (acyl-CoAs, ceramides, diacyglycerol, etc), which activate, among many other kinases, the critical protein kinase C-theta (PKCθ; Griffin et al., 1999; Yu et al., 2002). PKCθ-dependent phosphoinhibition of IRS1 at Ser1-101 (Li et al., 2004) is considered an important event that triggers lipid-induced IR. PKCθ expression levels are increased in the skeletal muscles of obese diabetics and hold an inverse relationship to insulin sensitivity (Schmitz-Peiffer et al., 1997; Yu et al., 2002), and PKCθ−/− null mice demonstrate a protective effect against IR despite a high-fat diet (Kim et al., 2004). These studies and many others have shaped the paradigm that IR is triggered when IRS1 is phosphoinhibited by kinases like PKCθ. However, some recent studies have revealed inconsistencies in this paradigm (summarized in Hoehn et al., 2008). Emerging evidence indicates that IRS1 is insufficient for orchestrating the insulin response (Krook et al., 1996) and that multiple receptor tyrosine kinases (RTKs) can trigger IR independent of IRS1 (Hoehn et al., 2008). These studies raise the possibility that major unidentified signaling nodes exist within the insulin signaling cascade, whose inhibition via the fatty acid/PKCθ pathway triggers IR.
In this study, we define a single, multimodular signal transducer, GIV, as a critical node in metabolic insulin signaling. GIV is a guanine nucleotide exchange factor (GEF) that activates Gαi1/2/3 (Garcia-Marcos et al., 2009); contains a SH2-like domain that directly binds InsR (Lin et al., 2014); is a direct substrate of InsR, which phosphorylates GIV at Y1764 (Lin et al., 2011); is a bona fide enhancer of the PI3K-Akt pathway downstream of InsR and other RTKs (Lin et al., 2011); and is a substrate for PKCθ. The latter phosphorylates and inhibits signaling via the GIV-Gαi axis (Lopez-Sanchez et al., 2013). Furthermore, a recent study has indicated that GIV may serve as a major regulator of the metabolic insulin response in skeletal muscle (Hartung et al., 2013); overexpression of GIV in myoblasts leads to hyperphosphorylation of IRS1 and enhanced glucose uptake, whereas depletion of GIV suppresses both. Despite these insights, the molecular mechanisms that enable GIV to enhance the metabolic insulin-IRS1 response in physiology or mechanisms that derail this pathway in the setting of IR remained unknown. On the basis of its ability to cross-talk with all these key mediators of metabolic insulin signaling, we asked whether GIV is a key determinant of insulin sensitivity in physiology and whether its phosphoregulation by PKCθ triggers IR.
RESULTS
Activation of Gαi by GIV-GEF is required for glucose uptake in skeletal muscles
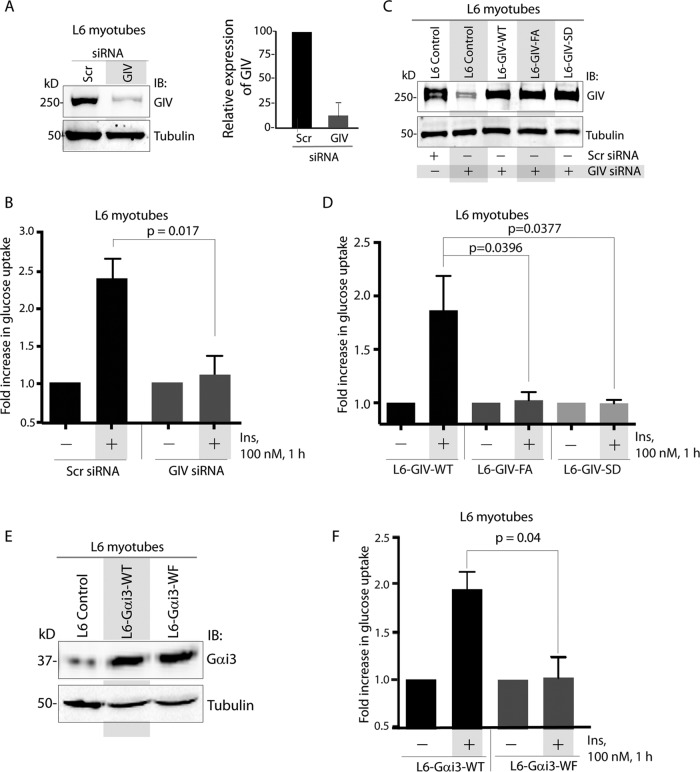
To determine the role of GIV-GEF in IR, we used differentiated L6 rat skeletal myotubes. Our rationale for this choice was guided by two facts: 1) although both adipocytes and skeletal muscles are sites for IR, full-length GIV is expressed more abundantly in skeletal muscles than in mature adipocytes (Uhlen et al., 2010); and 2) a recent study showed that levels of expression of GIV mRNA in skeletal muscle biopsies from normal subjects tracks with insulin sensitivity, as measured by a hyperinsulinemic–euglycemic clamp (Hartung et al., 2013). We found that depletion of GIV in L6 myotubes (by ∼80–85%; Figure 1A) reduced the efficiency of glucose uptake by ∼50% (Figure 1B), as determined by a well-established fluorometric assay (Yamamoto et al., 2006). This defect was rescued by stably expressing small interfering RNA (siRNA)-resistant wild-type GIV (GIV-WT) but not the GEF-deficient F1685A mutant of GIV (GIV-FA), which can neither bind nor activate Gαi (Garcia-Marcos et al., 2009; Figure 1, C and D). It is noteworthy that the levels of stable expression of GIV-WT or mutants in GIV-depleted L6 myotubes were similar to the levels of endogenous GIV in these cells (Figure 1C), indicating that the effects observed are not merely due to overexpression of GIV at nonphysiological levels. These findings indicate that GIV is required for glucose uptake in skeletal muscles and that its GEF domain is essential.
FIGURE 1:
Activation of Gαi by GIV-GEF is essential for glucose uptake in skeletal muscles. (A) Left, lysates of L6 myotubes treated either with control (Scr) or with GIV siRNA were analyzed for GIV and tubulin by IB. Right, bar graph displays efficiency of GIV depletion. (B) Control (Scr siRNA) and GIV-depleted (GIV siRNA) L6 myotubes were analyzed for glucose uptake after insulin stimulation by fluorometric assay. Bar graphs display fold change in uptake compared with starved controls (y-axis). Error bars represent mean ± SD; n = 3. (C) Control L6 myotubes or those stably expressing siRNA-resistant GIV-WT, GIV-FA, or GIV-SD were treated (+) or not (−) with either control (Scr) or GIV siRNA before lysis. Equal aliquots of whole-cell lysates were analyzed for GIV-FLAG expression by IB for GIV and tubulin. (D) L6 myotubes stably expressing siRNA-resistant GIV-WT, GIV-FA, or GIV-SD were depleted of endogenous GIV by siRNA as in C and analyzed for glucose uptake after insulin stimulation by fluorometric assay. Bar graphs display fold change in uptake compared with starved controls (y-axis). Error bars represent mean ± SD; n = 3. (E and F) L6 myotubes stably expressing Gαi3-WT and Gαi3-WF were analyzed for Gαi3 and tubulin by IB (E) and for glucose uptake (F). Bar graphs display fold change in uptake compared with starved controls (y-axis). Error bars represent mean ± SD; n = 3.
Next we asked whether phosphoinhibition of GIV's GEF at Ser-1689 by PKCθ (Lopez-Sanchez et al., 2013) also inhibits glucose uptake. We found that glucose uptake in cells expressing the constitutively phosphoinhibited S1689D mutant of GIV (GIV-SD) was half as efficient compared with those expressing GIV-WT (Figure 1, C and D), indicating that phosphoinhibition of GIV's GEF function by PKCθ impairs glucose uptake in response to insulin. These findings were reproduced in HeLa cells (Supplemental Figure S1), indicating that the effect of GIV-GEF we observe on glucose uptake may not be a restricted only to L6 myotubes but may represent a fundamental effect on IR.
To further pinpoint the impairment of Gi activation by GIV-GEF as the cause, we monitored glucose uptake in L6 myotubes stably expressing either wild-type (Gαi3-WT) or a dominant-negative W258F mutant of Gαi3, henceforth referred to as Gαi3-WF (Figure 1E), which cannot bind or be activated by GIV but localizes and interacts with Gβγ, G protein–coupled receptors (GPCRs), and Gαi regulators similar to Gαi3-WT (Garcia-Marcos et al., 2010). We analyzed Gαi3 (and not Gαi1/2), because it is the most abundant Gαi subunit expressed in skeletal muscles, as confirmed by proteomics (Hwang et al., 2010). Glucose uptake was reduced in cells expressing Gαi3-WF compared with those expressing Gαi3-WT (Figure 1F), confirming that GIV drives efficient glucose transport after insulin stimulation via its ability to activate Gαi proteins.
GIV binds ligand-activated InsRβ and modulates multiple tiers of metabolic insulin signaling via its GEF function
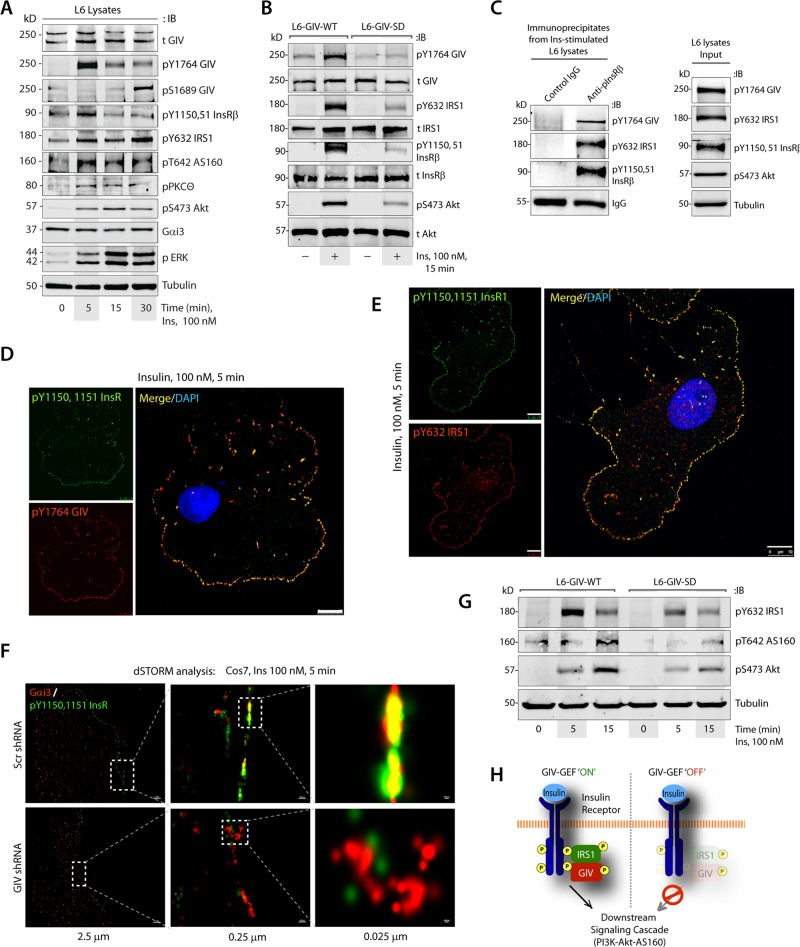
To determine how GIV's GEF function affects the insulin signaling cascade, we analyzed key components of metabolic insulin signaling in L6 myotubes responding to insulin. Immunoblotting (IB) for phosphoproteins revealed that insulin triggers activation of GIV at 5 min, coincides with peak autophosphorylation of InsRβ, and is followed by sustained phosphoactivation of IRS1 and Akt and phosphoinhibition of the GSV-associated Rab-GAP AS160 (Figure 2A); the latter is a key trigger step for exocytosis of GSVs (Miinea et al., 2005). Activation of PKCθ was initiated by 5 min and sustained up to 30 min, and inhibitory phosphorylation of GIV at S1689 by PKCθ peaked at 30 min. The time line of these events is consistent with the previously described role of this phospho event in the termination of GIV's GEF activity and disengaging GIV from Gαi (Lopez-Sanchez et al., 2013). A similar analysis comparing L6 myotubes expressing GIV-WT or GIV-SD revealed that phosphoinhibition of GIV's GEF activity by PKCθ affects several of these key upstream events. Compared with L6-GIV-WT cell lines, global suppression of the insulin response was encountered in L6-GIV-SD cells, starting with the most upstream event, that is, suppressed autophosphorylation of InsRβ at Y1150 and Y1151, which are essential for maximal phosphoactivation of substrate proteins (Flores-Riveros et al., 1989; Figure 2B). At the immediate postreceptor level, phosphoactivation of GIV and IRS1 was suppressed, and Akt phosphorylation downstream was impaired (Figure 2B). Similar findings were noted also in paired HeLa-GIV-WT versus HeLa-GIV-SD cells (Supplemental Figure S2A).
FIGURE 2:
GIV-GEF binds and enhances autophosphorylation of InsRβ and downstream metabolic insulin response. (A) Lysates of serum-starved L6 myotubes stimulated with insulin were analyzed for various components of metabolic insulin signaling by IB. (B) Lysates of serum-starved or insulin-stimulated L6 myotubes stably expressing GIV-WT or GIV-SD were analyzed for activation of GIV, IRS1, InsRβ, and Akt by IB. (C) Immunoprecipitation was carried out on lysates (right) of insulin-treated L6 myotubes with anti-pInsRβ or control immunoglobulin G (IgG). Bound immune complexes (left) were analyzed for activated GIV, IRS1, and InsRβ by IB. (D and E) Serum-starved Cos7 cells were stimulated with insulin, fixed, and subsequently stained for active GIV (pY1764-GIV; red, D), active IRS1 (pY632-IRS1; red, E), active InsRβ (pY1150/51-InsRβ; green), and 4′,6-diamidino-2-phenylindole (DAPI)/DNA (blue). Scale bar: 10 μm. (F) Serum-starved control (Scr shRNA) or GIV-depleted (GIV shRNA) Cos7 cells were stimulated with insulin, fixed, and stained for active InsRβ (pY1150/51-InsRβ; green) and Gαi3 (red) and analyzed by dSTORM microscopy. A high degree of colocalization was observed, as determined by the presence of yellow pixels in the merged images. (G) Lysates of starved and insulin-stimulated L6 myotubes stably expressing GIV-WT or GIV-SD were analyzed for activation of IRS1, AS160, Akt, and tubulin by IB. (H) Schematic illustrating how the presence or absence of a functional GIV-GEF, via which GIV links and activates Gi in the vicinity of InsRβ, dictates the intensity of metabolic insulin signaling, beginning with the activation and autophosphorylation of InsRβ.
Previous work showed that GIV's C-terminal SH2-like domain directly binds autophosphorylated cytoplasmic tails of multiple RTKs, including InsRβ (Lin et al., 2014; Midde et al., 2015). In L6 myotubes, active GIV[pY1764] coimmunoprecipitated with ligand-activated InsRβ-IRS1 complexes (Figure 2C). Active GIV[pY1764] also colocalized with the autophosphorylated InsRβ at PM microdomains (Figure 2D), where activated IRS1 adaptors coexist (Figure 2E). These findings suggest that the InsRβ-GIV-IRS1 complexes we observe in Figure 2C are likely assembled at the PM. To determine whether GIV links Gαi proteins to ligand-activated InsRβ at these PM microdomains, we used direct stochastic optical reconstruction microscopy (dSTORM). This mode of imaging achieves a spatial resolution of ∼25 nm, and a high degree of colocalization between endogenous proteins indicates they are likely to interact (Huang et al., 2010). In control cells, but not in GIV-depleted cells, Gαi3 and ligand-activated InsRβ showed a high degree of colocalization along the PM (Figure 2F, yellow pixels), indicating that active InsRβ and Gαi3 come within proximity to each other exclusively in the presence of GIV-GEF. As expected, the impairment of autophosphorylation of InsRβ in GEF-deficient L6-GIV-SD mutant cells was also accompanied by defects in downstream activation of Akt and phosphoinactivation of its target Rab-GAP, AS160 (Figure 2G and Supplemental Figure S2B). Taken together, these results demonstrate that GIV binds to ligand-activated InsRβ-IRS1 complexes on microdomains at the PM and links Gαi to such complexes. The presence or absence of a functional GIV-GEF, via which GIV links and activates Gi in the vicinity of RTKs, appears to be a key determinant of whether multiple tiers within the metabolic insulin signaling cascade are activated maximally, beginning with the autophosphorylation and activation of InsRβ (Figure 2H).
GIV directly binds and modulates the localization and functional phosphorylation of IRS1
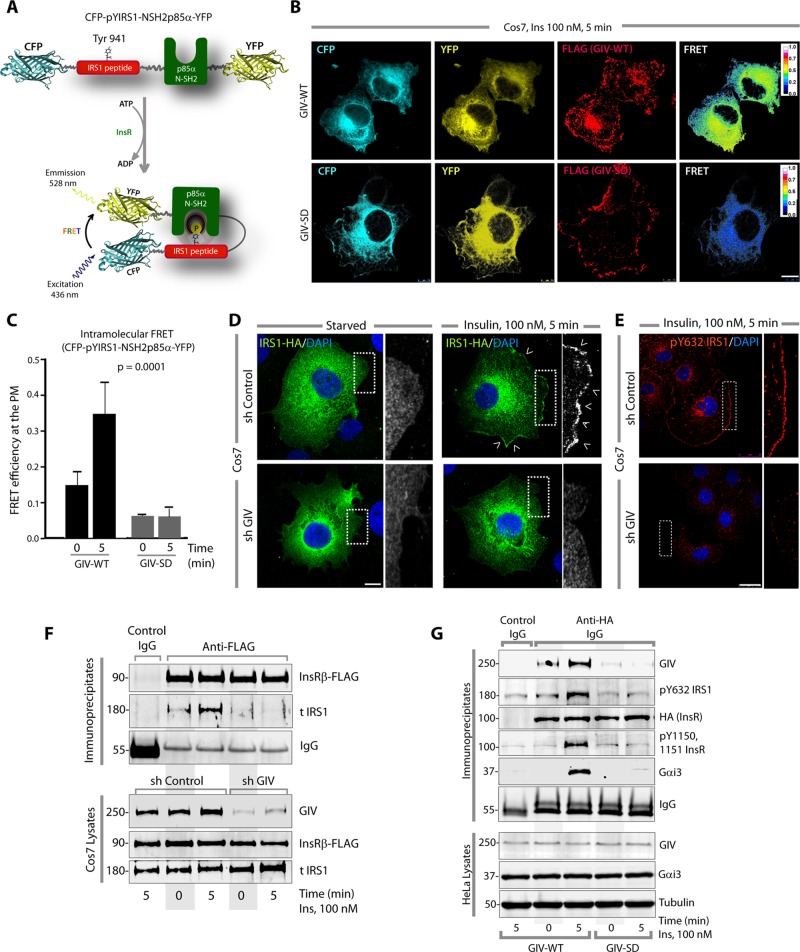
Next we investigated how GIV may affect the phosphorylation/activation of IRS1, which is a major adaptor for the metabolic insulin responses. Because the hypophosphorylation of IRS1 observed by immunoblotting lysates of L6 myotubes and HeLa cells expressing the GEF-deficient SD phosphomimetic mutant does not provide enough information about the spatial and temporal dynamics of IRS1 phospho-dephosphorylation in cells, we used a genetically encoded fluorescent biosensor, phocus-2nes (Sato et al., 2002) in fluorescence resonance energy transfer (FRET) studies. This biosensor shows energy transfer only when Y941 on IRS1 is phosphorylated and presents a docking site for the N-SH2 domain of p85α (PI3K), thereby providing a readout of the function of such phosphorylation (Figure 3A). In cells expressing GIV-WT, we observed a significant increase in FRET efficiency at/near the PM (F.E. 0.34 ± 0.08) within 5 min after insulin stimulation; however, in cells expressing GIV-SD, that response was blunted (F.E. 0.06 ± 0.03; Figure 3, B and C, and Supplemental Figure S3A), confirming that phosphoinhibition of GIV-GEF impairs functional phosphorylation of IRS1 at the PM. Because functional phosphorylation of IRS1 involves several steps, we asked whether GIV is required for the two earliest ones, that is, translocation of IRS1 from cytosol to the PM and its subsequent phosphorylation at that location in response to insulin. Compared with control cells, both steps were impaired in GIV-depleted cells (Figure 3, D and E, and Supplemental Figure S3, B and C). Coimmunoprecipitation studies on control or GIV-depleted cells further confirmed that recruitment of IRS1 to the activated InsRβ was impaired in the absence of GIV (Figure 3F). To further pinpoint whether GIV's GEF function is essential for the recruitment of IRS1 to ligand-activated InsR, we first looked for insulin-triggered translocation of IRS1 from the cytosol to the PM in Cos7 cells expressing WT or SD GIV mutant (Supplemental Figure S3D). We found that IRS1 localized to the PM in cells expressing GIV-WT exclusively after ligand stimulation, whereas localization at the PM was suppressed in cells expressing GIV-SD. Next we analyzed receptor-bound complexes by immunoprecipitation assays in GIV-depleted HeLa cells stably expressing WT or SD GIV mutant (Figure 3G). In HeLa-GIV-WT cells, ligand stimulation triggered robust autophosphorylation of InsR (pY1150, 1151), which coincided with the recruitment of pYIRS1, GIV, and Gαi3 (Figure 3G). Consistent with our prior observations in L6-GIV-SD cells (Figure 2B), autophosphorylation of InsR was suppressed also in HeLa-GIV-SD cells, and receptor-bound complexes (InsR-GIV-G protein or InsR-IRS1) were decreased. These results not only confirm the role of GIV's GEF function in enhancing the recruitment of IRS1 to ligand-activated InsR but also demonstrate the inhibitory effect of pS1689 GIV on both InsR-IRS1 and InsR-Gαi3 complexes.
FIGURE 3:
GIV-GEF directly binds and regulates the localization and activation of IRS1. (A) Schematic for the biosensor phocus-2nes is shown. Energy transfer from CFP to YFP occurs only when Y941 is phosphorylated and the N-SH2 domain of p85α binds the phosphotyrosine ligand. (B and C) Serum-starved Cos7 cells coexpressing phocus-2nes with either GIV-WT-FLAG or GIV-SD-FLAG were stimulated with insulin, fixed, stained for FLAG (far red), and analyzed for FRET using confocal microscopy. Images panels display (from left to right, B) CFP, YFP, FLAG (GIV), and intensities of acceptor emission due to FRET in each pixel 5 min after insulin stimulation. Image panels of serum-starved (0 min) cells are shown in Supplemental Figure S3A. Bar graph (C) displays the FRET efficiency observed in GIV-WT versus GIV-SD cells at 0 and 5 min. The analysis represents five regions of interest from 4 to 6 cells/experiment (three independent experiments). Error bars = mean ± SD. (D) Serum-starved control (sh Control) or GIV-depleted (sh GIV) Cos7 cells expressing IRS1-HA were stimulated with insulin, fixed, stained for HA (green) and DAPI/DNA (blue), and analyzed by confocal microscopy. Insets show the magnification of the boxed regions. Scale bar: 10 μm. Arrowheads denote PM. (E) Serum-starved control (sh Control) or GIV-depleted (sh GIV) Cos7 cells were stimulated with insulin, fixed, stained for endogenous pY632-IRS1 (red) and DAPI/DNA (blue), and analyzed by confocal microscopy. Insets show the magnification of the boxed regions. Scale bar: 10 μm. (F) Immunoprecipitation was carried out on lysates of starved or insulin-stimulated control (sh Control) or GIV-depleted (sh GIV) Cos7 cells expressing InsRβ-FLAG. Bound immune complexes were analyzed for IRS1, InsRβ (FLAG), and IgG by IB. IRS1 coimmunoprecipitated with InsRβ in control cells but not in GIV-depleted cells. (G) GIV-depleted HeLa cells stably expressing GIV-WT or GIV-SD were transiently transfected with InsR-HA, starved, and stimulated with 100 nM insulin for 5 min before lysis. InsR and receptor-bound complexes were immunoprecipitated by incubating equal aliquots of lysates with anti-HA mAb or contro IgG, followed by protein G beads. Immune complexes were analyzed for GIV, InsR (HA), ligand-activated InsR (pY1150, 1151 InsR), pY632 IRS1, and Gαi3 by IB. Equal loading of lysates was confirmed by analyzing GIV, Gαi3, and tubulin by IB. Maximal autophosphorylation of InsR and recruitment of GIV, IRS1, and Gαi3 to the receptor was observed in cells expressing GIV-WT exclusively after insulin stimulation, but not in cells expressing GIV-SD.
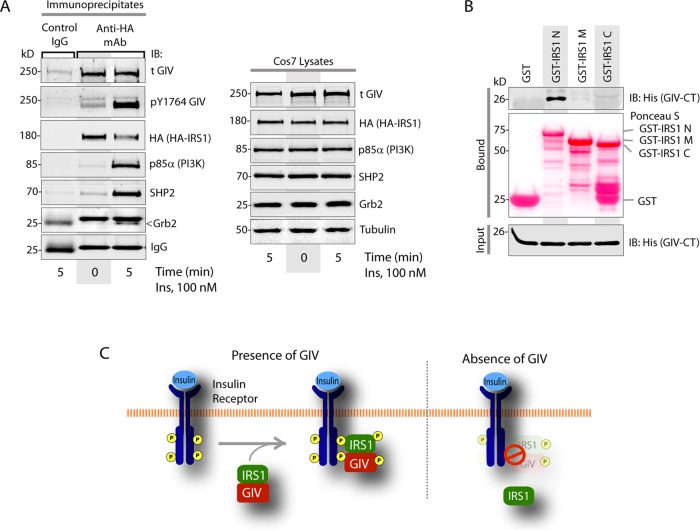
Because of the global effect of GIV depletion we observed on IRS1 localization, recruitment, and phosphoactivation, we next asked whether GIV binds IRS1. GIV coimmunoprecipitated with IRS1 before and after insulin stimulation (Figure 4A), indicating that the GIV-IRS1 interaction is constitutive. Pull-down assays with recombinant proteins showed that His-GIV-CT specifically bound the N-terminus of IRS1, demonstrating that the interaction is direct (Figure 4B). Furthermore, both WT and SD mutant GIV proteins bound IRS1 equally (Supplemental Figure S4, A and B), indicating that phosphorylation of GIV at S1689 by PKCθ does not impair GIV's ability to bind IRS1 and suggesting that GIV may bind IRS1 via a domain that is distinct from its GEF module. Despite the fact that GIV-SD retains its ability to bind IRS1, localization and activation of IRS1 were impaired in GIV-SD cells (Figure 3, B and C, and Supplemental Figure S3D), suggesting that the GIV-IRS1 interaction may serve a different role independent of the observed effects of phosphoinhibition of GIV-GEF on IRS1 signaling. We propose that the GIV-IRS1 interaction may serve as a scaffold for the stabilization of InsRβ(RTK)/GIV/IRS1 ternary complexes at the PM, within which GIV's GEF function may modulate the phosphoactivation of IRS1 downstream of growth factors (Figure 4C).
FIGURE 4:
GIV directly and constitutively binds IRS1. (A) Immunoprecipitation was carried out on lysates (right) of starved or insulin-treated Cos7 cells expressing IRS1-HA. Lysates and bound immune complexes (left) were analyzed for activated GIV (pY1764-GIV), total (t)GIV, IRS1 (HA), p85α, SHP2, and Grb2 by IB. (B) Pull-down assays were carried out with recombinant His-GIV-CT and GST-tagged domains of IRS1 (see the Supplemental Material) immobilized on glutathione beads. Bound (top) and input (bottom) proteins were analyzed for His-GIV-CT by IB with His mAb. (C) Schematic summarizing how the presence or absence of GIV affects localization and phosphoactivation of IRS1.
GIV-GEF is a target for the antagonist actions of fatty acids and insulin sensitizers
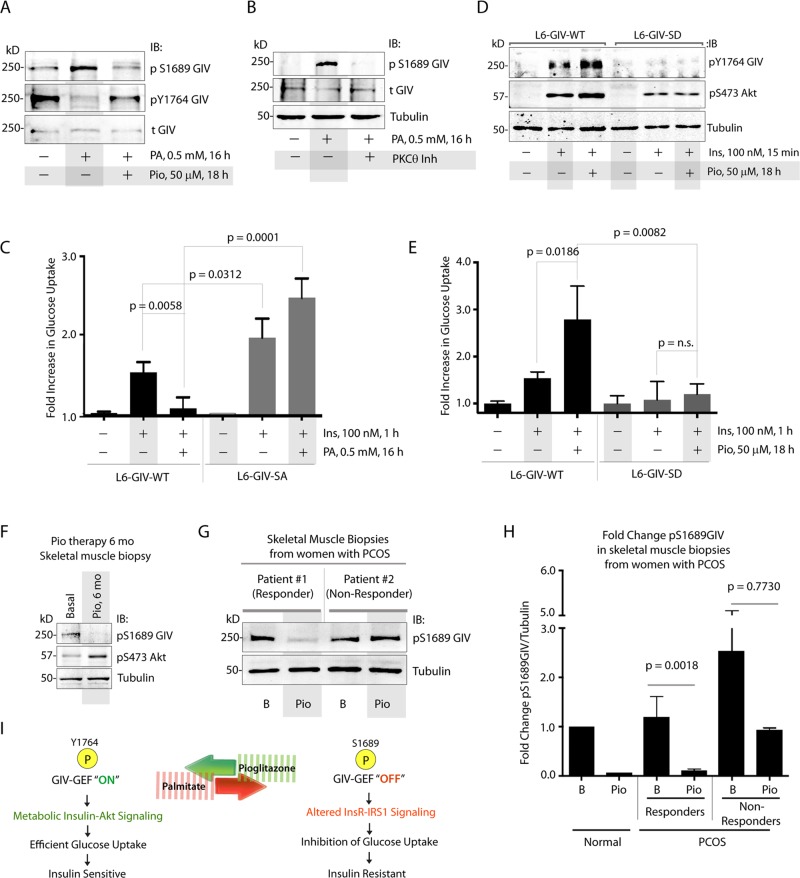
Because PKCθ is the kinase that orchestrates lipid-induced IR (Haasch et al., 2006), we next asked whether fatty acids induce IR in part by phosphoinhibition of GIV-GEF at S1689 by PKCθ. When we induced IR in L6 cells using albumin-conjugated sodium palmitate (PA), which is known to activate PKCθ (Griffin et al., 1999), we found that phosphorylation of GIV at S1689 was enhanced, GIV's ability to bind Gi was reduced, and phosphorylation of GIV at Y1764 was suppressed (Figure 5A and Supplemental Figure S5A), indicating that PA induces phosphoinhibition of GIV-GEF and concomitantly suppresses tyrosine-based signaling via GIV. PA requires PKCθ to exert such phosphoinhibition, because inhibition of PKCθ abolished phosphoinhibition of GIV-GEF (Figure 5B). When PA-treated, insulin-resistant L6 cells were incubated with pioglitazone (Pio), an insulin sensitizer in the thiazolidinedione (TZD) class of drugs, phosphorylation of GIV at S1689 was reduced, and tyrosine phosphorylation of GIV was enhanced, indicating that Pio antagonized both the effects of PA and effectively reversed the phosphoinhibition of GIV-GEF by PKCθ (Figure 5A). Consistent with its role as a true insulin sensitizer that improves insulin action in peripheral tissues, Pio also enhanced tyrosine phosphorylation of GIV and Akt signaling triggered by insulin in insulin-sensitive L6 cells never exposed to PA (Supplemental Figure S5, B and C).
FIGURE 5:
Phosphoinhibition of GIV-GEF by PKCθ is required for PA-induced IR and dephosphorylation of GIV-GEF is essential for the action of Pio. (A) Lysates of L6 myotubes treated (+) or not (−) with PA alone or a combination of PA and Pio were analyzed for phosphorylation of GIV at S1689 and Y1764 and total (t)GIV by IB. (B) Lysates of L6 myotubes treated (+) or not (−) with PA alone or a combination of PA and a pseudosubstrate PKCθ inhibitor were analyzed for phosphorylation of GIV at S1689 (pS1689 GIV), total (t)GIV, and tubulin by IB. (C) L6 myotubes stably expressing siRNA-resistant GIV-WT or GIV-SA were depleted of endogenous GIV by siRNA, treated with PA (+) or vehicle control (−), and subsequently analyzed for insulin-stimulated glucose uptake by fluorometric assay. Bar graph displays fold change in glucose uptake compared with starved controls (y-axis). Error bars represent mean ± SD; n = 3. (D) L6 myotubes stably expressing siRNA-resistant GIV-WT or GIV-SD were depleted of endogenous GIV by siRNA, treated (+) or not (−) with Pio, and subsequently stimulated with insulin before lysis. Lysates were analyzed for activation of GIV (pY1764 GIV) and Akt (pS473Akt) by IB. (E) L6 myotubes stably expressing siRNA-resistant GIV-WT or GIV-SD were depleted of endogenous GIV by siRNA, treated (+) or not (−) with Pio, and subsequently analyzed for insulin-stimulated glucose uptake by fluorometric assay. Bar graph displays fold change compared with starved controls (y-axis). Error bars represent mean ± SD; n = 3. (F) Equal aliquots of lysates of vastus lateralis muscle biopsies from obese T2DM subjects, obtained before (basal) or after 6 mo of Pio therapy, were analyzed for phosphoinhibition of GIV-GEF (pS1689 GIV) and phospho Akt (pS473 Akt) by IB. Representative samples are shown (n = 8). (G and H) Equal aliquots of lysates of vastus lateralis muscle biopsies from patients with PCOS, obtained before (basal) and after Pio therapy, were analyzed for pS1689GIV by IB. (G) A representative immunoblot of biopsies obtained from “responder” and “nonresponder” patients are shown (n = 8). Bar graph displays fold change in GIV phosphorylation at S1689 observed in normal and PCOS patients before and after Pio treatment (y-axis) (H). B, basal; Pio, pioglitazone treatment. Error bars represent mean ± SD. (I) Schematic illustrates our proposed model for GIV's role as a pivot for the antagonistic actions of fatty acids like palmitate that trigger IR (red arrow) and insulin sensitizers like Pio that reverse IR (green). Phosphorylation at S1689 is essential for PA to induce IR, whereas dephosphorylation is required for Pio to enhance tyrosine phosphorylation of IRS1 and GIV, restore Akt signaling, and reinstate insulin sensitivity.
We then investigated whether phosphoinhibition of GIV-GEF by PKCθ in L6 myotubes plays a role in mediating the antagonistic effects of PA and Pio in the induction and reversal of IR, respectively. PA induced IR in L6-GIV-WT cells, as determined by a blunted glucose-uptake response to insulin (Figure 5C). However, L6 cells expressing a nonphosphorylatable GIV S1689A mutant (L6-GIV-SA) were resistant to PA, that is, these cells remained sensitive to insulin regardless of PA treatment and demonstrated higher glucose uptake compared with L6-GIV-WT cells (Figure 5C). These results demonstrate that the selective inhibition of GIV-GEF by PKCθ via phosphorylation of a single Ser-1689 is an essential mechanism by which PA triggers IR in L6 myotubes. As for Pio, we found that it reinstated insulin signaling in PA-treated, insulin resistant L6-GIV-WT cells, as determined by restored tyrosine phosphorylation of GIV and Akt signaling (Figure 5D and Supplemental Figure S5D). However, L6 cells expressing a constitutively phosphoinhibited GIV SD mutant (L6-GIV-SD) were resistant to Pio, that is, these cells showed no discernible enhancement of signaling compared with L6-GIV-WT cells (Figure 5D and Supplemental Figure S5D). Furthermore, Pio reversed the PA-induced IR state in L6-GIV-WT cells, but not in L6-GIV-SD cells, as determined by glucose uptake after insulin stimulation (Figure 5E). Because Pio is known to improve insulin sensitivity in muscle tissue in part by antagonizing the activity of protein kinases such as PKCθ (Markova et al., 2010), our results demonstrate that reversal of phosphoinhibition of GIV-GEF by PKCθ on Ser-1689 is an essential mechanism via which Pio reverses IR and sensitizes L6 myotubes to the action of insulin. The inability to reverse such phosphoinhibition (as in the case of the GIV-SD mutant, which mimics a constitutive phosphoinhibited state) makes cells nonresponsive to the insulin-sensitizing actions of Pio.
The physiological significance of these observations in cultured L6 myotubes was confirmed by findings in patients with IR, in whom chronic treatment with Pio reduced the phosphoinhibition of GIV-GEF and enhanced phosphorylation of Akt in skeletal muscles (vastus lateralis) of obese T2DM patients (Figure 5F). Moreover, patients with polycystic ovarian syndrome (PCOS) in whom IR was clinically reversed by Pio therapy, that is, responders (as determined by 24-h glucose levels and glucose disposal rate [GDR] determined by a hyperinsulinemic–euglycemic clamp) showed a significant reduction in phosphoinhibition of GIV at S1689 in their muscles. By contrast, PCOS patients who failed the Pio treatment trial (i.e., nonresponders) had high pretreatment and/or posttreatment levels of phosphoinhibition of GIV-GEF (Figure 5, G and H). Taken together, our results demonstrate that a single phospho event (PKCθ-dependent phosphorylation of GIV at S1689), which selectively inhibits GIV-GEF and therefore abolishes activation of Gi by GIV, is a common pivot point for both PA and Pio to exert their antagonistic actions in IR. Phosphorylation at S1689 is essential for PA to induce IR, whereas dephosphorylation is required for Pio to enhance tyrosine phosphorylation of IRS1 and GIV, restore Akt signaling, and reinstate insulin sensitivity (Figure 5I).
Cell-permeant GIV-derived peptides can effectively reverse IR in skeletal muscle
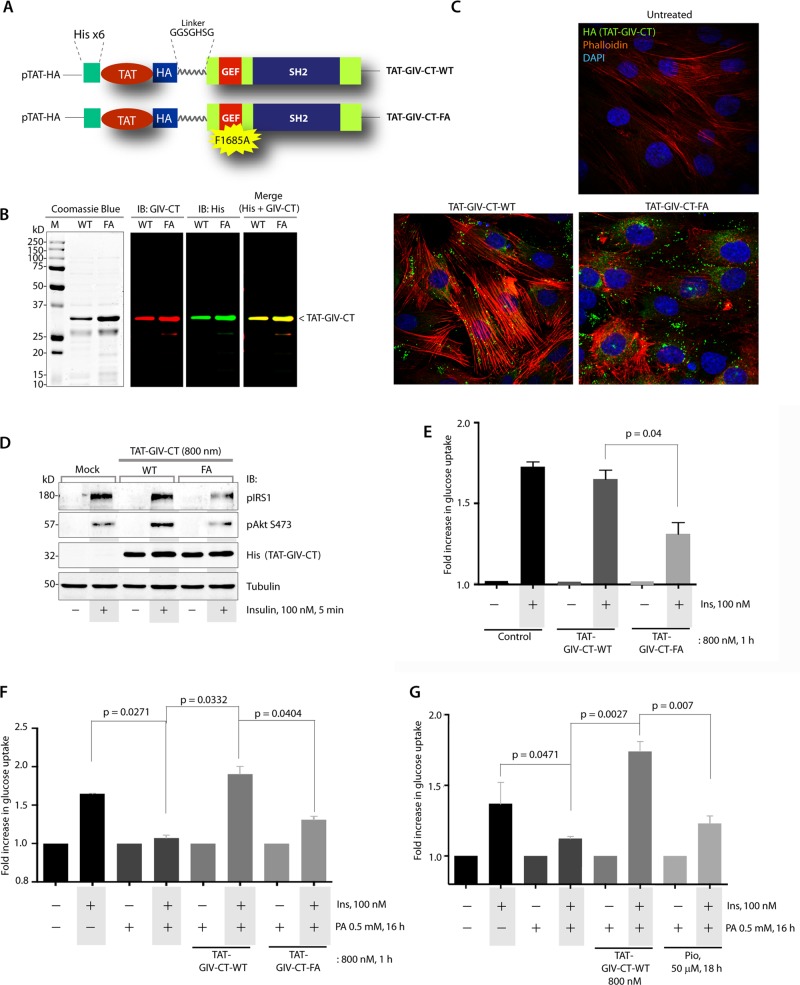
We next asked whether GIV-GEF can serve as a therapeutic target for exogenous modulation of IR. To investigate this, we used recently validated recombinant, cell-permeant TAT-tagged GIV-CT peptides (WT and GEF-deficient FA mutant peptides spanning GIV's GEF and SH2-like domains; Figure 6, A and B). These peptides offer a nongenetic approach for exogenous manipulation of GIV-GEF–dependent signaling programs and cellular phenotypes in diverse cells and a variety of pathophysiological processes (Ma et al., 2015). L6 myotubes homogeneously took up TAT peptides (∼90% efficiency of uptake; Figure 6C). Uptake of GIV-CT-WT peptides was associated with enhancement of stress-fiber formation and phosphorylation of IRS1 and Akt proteins in response to insulin (Figure 6, C and D). However, uptake of GIV-CT-FA peptides disrupted the actin stress fibers, as shown previously in other cell lines, and suppressed IRS1 and Akt phosphorylation in response to insulin. Consistent with these signaling programs, insulin-stimulated glucose uptake in the basal state was unaffected by GIV-CT-WT peptides but was significantly inhibited by GIV-CT-FA peptides (Figure 6E). GIV-CT-WT, but not the FA mutant peptides, effectively reversed PA-induced IR (Figure 6F), and ∼800 nM of WT peptide was as effective as 50 μM Pio in reversing such IR (Figure 6G). These studies demonstrate that cell-permeant GIV-CT peptides can enhance metabolic insulin signaling and reverse IR effectively in a GEF-dependent way.
FIGURE 6:
Cell-permeant TAT-GIV-CT-WT peptides, but not FA mutant peptides, effectively reverse lipid-induced IR in skeletal muscles. (A) Design of the cell-permeant TAT-GIV-CT peptides is shown. TAT-PTD was fused to His and HA tags and coupled via a linker (GGSGHSG) to the C-terminus of GIV (aa 1660–1870). (B) Purified recombinant TAT-GIV-CT peptides were analyzed by Coomassie blue staining and by IB with anti GIV-CT and anti-His antibodies. (C) L6 myotubes were treated with TAT-GIV-CT-WT or FA peptides and cultured overnight in low serum conditions (0.2% fetal bovine serum) before fixation. Fixed cells were stained for His (green), phalloidin (F-actin, red), and DAPI/DNA (blue) and analyzed by confocal microscopy. (D) L6 myotubes were treated with TAT-GIV-CT-WT or FA peptides, starved, and stimulated with insulin before lysis. Equal aliquots of lysates were analyzed for transduction of TAT-peptides with anti-His antibody, activation of IRS1 (pY632-IRS1) and Akt (pS473) by IB. (E) L6 myotubes were treated with TAT-GIV-CT-WT or FA peptides, starved, and subsequently analyzed for insulin-stimulated glucose uptake by fluorometric assay. Bar graph displays fold change in uptake compared with starved controls (y-axis). Error bars represent mean ± SD. (F) L6 myotubes were treated (+) or not (–) with PA to induce IR, then transduced with TAT-GIV-CT-WT or FA peptides, and subsequently analyzed for insulin-stimulated glucose uptake by fluorometric assay. Bar graph displays fold change in uptake compared with starved controls (y-axis). Error bars represent mean ± SD. (G) L6 myotubes were treated (+) or not (–) with PA to induce IR, then either treated with Pio or transduced with TAT-GIV-CT-WT peptides (as indicated), and subsequently analyzed for insulin-stimulated glucose uptake by fluorometric assay. Bar graph displays fold change in uptake compared with starved controls (y-axis). Error bars represent mean ± SD.
DISCUSSION
Phosphoinhibition of GIV-GEF by PKCθ triggers lipid-induced IR; dephosphorylation of GIV-GEF reinstates insulin sensitivity
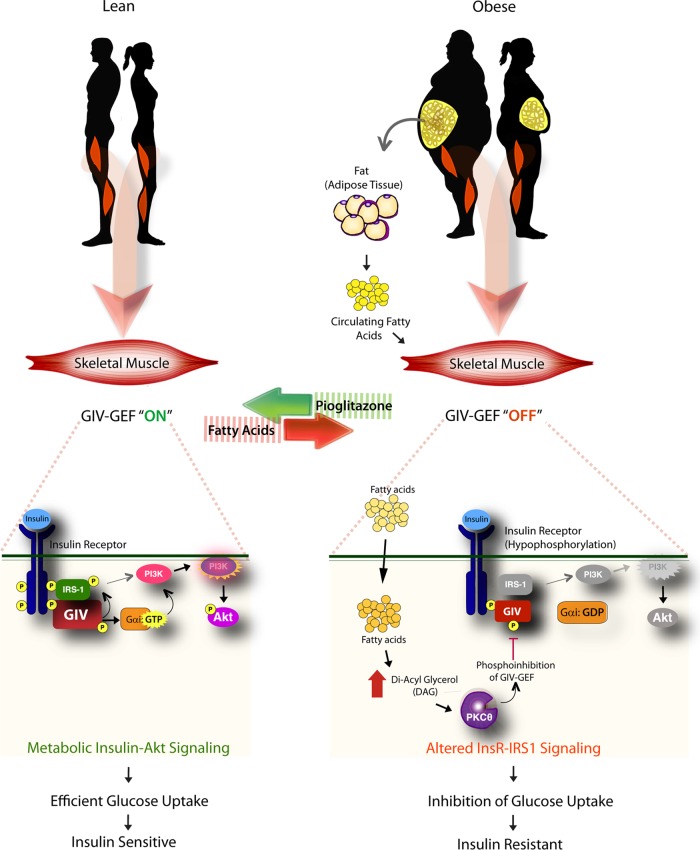
The fundamental discovery in this work is that GIV-GEF plays a major role as a dominant conduit for insulin response in the skeletal muscle, and identification of key phospho events that allow this GEF to serve as a decisive pivot/node for cellular insulin response in physiology and disease (see Figure 7). In lean individuals, insulin triggers activation of GIV by tyrosine phosphorylation (pY), GIV's GEF function is turned “on,” and Gαi is activated, metabolic insulin signaling is initiated through the InsR/IRS1/PI3K/Akt signaling axis, culminating in efficient exocytosis of GSVs and subsequent uptake of glucose. In the obese, circulating free fatty acids trigger the accumulation of diacyl glycerol (DAG) and activation of PKCθ in skeletal muscle, which in turn phosphorylates GIV's GEF motif at S1689 and selectively turns “off” the GEF function. Consequently, Gαi remains inactive, and a majority of the key elements of metabolic insulin signaling are suppressed, thereby triggering IR.
FIGURE 7:
Schematic summarizing how GIV-GEF is a pivotal node for metabolic insulin response in lean normals (left) and for lipid-induced IR in the obese (right). Left, in lean individuals, insulin triggers tyrosine phosphorylation and activation of GIV (pY1764), GIV's GEF function is “on” and Gαi is activated. Metabolic insulin signaling is enhanced through the InsR/IRS1/PI3K/Akt/AS160 signaling axis, resulting in efficient exocytosis of GSVs and rapid uptake of glucose. Right, in the obese, circulating free fatty acids trigger the accumulation of DAG, and PKCθ is activated. PKCθ phosphorylates GIV at S1689 and turns “off” its GEF function. Consequently, Gαi remains inactive, and the InsR/IRS1/PI3K/Akt/AS160 signaling cascade is suppressed, thereby triggering IR.
We also provide evidence that reversible phosphorylation of GIV at S1689 by PKCθ and the ability of this single phospho event to modulate the InsR-GIV-Gαi signaling axis are critical determinants of cellular insulin responses. We demonstrated that this phospho event alone is sufficient to mimic fatty acid–induced IR and that fatty acids require such phosphorylation to induce IR. Although this study dissected the interplay between GIV and PKCθ in lipid-induced IR, because GIV can intercept signaling downstream of multiple classes of receptors (GPCRs and RTKs) and non-RTKs alike (reviewed in Garcia-Marcos et al., 2015) and is an enhancer of STAT3 as well as its transcriptional target (Dunkel et al., 2012), it is possible that GIV is a central node for other major triggers of IR, that is, inflammation, suppression of adiponectin, leptin resistance, and so on, which also require activation of PKCθ (Itani et al., 2000; Lin et al., 2000; Shulman, 2000; Anderson et al., 2006) and/or the JAK-STAT3 pathway (Mashili et al., 2013; Wunderlich et al., 2013).
We also show that TZDs like Pio release the phosphoinhibition on GIV-GEF and restore its function. Such restoration was essential for TZD action, because TZDs failed to reverse IR and reinstate insulin sensitivity in cells expressing the constitutively phosphomimic GIV-S1689D mutant. Because chronic TZD therapy does not suppress PKCθ (Markova et al., 2010), the reduction in levels of GIV phosphorylation at S1689 we observed after TZD therapy is likely to be a consequence of dephosphorylation by one of the many S/T phosphatases that are activated by TZDs in a PPARγ-dependent manner (Altiok et al., 1997; Pugazhenthi and Khandelwal, 1998; Sharma et al., 2004; Cho et al., 2006). Regardless of the mechanism(s) involved, our results indicate that GIV is a major target of TZDs that can account, in part, for TZD action on skeletal muscle. We conclude that reversible phosphorylation at S1689 and inhibition of the GEF function, via which GIV activates Gαi, serves as a molecular switch for flipping skeletal muscles between insulin-sensitive and insulin-resistant states. Because GIV specifically binds Gαi and not Gαq/11 (Le-Niculescu et al., 2005), these findings do not account for the previously described role of yet another G protein, Gαq/11, in insulin response (Imamura et al., 1999).
GIV's GEF function modulates several tiers within the metabolic insulin signaling cascade
We demonstrated that activation of Gαi by GIV impacts many tiers within the insulin signaling cascade and that phosphoinhibition of GIV's GEF function antagonizes them all. We previously showed that GIV directly binds autophosphorylated cytoplasmic tails of ligand-activated InsR via its C-terminal SH2-like module (Lin et al., 2014). Both the SH2-like module and GIV's GEF functions are critical for coupling of G proteins to ligand-activated InsR (Garcia-Marcos et al., 2011; Lin et al., 2014; Midde et al., 2015). We show here that, at the level of the receptor, activation of Gαi via GIV's GEF motif is required for maximal autophosphorylation and activation of InsRβ and recruitment and phosphoactivation of its major substrate, IRS1. The sites of autophosphorylation on InsRβ that GIV enhanced, Y1150 and Y1151, are required for maximal activation of the InsRβ kinase (White et al., 1988), and a failure to activate InsRβ kinase in skeletal muscles has been implicated in IR (Maegawa et al., 1991; Nolan et al., 1994; Goodyear et al., 1995). Although it is unclear how activation of Gαi by GIV may enhance receptor autophosphorylation, it is not entirely surprising, because activation of Gαi has previously been implicated in the enhancement of InsR autophosphorylation (Kreuzer et al., 2004) and because we previously showed that activation of Gαi by GIV's GEF function can similarly enhance autophosphorylation of yet another RTK, EGFR (Ghosh et al., 2010). In both instances, suppression of protein tyrosine phosphatases (PTPs) has been implicated as the mechanism for enhanced receptor autophosphorylation (Moxham and Malbon, 1996; Lin et al., 2014). Because GIV directly binds ligand-activated InsRβ (Lin et al., 2014) and triggers the formation of InsRβ-Gαi complexes at the PM, it is possible that the formation of such InsRβ-GIV-Gαi complexes suppresses the recruitment and/or activation of key PTPases. We conclude that the GIV-Gαi axis enhances cellular insulin response by increasing InsRβ kinase activity and autophosphorylation, two upstream events in insulin signaling.
At the immediate postreceptor level, we demonstrate that GIV binds and modulates the functions of IRS1. Activation of Gαi by GIV enhanced the recruitment of IRS1 to the ligand-activated receptors at the PM, triggered robust tyrosine phosphorylation at Y632 and Y941 on IRS1, and enhanced the formation of IRS1-p85α(PI3K) complexes. Unlike the ligand-dependent nature of InsRβ-GIV or InsRβ-IRS1 (Sun et al., 1991) interactions, the GIV-IRS1 interaction was constitutive. We also provide evidence that this binding was direct and that it involved the C-terminal region of GIV and the N-terminal region of IRS1. The latter contains a phosphotyrosine-binding domain (PTB) that is responsible for the direct interaction of IRS1 with InsRβ (Eck et al., 1996). Our findings of InsRβ-GIV-IRS1 complexes upon insulin stimulation suggest that GIV may bind IRS1 at a distinct site where the autophosphorylated cytoplasmic tail of InsRβ docks within the PTB. The enhanced recruitment of IRS1 to InsRβ in the presence of GIV suggests that GIV may serve as a signal amplifier at the immediate postreceptor level by facilitating the recruitment of more IRS1 adaptors per activated InsR. Based on recent experimental evidence that questions the exclusivity of IRS1 for InsRβ (Knowlden et al., 2008) and that shows GIV is capable of binding multiple RTKs (e.g., EGFR, PDGFR, VEGFR; Lin et al., 2014; Lopez-Sanchez et al., 2014), it is possible that GIV provides the necessary molecular basis for IRS1 to serve as a common conduit for metabolic response observed downstream of receptors other than InsRβ.
Although the precise mechanism of GIV-IRS1 interaction remains uncertain, this interaction adds GIV to the lengthy list of proteins that IRS1 scaffolds within the insulin signaling cascade (White, 2006). The finding that GIV enhanced tyrosine phosphorylation of IRS1 is consistent with the concomitant increase in the kinase activity of InsRβ and enhanced recruitment of IRS1 to the PM, the latter is a prerequisite for maximal tyrosine phosphorylation of IRS1 (Myers et al., 1995; Voliovitch et al., 1995). We conclude that GIV is required for maximal PM recruitment and tyrosine phosphorylation of IRS1, both key events implicated in metabolic insulin signaling via IRS1. In doing so, and by virtue of its ability to directly bind and bring together several other components of the metabolic insulin response (InsR, IRS1, G proteins, actin, PI3K, Akt), GIV serves as an integral hub at the immediate postreceptor that fine-tunes IRS1-dependent metabolic insulin signaling.
Further downstream, the PI3K-Akt signaling pathway was maximally enhanced in the presence of an intact GIV-GEF, and a major pathway downstream of Akt was triggered, that is, phosphoinhibition of the Rab-GAP AS160. Prior studies have demonstrated that docking of GSVs at the PM requires activation of Rab proteins (Miinea et al., 2005; Sun et al., 2010; Lansey et al., 2012) in response to insulin (Bai et al., 2007). By triggering the phosphoinhibition of Rab-GAP AS160, GIV's GEF function is likely to affect the exocytosis of GSVs via potentiation of Rab GTPases. We conclude that GIV functionally interacts with and enhances key signaling events that can also coordinate membrane trafficking within the insulin response cascade, and in doing so, it delineates a molecular basis for the observed engagement between these events during insulin-triggered glucose uptake into cells (Leto and Saltiel, 2012).
Although this study has specifically dissected the role of GIV's GEF function in coordinating key signaling events that comprise the metabolic insulin response, it is notable that the GEF motif merely represents a stretch of ∼30–35 aa within a 1871-aa-long, multimodular protein made up of several other key functional modules that may take part in other key aspects of glucose uptake. One such well-defined module is GIV's SH2-like domain, which is necessary and sufficient for GIV to directly bind the autophosphorylated cytoplasmic tail of InsR; without a functional SH2-like domain, GIV can neither bind InsR nor facilitate the formation of InsR–G protein complexes (Lin et al., 2014). Another such module, whose boundaries remain to be defined, but which appears to be functionally distinct from the GEF module, is a region within GIV's C-terminus that directly binds IRS1 (Figure 4B); it is possible that selective inhibition of GIV-IRS1 interaction may also impair the metabolic insulin response and glucose uptake. Our own recent findings that GIV regulates cargo trafficking from the ER-Golgi intermediate compartment (ERGIC) to the Golgi (Lo et al., 2015) raises the possibility that GIV may also play a role in regulating GLUT4 trafficking from the Golgi to GSVs. Additionally, GIV is also known to regulate clathrin-mediated endocytosis and endocytic trafficking (Beas et al., 2012; Weng et al., 2014), two processes closely intertwined with and key determinants of the kinetics of GLUT4 trafficking, glucose uptake, and down-regulation of insulin receptor signaling. Because the actin cytoskeleton has also been described as a tether for GSVs (Stockli et al., 2011), it is possible that another previously characterized module that enables GIV to remodel the cortical actin cytoskeleton further aids in GSV exocytosis and glucose uptake (Enomoto et al., 2005; Ghosh et al., 2010). Thus, it is likely that many of GIV's modules, not just its C-terminal GEF motif, may play a role in integrating signaling events with vesicular trafficking and cytoskeletal changes to orchestrate glucose uptake after insulin stimulation.
Selective modulation of GIV-GEF emerges as a therapeutic strategy for reversal of IR
We found that cell-penetrable GIV peptides were as effective as TZDs in their ability to reverse fatty acid–induced IR in a GEF-dependent way, and activation of Gαi via GIV's GEF thus mimics the action and matches the potency of TZDs. Because postprandial lipotoxicity can also suppress GIV expression in skeletal muscles (Supplemental Figure S6), our results using cell-permeant peptides suggest that replenishing GIV-CT (with active GEF) by gene therapy may be a viable strategy for the treatment of IR. Other strategies include agonists of GIV's GEF function, antagonists of the inhibitory phospho event on GIV triggered by PKCθ, or activation of phosphatases that dephosphorylate GIV—all approaches that may serve as more refined, effective, and precise therapeutic strategies to reverse IR in skeletal muscle. GIV expressed in adipocytes is also likely to enhance the metabolic insulin response in adipose tissue, the second major site of IR. However, phosphoinhibition of GIV-GEF by PKCθ is unlikely to be a trigger for IR, because this kinase is undetectable in adipocytes (Fleming et al., 1998). Instead, mechanisms such as transcriptional repression, single nucleotide polymorphisms (SNPs), or posttranslational modifications (splice variants) that reduce the levels of full-length GIV may play a role. Further studies are required to determine how IR is triggered in adipocytes and whether the GIV-targeted approaches we show here can reverse IR also in the adipose tissue.
In conclusion, we have defined activation of Gαi by GIV's GEF function as a central node that coordinately enhances the physiological insulin response and how its deregulation heralds IR. Because this node also serves as the point of convergence for the antagonistic actions of fatty acids and insulin sensitizers, selective modulation of this node emerges as a promising and precise strategy to treat T2DM and other conditions in which IR plays a central pathophysiological role.
MATERIALS AND METHODS
Detailed methods are presented in the Supplemental Materials.
Cell culture, transfection, IB, immunofluorescence, and protein–protein interaction assays
These assays were carried out exactly as described before (Ghosh et al., 2008, 2010). All Odyssey images were processed using ImageJ software (National Institutes of Health [NIH], Bethesda, MD) and were assembled for presentation using Photoshop and Illustrator software (Adobe Systems, San Jose, CA).
dSTORM and FRET imaging
Direct Stochastic Optical Reconstruction Microscopy imaging was performed to reveal the interaction endogenous Gαi3 and active InsRβ at molecular level (Huang et al., 2010). Control short hairpin RNA (shRNA) and GIV shRNA Cos7 stable cells were starved and stimulated with 100 nM insulin and stained with anti-Gαi3 (1:30; Calbiochem, San Diego, CA) and phosho-InRβ antibodies (1:100; Santa Cruz Biotechnologies, Dallas, TX).
FRET assays were performed using the intracellular phosphorylation biosensors custom (phocus-2nes) designed by Yoshio Umezawa's group at the University of Tokyo (Sato et al., 2002). GIV shRNA Cos7 stable cells were transfected with phocus-2nes and GIV-WT-FLAG or GIV-S1689D-FLAG. Cells were starved and stimulated with insulin and were stained with anti-FLAG antibody following the standard immunofluorescence protocol.
Patient samples
Biopsies of vastus lateralis muscle used for GIV phosphorylation analysis were collected in the Special Diagnostic and Treatment Unit of the Veterans Affairs Medical Center (San Diego, CA) and the General Clinical Research Center, University of California, San Diego (UCSD). Muscle samples were collected from healthy, normal, cycling women or women with PCOS before and after a course of treatment with Pio (45 mg/d, for 6 mo; Aroda et al., 2009). Collection and storage of muscle samples and measurement of eGDR were performed as described previously (Thorburn et al., 1990). The experimental protocol was approved by the Human Research Protection Program of the UCSD.
Data analysis and statistics
All experiments were repeated at least three times, and results were presented either as one representative experiment or as mean ± SD or SEM. Statistical significance was assessed with the two-tailed Student's t test.
Supplementary Material
Acknowledgments
We thank Marilyn Farquhar, Gordon Gill, Alan Saltiel, and Jerry Olefsky (UCSD) for thoughtful comments along the way and during the preparation of this article and Kersi Pestonjamasp for assistance with dSTORM microscopy at UCSD Moore's Cancer Center Microscopy Shared Facility (supported by NIH Grant P30 CA23100). This work was funded by the NIH (R01CA160911) and the Burroughs Wellcome Fund (CAMS award to P.G.). G.S.M was supported by the Doris Duke Charitable Foundation (DDCF grant 2013073 to P.G.) and I.L.-S. by a fellowship from the American Heart Association (AHA 14POST20050025). T.P.C. and R.R.H. were supported by grants from the American Diabetes Association and the Medical Research Service, Department of Veterans Affairs, VA San Diego Healthcare System (R.R.H.). N.K. was supported by a Cancer Cell Biology Training Grant from the National Cancer Institute (5T32CA067754-17).
Abbreviations used:
- DAG
diacyl glycerol
- DAPI
4′,6-diamidino-2-phenylindole
- F.E.
FRET efficiency
- FRET
fluorescence resonance energy transfer
- GAP
GTPase-activating protein
- GDR
glucose disposal rate
- GEF
guanine nucleotide exchange factor
- GLUT4
glucose transporter 4
- GPCR
G protein–coupled receptor
- GSV
GLUT4 storage vesicle
- IB
immunoblotting
- IgG
immunoglobulin G
- IR
insulin resistance
- IRS1
insulin receptor substrate 1
- PA
palmitate
- PCOS
polycystic ovarian syndrome
- phocus-2nes
phosphorylation biosensors custom
- PI3K
phosphoinositide 3 kinase
- Pio
pioglitazone
- PKCθ
protein kinase C-theta
- PM
plasma membrane
- PTB
phosphotyrosine-binding domain
- PTP
protein tyrosine phosphatase
- RTK
receptor tyrosine kinase
- SH2
Src homology 2
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- T2DM
type 2 diabetes mellitus
- TZDs
thiazolidinediones
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0553) on September 16, 2015.
REFERENCES
- Altiok S, Xu M, Spiegelman BM. PPARγ induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K, Fitzgerald M, Dupont M, Wang T, Paz N, Dorsch M, Healy A, Xu Y, Ocain T, Schopf L, et al. Mice deficient in PKC theta demonstrate impaired in vivo T cell activation and protection from T cell-mediated inflammatory diseases. Autoimmunity. 2006;39:469–478. doi: 10.1080/08916930600907954. [DOI] [PubMed] [Google Scholar]
- Aroda VR, Ciaraldi TP, Burke P, Mudaliar S, Clopton P, Phillips S, Chang RJ, Henry RR. Metabolic and hormonal changes induced by pioglitazone in polycystic ovary syndrome: a randomized, placebo-controlled clinical trial. J Clin Endocrinol Metab. 2009;94:469–476. doi: 10.1210/jc.2008-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Wang Y, Fan J, Chen Y, Ji W, Qu A, Xu P, James DE, Xu T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab. 2007;5:47–57. doi: 10.1016/j.cmet.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Beas AO, Taupin V, Teodorof C, Nguyen LT, Garcia-Marcos M, Farquhar MG. Gαs promotes EEA1 endosome maturation and shuts down proliferative signaling through interaction with GIV (Girdin) Mol Biol Cell. 2012;23:4623–4634. doi: 10.1091/mbc.E12-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AM. Inflammation, thrombosis and acute coronary syndromes. Diabetes Vasc Dis Res. 2005;2:113–121. doi: 10.3132/dvdr.2005.018. [DOI] [PubMed] [Google Scholar]
- Chen JF, Guo JH, Moxham CM, Wang HY, Malbon CC. Conditional, tissue-specific expression of Q205L Gαi2 in vivo mimics insulin action. J Mol Med (Berl) 1997;75:283–289. doi: 10.1007/s001090050113. [DOI] [PubMed] [Google Scholar]
- Cho DH, Choi YJ, Jo SA, Ryou J, Kim JY, Chung J, Jo I. Troglitazone acutely inhibits protein synthesis in endothelial cells via a novel mechanism involving protein phosphatase 2A-dependent p70 S6 kinase inhibition. Am J Physiol Cell Physiol. 2006;291:C317–C326. doi: 10.1152/ajpcell.00491.2005. [DOI] [PubMed] [Google Scholar]
- Ciaraldi TP, Maisel A. Role of guanine nucleotide regulatory proteins in insulin stimulation of glucose transport in rat adipocytes. Influence of bacterial toxins. Biochem J. 1989;264:389–396. doi: 10.1042/bj2640389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel Y, Ong A, Notani D, Mittal Y, Lam M, Mi X, Ghosh P. STAT3 protein up-regulates Gα-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. J Biol Chem. 2012;287:41667–41683. doi: 10.1074/jbc.M112.390781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, Murakumo Y, Usukura J, Kaibuchi K, Takahashi M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Fleming I, MacKenzie SJ, Vernon RG, Anderson NG, Houslay MD, Kilgour E. Protein kinase C isoforms play differential roles in the regulation of adipocyte differentiation. Biochem J. 1998;333:719–727. doi: 10.1042/bj3330719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Riveros JR, Sibley E, Kastelic T, Lane MD. Substrate phosphorylation catalyzed by the insulin receptor tyrosine kinase. Kinetic correlation to autophosphorylation of specific sites in the beta subunit. J Biol Chem. 1989;264:21557–21572. [PubMed] [Google Scholar]
- Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell. 2011;22:673–686. doi: 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, Ear J, Farquhar MG. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J Biol Chem. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for Gαi with a unique motif that regulates Akt signaling. Proc Natl Acad Sci USA. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M, Ghosh P, Farquhar MG. GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J Biol Chem. 2015;290:6697–6704. doi: 10.1074/jbc.R114.613414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Beas AO, Bornheimer SJ, Garcia-Marcos M, Forry EP, Johannson C, Ear J, Jung BH, Cabrera B, Carethers JM, Farquhar MG. A Gαi-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol Biol Cell. 2010;21:2338–2354. doi: 10.1091/mbc.E10-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Gαi3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla A, Klement K, Nurnberg B. The heterotrimeric G protein G(i3) regulates hepatic autophagy downstream of the insulin receptor. Autophagy. 2007;3:393–395. doi: 10.4161/auto.4256. [DOI] [PubMed] [Google Scholar]
- Goodyear LJ, Giorgino F, Sherman LA, Carey J, Smith RJ, Dohm GL. Insulin receptor phosphorylation, insulin receptor substrate-1 phosphorylation, and phosphatidylinositol 3-kinase activity are decreased in intact skeletal muscle strips from obese subjects. J Clin Invest. 1995;95:2195–2204. doi: 10.1172/JCI117909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin ME, Marcucci MJ, Cline GW, Bell K, Barucci N, Lee D, Goodyear LJ, Kraegen EW, White MF, Shulman GI. Free fatty acid-induced insulin resistance is associated with activation of protein kinase C theta and alterations in the insulin signaling cascade. Diabetes. 1999;48:1270–1274. doi: 10.2337/diabetes.48.6.1270. [DOI] [PubMed] [Google Scholar]
- Haasch D, Berg C, Clampit JE, Pederson T, Frost L, Kroeger P, Rondinone CM. PKCθ is a key player in the development of insulin resistance. Biochem Biophys Res Commun. 2006;343:361–368. doi: 10.1016/j.bbrc.2006.02.177. [DOI] [PubMed] [Google Scholar]
- Hartung A, Ordelheide AM, Staiger H, Melzer M, Haring HU, Lammers R. The Akt substrate Girdin is a regulator of insulin signaling in myoblast cells. Biochim Biophys Acta. 2013;1833:2803–2811. doi: 10.1016/j.bbamcr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Hoehn KL, Hohnen-Behrens C, Cederberg A, Wu LE, Turner N, Yuasa T, Ebina Y, James DE. IRS1-independent defects define major nodes of insulin resistance. Cell Metab. 2008;7:421–433. doi: 10.1016/j.cmet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Hojlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Vollenweider P, Egawa K, Clodi M, Ishibashi K, Nakashima N, Ugi S, Adams JW, Brown JH, Olefsky JM. G alpha-q/11 protein plays a key role in insulin-induced glucose transport in 3T3-L1 adipocytes. Mol Cell Biol. 1999;19:6765–6774. doi: 10.1128/mcb.19.10.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itani SI, Zhou Q, Pories WJ, MacDonald KG, Dohm GL. Involvement of protein kinase C in human skeletal muscle insulin resistance and obesity. Diabetes. 2000;49:1353–1358. doi: 10.2337/diabetes.49.8.1353. [DOI] [PubMed] [Google Scholar]
- Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh Y, Ishizuka T, Morita H, Ishizawa M, Miura A, Kajita K, Kimura M, Suzuki T, Sakuma H, Yasuda K. Effect of pertussis toxin on insulin-induced signal transduction in rat adipocytes and soleus muscles. Cell Signal. 2000;12:223–232. doi: 10.1016/s0898-6568(99)00081-9. [DOI] [PubMed] [Google Scholar]
- Kim JK, Fillmore JJ, Sunshine MJ, Albrecht B, Higashimori T, Kim DW, Liu ZX, Soos TJ, Cline GW, O’Brien WR, et al. PKC-theta knockout mice are protected from fat-induced insulin resistance. J Clin Invest. 2004;114:823–827. doi: 10.1172/JCI22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlden JM, Jones HE, Barrow D, Gee JM, Nicholson RI, Hutcheson IR. Insulin receptor substrate-1 involvement in epidermal growth factor receptor and insulin-like growth factor receptor signalling: implication for Gefitinib (“Iressa”) response and resistance. Breast Cancer Res Treat. 2008;111:79–91. doi: 10.1007/s10549-007-9763-9. [DOI] [PubMed] [Google Scholar]
- Kreuzer J, Nurnberg B, Krieger-Brauer HI. Ligand-dependent autophosphorylation of the insulin receptor is positively regulated by Gi-proteins. Biochem J. 2004;380:831–836. doi: 10.1042/BJ20031659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook A, Moller DE, Dib K, O’Rahilly S. Two naturally occurring mutant insulin receptors phosphorylate insulin receptor substrate-1 (IRS-1) but fail to mediate the biological effects of insulin. Evidence that IRS-1 phosphorylation is not sufficient for normal insulin action. J Biol Chem. 1996;271:7134–7140. doi: 10.1074/jbc.271.12.7134. [DOI] [PubMed] [Google Scholar]
- Krupinski J, Rajaram R, Lakonishok M, Benovic JL, Cerione RA. Insulin-dependent phosphorylation of GTP-binding proteins in phospholipid vesicles. J Biol Chem. 1988;263:12333–12341. [PubMed] [Google Scholar]
- Lansey MN, Walker NN, Hargett SR, Stevens JR, Keller SR. Deletion of Rab GAP AS160 modifies glucose uptake and GLUT4 translocation in primary skeletal muscles and adipocytes and impairs glucose homeostasis. Am J Physiol Endocrinol Metabol. 2012;303:E1273–E1286. doi: 10.1152/ajpendo.00316.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J Biol Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- Le Roith D, Zick Y. Recent advances in our understanding of insulin action and insulin resistance. Diabetes Care. 2001;24:588–597. doi: 10.2337/diacare.24.3.588. [DOI] [PubMed] [Google Scholar]
- Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- Li Y, Soos TJ, Li X, Wu J, Degennaro M, Sun X, Littman DR, Birnbaum MJ, Polakiewicz RD. Protein kinase C theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101) J Biol Chem. 2004;279:45304–45307. doi: 10.1074/jbc.C400186200. [DOI] [PubMed] [Google Scholar]
- Lin C, Ear J, Midde K, Lopez-Sanchez I, Aznar N, Garcia-Marcos M, Kufareva I, Abagyan R, Ghosh P. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol Biol Cell. 2014;25:3654–3671. doi: 10.1091/mbc.E14-05-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Ear J, Pavlova Y, Mittal Y, Kufareva I, Ghassemian M, Abagyan R, Garcia-Marcos M, Ghosh P. Tyrosine phosphorylation of the Gα-interacting protein GIV promotes activation of phosphoinositide 3-kinase during cell migration. Sci Signal. 2011;4:ra64. doi: 10.1126/scisignal.2002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, O’Mahony A, Mu Y, Geleziunas R, Greene WC. Protein kinase C-θ participates in NF-κB activation induced by CD3-CD28 costimulation through selective activation of IκB kinase β. Mol Cell Biol. 2000;20:2933–2940. doi: 10.1128/mcb.20.8.2933-2940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo IC, Gupta V, Midde KK, Taupin V, Lopez-Sanchez I, Kufareva I, Abagyan R, Randazzo PA, Farquhar MG, Ghosh P. Activation of Gαi at the Golgi by GIV/Girdin imposes finiteness in Arf1 signaling. Dev Cell. 2015;33:189–203. doi: 10.1016/j.devcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Dunkel Y, Roh YS, Mittal Y, De Minicis S, Muranyi A, Singh S, Shanmugam K, Aroonsakool N, Murray F, et al. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat Commun. 2014;5:4451. doi: 10.1038/ncomms5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Garcia-Marcos M, Mittal Y, Aznar N, Farquhar MG, Ghosh P. Protein kinase C-theta (PKCtheta) phosphorylates and inhibits the guanine exchange factor, GIV/Girdin. Proc Natl Acad Sci USA. 2013;110:5510–5515. doi: 10.1073/pnas.1303392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma GS, Aznar N, Kalogriopoulos N, Midde KK, Lopez-Sanchez I, Sato E, Dunkel Y, Gallo RL, Ghosh P. Therapeutic effects of cell-permeant peptides that activate G proteins downstream of growth factors. Proc Natl Acad Sci USA. 2015;112:E2602–E2610. doi: 10.1073/pnas.1505543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa H, Shigeta Y, Egawa K, Kobayashi M. Impaired autophosphorylation of insulin receptors from abdominal skeletal muscles in nonobese subjects with NIDDM. Diabetes. 1991;40:815–819. doi: 10.2337/diab.40.7.815. [DOI] [PubMed] [Google Scholar]
- Markova I, Zidek V, Musilova A, Simakova M, Mlejnek P, Kazdova L, Pravenec M. Long-term pioglitazone treatment augments insulin sensitivity and PKC-epsilon and PKC-theta activation in skeletal muscles in sucrose fed rats. Physiol Res. 2010;59:509–516. doi: 10.33549/physiolres.931852. [DOI] [PubMed] [Google Scholar]
- Mashili F, Chibalin AV, Krook A, Zierath JR. Constitutive STAT3 phosphorylation contributes to skeletal muscle insulin resistance in type 2 diabetes. Diabetes. 2013;62:457–465. doi: 10.2337/db12-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midde KK, Aznar N, Laederich MB, Ma GS, Kunkel MT, Newton AC, Ghosh P. Multimodular biosensors reveal a novel platform for activation of G proteins by growth factor receptors. Proc Natl Acad Sci USA. 2015;112:E937–E946. doi: 10.1073/pnas.1420140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miinea CP, Sano H, Kane S, Sano E, Fukuda M, Peranen J, Lane WS, Lienhard GE. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxham CM, Malbon CC. Insulin action impaired by deficiency of the G-protein subunit Giα2. Nature. 1996;379:840–844. doi: 10.1038/379840a0. [DOI] [PubMed] [Google Scholar]
- Myers MG, Jr, Grammer TC, Brooks J, Glasheen EM, Wang LM, Sun XJ, Blenis J, Pierce JH, White MF. The pleckstrin homology domain in insulin receptor substrate-1 sensitizes insulin signaling. J Biol Chem. 1995;270:11715–11718. doi: 10.1074/jbc.270.20.11715. [DOI] [PubMed] [Google Scholar]
- Nolan JJ, Freidenberg G, Henry R, Reichart D, Olefsky JM. Role of human skeletal muscle insulin receptor kinase in the in vivo insulin resistance of noninsulin-dependent diabetes mellitus and obesity. J Clin Endocrinol Metab. 1994;78:471–477. doi: 10.1210/jcem.78.2.8106637. [DOI] [PubMed] [Google Scholar]
- O’Brien RM, Houslay MD, Milligan G, Siddle K. The insulin receptor tyrosyl kinase phosphorylates holomeric forms of the guanine nucleotide regulatory proteins Gi and Go. FEBS Lett. 1987;212:281–288. doi: 10.1016/0014-5793(87)81361-3. [DOI] [PubMed] [Google Scholar]
- Pessin JE, Saltiel AR. Signaling pathways in insulin action: molecular targets of insulin resistance. J Clin Invest. 2000;106:165–169. doi: 10.1172/JCI10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugazhenthi S, Khandelwal RL. Insulin action on protein phosphatase-1 activation is enhanced by the antidiabetic agent pioglitazone in cultured diabetic hepatocytes. Mol Cell Biochem. 1998;182:185–191. [PubMed] [Google Scholar]
- Rothenberg PL, Kahn CR. Insulin inhibits pertussis toxin-catalyzed ADP-ribosylation of G-proteins. Evidence for a novel interaction between insulin receptors and G-proteins. J Biol Chem. 1988;263:15546–15552. [PubMed] [Google Scholar]
- Rundek T, Gardener H, Xu Q, Goldberg RB, Wright CB, Boden-Albala B, Disla N, Paik MC, Elkind MS, Sacco RL. Insulin resistance and risk of ischemic stroke among nondiabetic individuals from the northern Manhattan study. Arch Neurol. 2010;67:1195–1200. doi: 10.1001/archneurol.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sato M, Ozawa T, Inukai K, Asano T, Umezawa Y. Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]
- Schmitz-Peiffer C, Browne CL, Oakes ND, Watkinson A, Chisholm DJ, Kraegen EW, Biden TJ. Alterations in the expression and cellular localization of protein kinase C isozymes epsilon and theta are associated with insulin resistance in skeletal muscle of the high-fat-fed rat. Diabetes. 1997;46:169–178. doi: 10.2337/diab.46.2.169. [DOI] [PubMed] [Google Scholar]
- Sharma C, Pradeep A, Pestell RG, Rana B. Peroxisome proliferator-activated receptor gamma activation modulates cyclin D1 transcription via beta-catenin-independent and cAMP-response element-binding protein-dependent pathways in mouse hepatocytes. J Biol Chem. 2004;279:16927–16938. doi: 10.1074/jbc.M309045200. [DOI] [PubMed] [Google Scholar]
- Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zheng X, Malbon CC, Wang H. Gαi2 enhances in vivo activation of and insulin signaling to GLUT4. J Biol Chem. 2001;276:34651–34658. doi: 10.1074/jbc.M105894200. [DOI] [PubMed] [Google Scholar]
- Stockli J, Fazakerley DJ, James DE. GLUT4 exocytosis. J Cell Sci. 2011;124:4147–4159. doi: 10.1242/jcs.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bilan PJ, Liu Z, Klip A. Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci USA. 2010;107:19909–19914. doi: 10.1073/pnas.1009523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- Thorburn AW, Gumbiner B, Bulacan F, Wallace P, Henry RR. Intracellular glucose oxidation and glycogen synthase activity are reduced in non-insulin-dependent (type II) diabetes independent of impaired glucose uptake. J Clin Invest. 1990;85:522–529. doi: 10.1172/JCI114468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Voliovitch H, Schindler DG, Hadari YR, Taylor SI, Accili D, Zick Y. Tyrosine phosphorylation of insulin receptor substrate-1 in vivo depends upon the presence of its pleckstrin homology region. J Biol Chem. 1995;270:18083–18087. doi: 10.1074/jbc.270.30.18083. [DOI] [PubMed] [Google Scholar]
- Weng L, Enomoto A, Miyoshi H, Takahashi K, Asai N, Morone N, Jiang P, An J, Kato T, Kuroda K, et al. Regulation of cargo-selective endocytosis by dynamin 2 GTPase-activating protein girdin. EMBO J. 2014;33:2098–2112. doi: 10.15252/embj.201488289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF. Regulating insulin signaling and beta-cell function through IRS proteins. Canadian J Physiol Pharmacol. 2006;84:725–737. doi: 10.1139/y06-008. [DOI] [PubMed] [Google Scholar]
- White MF, Shoelson SE, Keutmann H, Kahn CR. A cascade of tyrosine autophosphorylation in the beta-subunit activates the phosphotransferase of the insulin receptor. J Biol Chem. 1988;263:2969–2980. [PubMed] [Google Scholar]
- Wunderlich CM, Hovelmeyer N, Wunderlich FT. Mechanisms of chronic JAK-STAT3-SOCS3 signaling in obesity. Jak-Stat. 2013;2:e23878. doi: 10.4161/jkst.23878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Sato T, Kawasaki K, Murosaki S, Yamamoto Y. A nonradioisotope, enzymatic assay for 2-deoxyglucose uptake in L6 skeletal muscle cells cultured in a 96-well microplate. Analytical Biochem. 2006;351:139–145. doi: 10.1016/j.ab.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Yu C, Chen Y, Cline GW, Zhang D, Zong H, Wang Y, Bergeron R, Kim JK, Cushman SW, Cooney GJ, et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.