Dynein light chain 1 (LC1) of the outer arm dynein (OAD) complex associates with the microtubule-binding domain (MTBD) of γ heavy chain inside the complex. LC1 is considered to regulate the OAD activity and ciliary/flagellar motion by modulating γ MTBD's affinity to the B-tubule of the doublet microtubule in the axoneme.
Abstract
The outer arm dynein (OAD) complex is the main propulsive force generator for ciliary/flagellar beating. In Chlamydomonas and Tetrahymena, the OAD complex comprises three heavy chains (α, β, and γ HCs) and >10 smaller subunits. Dynein light chain-1 (LC1) is an essential component of OAD. It is known to associate with the Chlamydomonas γ head domain, but its precise localization within the γ head and regulatory mechanism of the OAD complex remain unclear. Here Ni-NTA-nanogold labeling electron microscopy localized LC1 to the stalk tip of the γ head. Single-particle analysis detected an additional structure, most likely corresponding to LC1, near the microtubule-binding domain (MTBD), located at the stalk tip. Pull-down assays confirmed that LC1 bound specifically to the γ MTBD region. Together with observations that LC1 decreased the affinity of the γ MTBD for microtubules, we present a new model in which LC1 regulates OAD activity by modulating γ MTBD's affinity for the doublet microtubule.
INTRODUCTION
Cilia and flagella are microtubule (MT)-based organelles that protrude from eukaryotic cells. The axoneme of the cilia and flagella has an identical “9+2 structure” in which two central microtubules are surrounded by nine doublet microtubules (DMTs; Bui et al., 2008; Pigino et al., 2012). Cilia/flagella are essential in lower eukaryotes like Chlamydomonas and Tetrahymena for cell motility and are also important in higher eukaryotes for sperm motility, generation of fluid flow in the trachea and the brain ventricle, and left–right determination during embryonic development (Sanderson et al., 1981; Nonaka et al., 1998; Sawamoto et al., 2006). Defects in various kinds of cilia/flagella proteins lead to a variety of human diseases called ciliopathies (Kurkowiak et al., 2015).
The bending motion of cilia/flagella is driven by MT minus end–directed motor proteins—axonemal dyneins. Axonemal dyneins are docked periodically on the DMTs and are divided into two classes: outer arm dynein (OAD) and inner arm dynein (IAD). The OAD complex generates ∼80% of the propulsive force of cilia/flagella (Brokaw et al., 1994). The OAD complex is a huge (∼2 MDa) multiprotein complex composed of three heavy chains (α, β, and γ HCs), two intermediate chains (ICs), and 11 light chains (LCs; nomenclature in this work is based on the Chlamydomonas OAD, unless otherwise noted). Each of the three HCs is composed of a tail domain (corresponding to the N-terminal one-third sequence) and a head domain (the C-terminal two-thirds). Three HCs form a heterotrimer through their tail domain; the OAD complex thus appears as a three-headed structure (Goodenough and Heuser, 1984; Toyoshima, 1987). Based on recent x-ray crystallography studies, the head domain can be divided into several regions, including the linker, the AAA+ ring, the stalk, the strut/buttress, and the C-sequence (Carter et al., 2011; Kon et al., 2011, 2012; Schmidt et al., 2012). The stalk, extending from the AAA+ ring, is composed of a 15-nm-long antiparallel coiled-coil domain (consisting of coiled coil 1 [CC1] and coiled coil 2 [CC2]) with a microtubule-binding domain (MTBD) at the tip (Gee et al., 1997; Koonce, 1997). Dynein's interaction with microtubules is considered to be modulated by the registry shift in the stalk coiled coil and consequent conformational change in the MTBD, which are coupled with the ATPase cycle in the AAA+ ring (Gibbons et al., 2005; Redwine et al., 2012). In the axonemes of cilia/flagella, the OAD complex is tethered to the A-tubule of the DMT via its tail domain and interacts with the adjacent B-tubule of DMT at its MTBD (Movassagh et al., 2010; Lin et al., 2014).
Dynein light chain-1 (LC1) is a 22-kDa light chain of the OAD complex and is widely conserved among many species. LC1 is found to be essential for proper beating of cilia/flagella in many species, including Chlamydomonas reinhardtii, Schmidtea mediterranea, Paramecium tetraurelia, and Trypanosoma brucei. Knockdown of LC1 and/or expression of mutant forms of LC1 in these organisms results in a variety of abnormal phenotypes, including slower swimming, attenuation of beat frequency, and failure of assembly of the OAD complex (Baron et al., 2007; Patel-King and King, 2009; Rompolas et al., 2010; Kutomi et al., 2012). Mutations in human LC1 lead to primary ciliary dyskinesia (Horváth et al., 2005; Mazor et al., 2011). Despite its importance in vivo, the mechanism of LC1's regulation of OAD activity is not clearly understood, partially because the localization of LC1 within the OAD complex has not been precisely determined. From cross-linking experiment results, LC1 was found to bind to the head domain of the γ HC (Benashski et al., 1999). Although LC1 was assumed to bind to the AAA+ ring of the γ head domain (Patel-King and King, 2009), structural evidence has not been obtained.
Here we use electron microscopy (EM) and biochemical analysis to identify the localization of LC1 within the OAD complex. We find that one of the stalk tips in the three heads appears larger than the other two. Nickel–nitriloacetic acid (NTA)–nanogold labeling of histidine (His)-tagged LC1 shows that the larger stalk tip holds LC1. Together with the data on the stoichiometry of binding, mapping of the binding region, and LC1's effects on microtubule binding, our present findings suggest a new model in which one copy of LC1 localizes to the γ MTBD region and regulates its affinity for the B-tubule of the DMT.
RESULTS
LC1 associates with the stalk tip
LC1 was found to associate with the γ head domain by cross-linking experiments (Benashski et al., 1999), but no direct structural evidence has been obtained. To determine directly the localization of LC1 within the OAD complex, we sought to label His-tagged recombinant LC1 with Ni-NTA-nanogold particles and observe them in the EM (Kitai et al., 2011; Acar et al., 2013). The His-tagged LC1 constructs were expressed in wild-type strains of Chlamydomonas and Tetrahymena because no LC1-null mutant is available in any organism. In Chlamydomonas, the His tag was introduced to the C-terminus of LC1 (LC1-C-His; Figure 1A). In Tetrahymena, a green fluorescent protein (GFP) containing a His tag and a streptavidin-binding peptide tag (referred to as hsGFP-tag) was constructed and introduced into the C-terminus of LC1 (LC1-C-hsGFP) by homologous recombination (Supplemental Figures S1B and S3A). The GFP tag was used to simplify the phenotypic assortment process of the Tetrahymena macronucleus (see Supplemental Materials and Methods for details). In both Chlamydomonas and Tetrahymena, recombinant LC1 was incorporated into the OAD complex and cilia/flagella, as confirmed by GFP fluorescence and/or Western blots (Supplemental Figure S1). Recombinant Chlamydomonas with His-tagged LC1 showed a similar motility phenotype to that of wild type (Supplemental Figure S2), and thus the recombinant LC1 constructs are believed to be functional in vivo. The His tag in OAD was clearly accessible to Ni-NTA, since the recombinant OAD complexes could be purified using Ni-NTA resin (Supplemental Figure S1, A and C).
FIGURE 1:
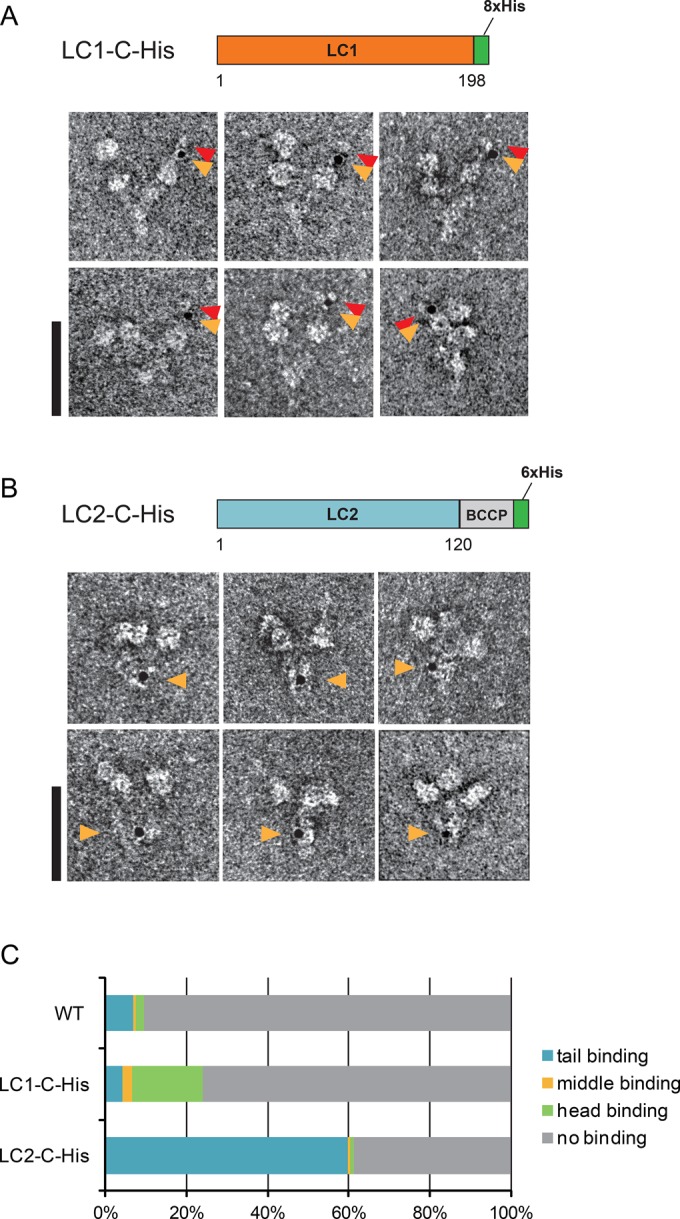
Ni-NTA-nanogold labeling of recombinant Chlamydomonas OAD complex. (A) Diagram of the LC1-C-His construct and negatively stained EM images of the gold-labeled LC1-C-His OAD complex. Orange arrowheads indicate gold particles. Gold-labeled stalk tips sometimes had extra densities (indicated by red arrowheads). Bar, 50 nm. (B) Schematic diagram of the LC2-C-His construct and negatively stained EM images of the gold-labeled LC2-C-His OAD complex. Orange arrowheads show gold particles. Bar, 50 nm. (C) Fractions of the Ni-NTA-nanogold binding. Negatively stained OAD molecules were classified as tail binding, middle binding, head binding, or no binding. Molecules gold-labeled at a position between the tail and the head were classified as middle binding.
In EM, the purified recombinant OAD complex exhibited a three-headed structure (Figure 1, A and B), as previously reported for wild-type OAD (Goodenough and Heuser, 1984; Toyoshima, 1987). First, as a control to assess whether Ni-NTA-nanogold labeling is applicable to the OAD complex, we used recombinant Chlamydomonas OAD containing LC2–biotin carboxyl carrier protein–His (referred to as LC2-C-His; Furuta et al., 2009). In the LC2-C-His OAD complex, gold particles were frequently found inside the tail domain (Figure 1B, orange arrowheads). This result is consistent with a previous model in which LC2 was localized to the tail domain (DiBella et al., 2001). The gold binding was believed to be specific because the binding ratio with LC2-C-His (61.2%, 134 of 219 molecules) was much higher than the binding ratio with the wild-type OAD complex without a His tag (9.4%, 61 of 650 molecules), and gold particles bound to the tail domain more specifically compared with the binding locations in the wild-type OAD complex (Figure 1C). From these results, we concluded that Ni-NTA-nanogold labeling could be used for localization in the OAD complex.
Next we performed Ni-NTA-nanogold labeling EM for OAD with LC1-C-His (Supplemental Figure S1Aii). Although the labeling ratio of the LC1-C-His OAD complex (24.1%, 233 of 965 molecules) was lower than that of the LC2-C-His OAD complex, it was higher than that of the wild-type OAD complex (Figure 1C). The relatively low labeling ratio observed is probably due to the presence of wild-type LC1 in the purified OAD sample (Supplemental Figure S1Aii). For the LC2-C-His OAD complex, LC2 was replaced by the recombinant form because His-tagged LC2 was expressed in an LC2-null mutant strain (oda12). The ratio of endogenous to His-tagged LC1 in purified LC1-C-His OAD samples was ∼2:1, as judged by Western blot analysis (Supplemental Figure S1Aii). For LC1-C-His, we found that one of the head domains was specifically labeled with nanogold particle (Figure 1A, orange arrowheads). Because gold particles were rarely bound to the head domain in the control (LC2-C-His and wild type; Figure 1C), gold particle binding to the head domain of LC1-C-His OAD is believed to represent specific binding to LC1. We carefully examined the gold particle binding pattern in LC1-C-His and found that the gold particles were frequently distant from the AAA+ ring, near the stalk tip (Figure 1A). Similar localization of Ni-NTA-nanogold particles was observed in the recombinant Tetrahymena OAD complex with LC1-C-hsGFP (Supplemental Figure S3A). These results were in striking contrast with the previous proposal that LC1 was associated with the AAA+ ring (Benashski et al., 1999; Patel-King and King, 2009). To ensure that the gold binding to the stalk tip was not due to an artifact caused by the flexibility of the Ni-NTA-nanogold labeling, we constructed a Tetrahymena strain (βHC-C-hGFP) that expresses β HC carrying an hGFP (GFP containing His tag) at the C-terminus (Supplemental Figure S1, B and C). This construct was used as a control for a His tag present inside the AAA+ ring, since the C-terminus of the HC was found to reside inside the AAA+ ring (Carter et al., 2011; Kon et al., 2011, 2012; Schmidt et al., 2012). Labeling of the βHC-C-hGFP OAD complex with Ni-NTA-nanogold particles resulted in localization of gold particles at the AAA+ ring edge, not at the stalk tip (Supplemental Figure S3B). Thus the gold particle binding to the stalk tip appeared to reflect the localization of LC1 outside of the AAA+ ring, most likely at the stalk tip.
The γ stalk has a bulged tip due to the attached LC1
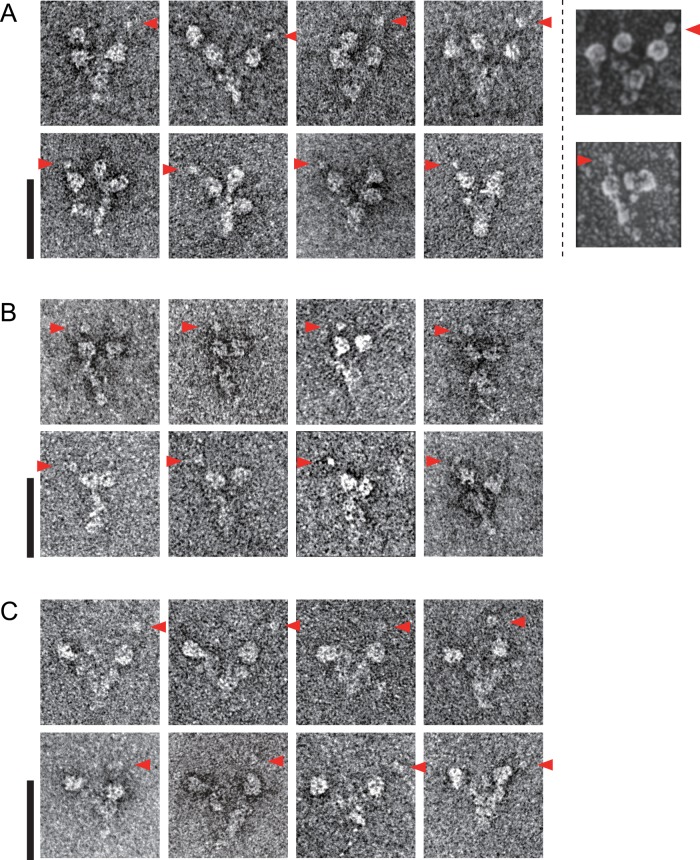
In the course of EM observation of the gold-labeled OAD complex with His-tagged LC1, we noticed that the gold-labeled stalk tip sometimes possessed extra density and looked bulged compared with the other HC stalk tips (Figure 1A, red arrowheads). To investigate whether this bulged appearance of one stalk tip was an inherent feature of the OAD complex or an artifact due to protein tagging of LC1 and/or gold labeling, we also observed native Chlamydomonas and Tetrahymena OAD complexes by negative-staining EM. We found one of the three stalk tips appeared more prominent than the others even in the native OAD complex without an attached gold particle (Figure 2A and Supplemental Figure S4A). The large stalk tip was not always visible by negative staining, possibly because it was sensitive to the staining condition. However, this feature could be observed in previous rotary shadowed EM images (Figure 2A, rightmost; Goodenough and Heuser, 1984). Therefore we concluded that the presence of one prominent stalk tip was an intrinsic feature of the OAD complex.
FIGURE 2:
Negatively stained EM images of Chlamydomonas OAD complex. (A) The wild- type (three-headed) Chlamydomonas OAD complex. Previous rotary shadowing EM images are shown in the rightmost column for comparison. (The rotary shadowed EM images were reproduced with permission from Goodenough and Heuser, 1984; copyright Elsevier.) (B) The oda11 (βγ two-headed) Chlamydomonas OAD complex. (C) The oda4-s7 (αγ two-headed) Chlamydomonas OAD complex. Large stalk tips are indicated by red arrowheads. Bars, 50 nm.
To identify which of the three HCs had a large stalk tip, we purified Chlamydomonas mutant OAD complexes lacking either the α HC (βγ two-headed) or the β HC (αγ two-headed) from strains oda11 and oda4-s7, respectively. In both mutant OADs, one of the two heads possessed a large stalk tip (Figure 2, B and C). Single-particle analysis of oda11 βγ heads also confirmed that one of the stalk tips was prominent (Supplemental Figure S4B). Because both of these mutant OADs retain the γ HC, we assigned that the γ stalk tip was the one that appeared prominent. In a recombinant strain oda11×LC1-C-His, the larger stalk tip of the OAD complex was labeled by Ni-NTA-nanogold (Supplemental Figure S4C). From these results, we concluded that the γ stalk tip attached LC1 and appeared larger than other stalk tips.
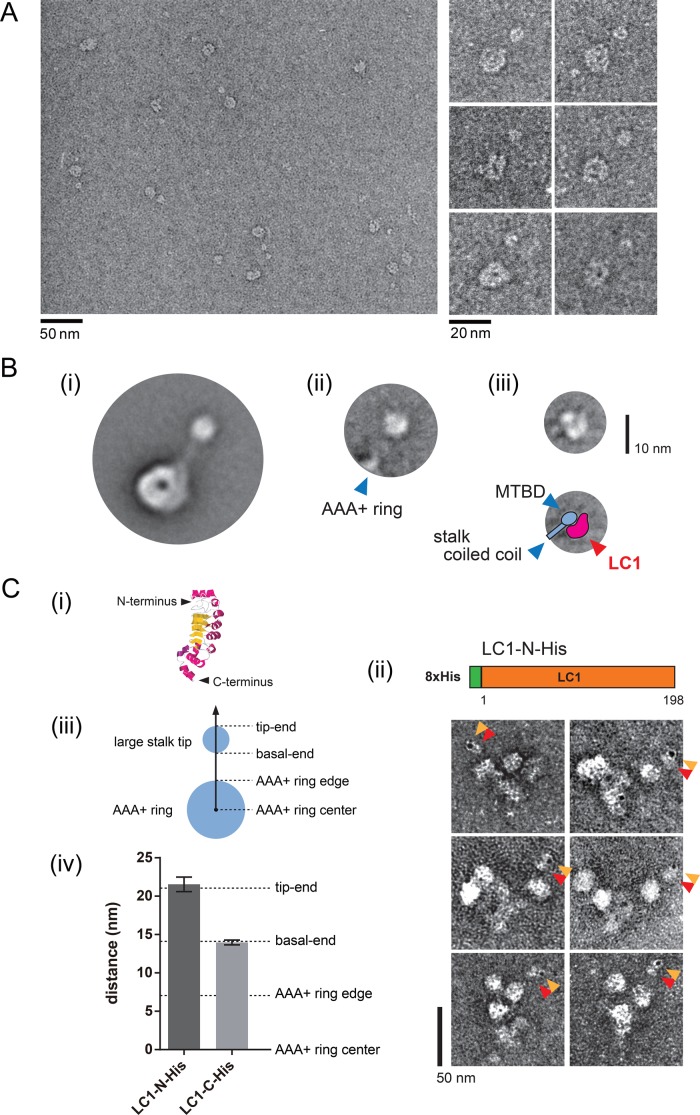
We tried to dissect the architecture of the γ stalk tip by single-particle analysis. In the averaged image of the oda11 OAD (βγ) heads, the detailed structure of the stalk tip was obscured (Supplemental Figure S4B). This might be because of the variety of head angles relative to the carbon grid due to the presence of the tail domain and the other head domain. Thus a single-headed fragment is appropriate for observation with little steric interference. Tetrahymena DYH3 head, which is equivalent to the Chlamydomonas γ head, could be isolated by chymotryptic digestion from the Tetrahymena OAD complex, and it retains the single-headed structure in EM (Toyoshima, 1987; Yamaguchi et al., 2015). Purified Tetrahymena DYH3 head fragments were more homogenous in negative-staining EM than the oda11 heads (Figure 3A). Association of endogenous LC1 with the obtained Tetrahymena DYH3 head fragment was verified by Western blot analysis (Supplemental Figure S5A). Using the Tetrahymena DYH3 head fragments, reasonable class average was obtained (Figure 3Bi). As for the AAA+ ring, the averaged Tetrahymena DYH3 head image was similar to the previously reported “right view” of the Dictyostelium cytoplasmic dynein head (Roberts et al., 2009). However, there was a globular structure at the tip of the averaged Tetrahymena DYH3 head image (Figure 3Bi). The images were subsequently classified and aligned according to the stalk region by applying a binary mask (Supplemental Figure S5B). In the resultant class average, the stalk-tip region was found to be composed of two substructures (Figure 3Bii and Supplemental Figure S5B). However, in the averaged image, only the base region of the stalk was visible, and the details of the stalk-tip region were obscured (Figure 3Bii). Therefore we further analyzed the stalk-tip region by performing single-particle analysis on the images of the Tetrahymena DYH3 stalk tip alone (Supplemental Figure S5C). The averaged stalk-tip structures appeared heterogeneous (Supplemental Figure S5C), but a class-averaged image similar to Figure 3Bii was obtained (Figure 3Biii). The overall shape and size of the averaged stalk tip in Figure 3Biii were round and with a 7-nm diameter, but it was composed of a smaller round substructure (∼3 nm in diameter) and an elliptic substructure (long axis of ∼7 nm and short axis of ∼3 nm). The smaller round substructure was connected to the stalk coiled coil and believed to correspond to the MTBD. The elliptic substructure was similar in size and shape to the reported NMR structure of LC1 (Wu et al., 2000; Patel-King and King, 2009), suggesting that this additional elliptic structure corresponded to LC1.
FIGURE 3:
Structural analysis of the stalk tip. (A) EM images of the Tetrahymena DYH3 head fragment. General view (left) and selected EM images (right) of negatively stained DYH3 head fragment (which corresponds to the Chlamydomonas γ head). (B) Single-particle analysis of the Tetrahymena DYH3 head fragment. (i) Major class average of the Tetrahymena DYH3 head. A round structure was found at the stalk tip. (ii) Representative class average of the stalk region. The major class in (i) was subclassified and aligned according to the stalk region by applying a mask to most of the AAA+ ring region. See also Supplemental Figure S5B. Location of the edge of the AAA+ ring is shown by blue arrowhead. (iii) Representative class average of the stalk-tip region, showing substructures. Only the stalk-tip regions of the Tetrahymena DYH3 head were used for the averaging in (iii). See also Supplemental Figure S5C. Schematic diagram of our interpretation is shown below. Locations of MTBD and stalk coiled coil are indicated by blue arrowheads. An additional structure, possibly corresponding to LC1, is indicated by the red arrowhead. (C) Localization of the N- and C-termini of LC1 in a large stalk tip. (i) The previously reported NMR structure of LC1 (Protein Data Bank ID: 1M9L). The N-terminus and C-terminus of LC1 are indicated. (ii) Schematic diagram of the Chlamydomonas LC1-N-His construct and EM images of the gold-labeled LC1-N-His OAD complex. Orange arrowheads indicate gold particles, and red arrowheads indicate large stalk tips. (iii) Schematic diagram of the AAA+ ring and the large stalk tip. (iv) Comparison of the gold particle locations in the LC1-N-His and LC1-C-His OAD complexes. The distance between the center of the AAA+ ring and that of the gold particle was measured for the OAD complexes with LC1-N-His and LC1-C-His. The distances from the AAA+ ring center to the edge of the AAA+ ring, basal end, and tip end of the large stalk tip were measured as in the axis in C (iii) from EM images without gold particles and are indicated by dashed lines in the graph in (iv). The values were as follows: LC1-N-His, 21.5 ± 0.9 nm (n = 26); LC1-C-His, 14.0 ± 0.3 nm (n = 69); AAA+ ring edge, 7.0 ± 0.2 nm (n = 21); basal end, 14.1 ± 0.6 nm (n = 27); tip end, 21.1 ± 0.6 nm (n = 27) (mean ± SE).
The N- and C-termini are located at opposite sides of the long axis of LC1 (Figure 3Ci; Wu et al., 2000; Patel-King and King, 2009). To determine the orientation of LC1 inside the large stalk tip, we made an LC1 construct with a His tag attached at the N-terminus (LC1-N-His; Figure 3Cii). In the LC1-C-His OAD complex, the gold particles were found at the basal end of the structure (Figure 1A). In contrast, in the LC1-N-His OAD complex, the gold particles were bound at the tip end of the structure (Figure 3Cii). To quantify these observation, we measured the center distances of the AAA+ ring and gold particles for the LC1-N-His and LC1-C-His OADs. The mean value of center distances of LC1-N-His was significantly larger than that of LC1-C-His (Figure 3Civ). Subsequently we measured the distances from the center of the AAA+ ring to the basal end and the tip end of the large stalk tip (Figure 3Civ) and confirmed that LC1 located inside the large stalk tip with its N-terminus at the tip end and the C-terminus at the basal end (Figure 3Ciii).
One copy of LC1 binds specifically to the γ stalk region
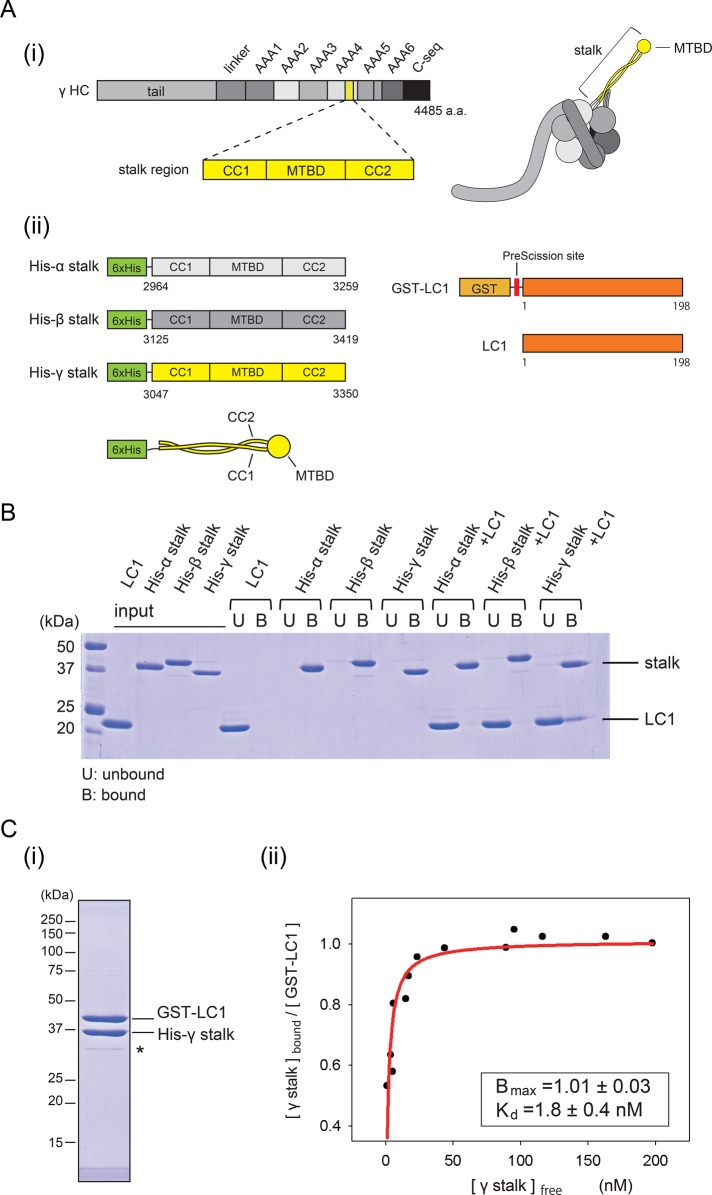
To analyze further the interaction between the stalk and LC1, we used bacterially expressed proteins for biochemical analyses. We made His-tagged stalk fragments from Chlamydomonas α, β, and γ HC (hereinafter referred to as His-α stalk, His-β stalk, and His-γ stalk, respectively) and Chlamydomonas full-length LC1 constructs (GST-LC1 and LC1; Figure 4A). First, we checked the specificity of the binding by pull-down assay (Figure 4B). His-tagged stalk constructs were bound to Ni-NTA magnetic beads and detected in the bound fractions. In the absence of the stalk, LC1 was detected only in the unbound fraction. When LC1 was incubated with the stalk constructs, LC1 was detected in the bound fraction only with the His-γ stalk.
FIGURE 4:
Binding properties of LC1 to the stalk. (A) Constructs used for binding assay. (i) Schematic diagram of the γ HC sequence and protein structure. The stalk region is highlighted in yellow. (ii) Schematic diagrams of the stalk fragments and the LC1 constructs. His-α stalk, His-β, stalk and His-γ stalk are His-tagged stalk fragments constructed from Chlamydomonas α, β, and γ HCs, respectively. GST-LC1 designates GST-tagged, full-length Chlamydomonas LC1. LC1 denotes GST-tag removed LC1 by PreScission protease. The size of each construct is not drawn to scale. (B) Binding of LC1 and the stalk. Pull-down assays exploiting the His tag in the stalk constructs, analyzed by SDS–PAGE. The unbound fraction is indicated by U and the bound fraction by B above the image of the gel. (C) Stoichiometry and affinity of the binding of the γ stalk and LC1. (i) Result of tandem affinity purification. Affinity purification was sequentially performed using the His tag and GST tag. The intensity of the band possibly corresponding to a degradation product (indicated by an asterisk) was <5% of the two major bands and therefore does not significantly influence the result. (ii) Quantitative GST pull-down assay of His-γ stalk and GST-LC1. Increasing concentrations of the γ stalk were incubated with a fixed concentration of GST-LC1 and subjected to GST pull-down assay. The free γ stalk concentration in the unbound fraction and the molar ratio of γ stalk to GST-LC1 in the bound fraction were determined and plotted. The plot was fitted with the equation Y = Bmax × X/(Kd + X), where Bmax is the maximal binding ratio and Kd is the concentration at half-maximal binding. The binding was saturated at Bmax = 1.0 (molar ratio), with Kd = 1.8 nM (R2 = 0.81).
Next we examined the stoichiometry of the binding of LC1 to the γ stalk. By sequential purification using the His-tag and GST-tag, we purified His-γ stalk and GST-LC1 as a complex (Figure 4Ci). The molar ratio of His-γ stalk relative to GST-LC1 in the complex was determined to be 0.91 from the band intensities, considering the relationship between the band intensities and the molar concentration (see details for Supplemental Figure S6A). To determine further the stoichiometry, we performed a quantitative glutathione S-transferase (GST) pull-down assay using His-γ stalk and GST-LC1 prepared separately. The molar ratio of His-γ stalk to GST-LC1 was 1.0, with Kd = 1.8 nM at saturation (Figure 4Cii). From these results, we concluded that one copy of LC1 is bound to the γ stalk region. The very low Kd value suggests that the binding of LC1 to the γ stalk region is highly stable.
Binding of the His-γ stalk and GST-LC1 was also tested in high-salt conditions, and a lower degree of binding was detected even in the presence of 600 mM NaCl (Figure 5, B and C). This is consistent with the fact that LC1 was copurified with the OAD complex extracted under high-salt conditions in our preparations (Supplemental Figure S1, Aiii and Cii), as well as in previous reports (Benashski et al., 1999; Patel-King and King, 2009).
FIGURE 5:
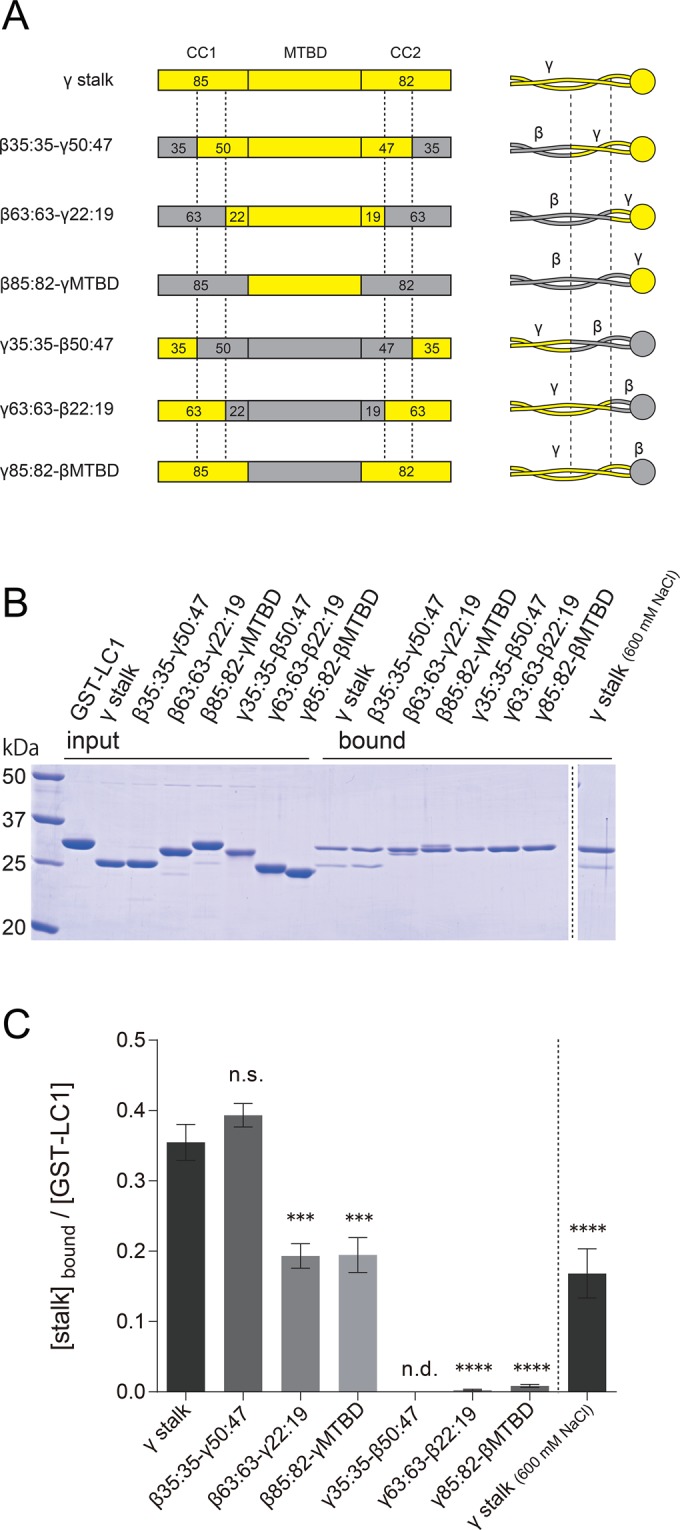
Mapping of the LC1 binding region within the γ stalk. (A) Schematics of the sequences and structures of chimeric stalk constructs. Chimeric stalk constructs of γ HC (yellow regions) and β HC (gray regions) were designed and used for GST pull-down assays with GST-LC1. The junction sites in the stalk coiled coil are indicated by dashed lines. The nomenclature is based on the number of amino acid residues in CC1 and CC2, and the former and the latter parts of the name represent the numbers in the base and tip regions. For example, in β35:35-γ50:47, the base region (35 aa of CC1 and 35 aa of CC2) is β HC sequence, and the tip region (50 aa of CC1, MTBD, and 47 aa of CC2) is γ HC sequence. See also Supplemental Figure S7. The His tags at the N-termini, for purification purposes, are omitted from the diagram. (B) SDS–PAGE of GST pull-down assays of chimeric stalk constructs and GST-LC1. Binding of the stalk constructs and GST-LC1 was carried out in PBS containing 140 mM (left) and 600 mM (right) NaCl. (C) Quantification of the binding to GST-LC1. Ratios of stalk constructs and GST-LC1 in the bound fraction were calculated for each construct from SDS–PAGE results. GST pull-down assay was carried out in a concentration range in which the binding was not saturated. The mean values were calculated from three independent experiments, and error bars represent the SE. Statistical significances compared with the γ stalk (140 mM NaCl) were determined using one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test (***p < 0.001, ****p < 0.0001; n.s., not significant; n.d., not detected).
The γ MTBD region is sufficient for the binding of LC1
Because LC1 can bind to the stalk region of γ HC but not to that of β HC, there must be an LC1-binding region in the γ HC stalk. To determine this specific region, we designed a series of chimeric stalk constructs of γ HC and β HC. To avoid possible disruption of the stalk coiled coil and/or the MTBD structure, we fused the β HC sequence and the γ HC sequence at corresponding sites, instead of making deletion or truncation constructs (Figure 5A and Supplemental Figure S7). The constructs are divided into two types: constructs with γ MTBD (β35:35-γ50:47, β63:63-γ22:19, β85:82-γMTBD) and constructs with β MTBD (γ35:35-β50:47, γ63:63-β22:19, γ85:82-βMTBD). The name of the constructs shows the number of amino acid residues of CC1 and CC2 in each construct. These constructs were incubated with GST-LC1 and subjected to GST pull-down assays. We found that constructs with the γ MTBD bound to GST-LC1, but constructs with the β MTBD did not (Figure 5, B and C). It is noteworthy that the construct β85:82-γMTBD, having a γ HC sequence only in the MTBD region, bound to the GST-LC1, but the construct γ85:82-βMTBD, having the whole γ stalk sequence except for the MTBD region, did not. For the construct (β35:35-γ50:47) with the region corresponding to 3082–3109 amino acids (aa) in CC1 and 3288–3315 aa in CC2 of the γ stalk coiled coil (indicated by dashed lines in Figure 5A; see also Supplemental Figure S7A), binding to LC1 was at the same level as for the wild-type γ stalk (Figure 5C). In contrast, ∼50% level of binding was observed for the constructs without this region (β63:63-γ22:19 and β85:82-γMTBD; Figure 5C). Because this region alone cannot accomplish LC1 binding (see data for γ63:63-β22:19 and γ85:82-βMTBD in Figure 5), this coiled-coil region of the γ HC is considered to promote the binding between LC1 and the γ MTBD region. The exact underlying mechanism is not clear, but a conformational change of coiled coil might affect γ MTBD's configuration and consequently the affinity of γ MTBD to LC1. With this in mind, to verify further that the γ MTBD region is sufficient for LC1 binding, we constructed a fragment containing only the γ MTBD region, which should no longer be affected by the stalk coiled-coil conformation. By GST-pull down assay, the γ MTBD fragment was also found to bind to GST-LC1 (Supplemental Figure S6B). From these results, we concluded that the γ MTBD region is responsible and sufficient for binding to LC1.
LC1 decreases the affinity of the γ stalk for the microtubule
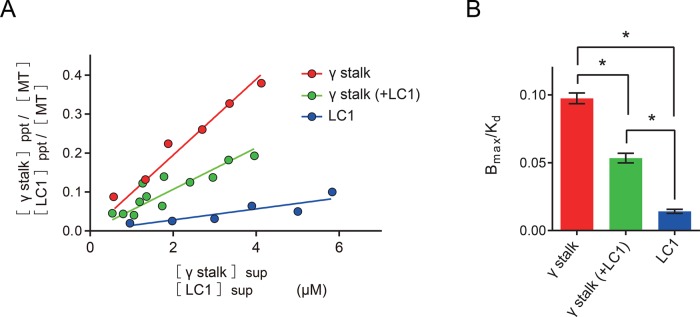
The MTBD is the region that serves as the binding interface with the microtubule (Mizuno et al., 2004; Carter et al., 2008; Redwine et al., 2012). Because LC1 was found to bind to the γ MTBD region, we speculated that LC1 might affect the binding of the γ stalk to the microtubule. To test this, we conducted a microtubule copelleting assay (Supplemental Figure S8). The affinity of the γ stalk–region fragment (His-γ stalk) in the copelleting assay with microtubules was Bmax/Kd = 0.097 (Figure 6 and Table 1). Assuming that the dynein stalk binds to the tubulin dimer in a 1:1 molar ratio at saturation (Mizuno et al., 2004; Carter et al., 2008; Redwine et al., 2012), Kd for the microtubule–γ stalk interaction was estimated to be 10 μM (Table 1). This Kd value is reasonable compared with the reported Kd values for other kinds of dynein stalk and MTBD fragments (Mizuno et al., 2004; Gibbons et al., 2005; Shimizu et al., 2008; McNaughton et al., 2010; Kato et al., 2014), and thus the assumption of Bmax = 1 is appropriate. Next, to reveal LC1's effect on the microtubule binding of the γ stalk, we first purified the γ stalk–LC1 complex using successive affinity chromatography, taking advantage of His and GST tags (note that the GST tag was removed from LC1 in the final step of purification). The purified γ stalk–LC1 complex also bound to microtubule, but its affinity for microtubules was decreased to about half of that of the γ stalk (Bmax/Kd = 0.053; Kd = 19 μM; Figure 6 and Table 1). The binding of LC1 to microtubules was also tested, and LC1 was found to bind to microtubules as previously reported (Patel-King and King, 2009). However, LC1's affinity for microtubules was found to be significantly lower than that of the γ stalk and the γ stalk-LC1 complex (Bmax/Kd = 0.014; Figure 6 and Table 1). Although we were unable to determine the Bmax and Kd of LC1 because of the extremely low affinity, we estimate that the overall affinity of LC1 to microtubules is less than one-seventh of that of the γ stalk.
FIGURE 6:
Binding of γ stalk and LC1 to the microtubule. (A) The molar ratios of γ stalk and tubulin dimer in the precipitated fractions (ppt) are plotted against the free γ stalk concentrations in the supernatant fraction (sup) for γ stalk (red) and γ stalk-LC1 complex (green). The molar ratios of LC1 and tubulin dimer in the precipitated fraction and the free LC1 concentrations in the supernatant fraction are plotted for LC1 (blue). See also Supplemental Figure S8. When X < Kd, Y = BmaxX/(Kd + X) can be approximated by Y = BmaxX/Kd. Thus the slopes (Bmax/Kd) were determined from the linear curve fitting. (B) Comparison of affinities (Bmax/Kd) for microtubules. Error bars represent the SE in the fitting. The values of Bmax/Kd are also summarized in Table 1. Statistical analyses were performed using one-way ANOVA, followed by Tukey's multiple comparisons test (*p < 0.0001).
TABLE 1:
Summary of affinities for microtubules.
| Construct | Bmax/Kd | Kd (μM) |
|---|---|---|
| γ Stalk | 0.097 | 10 |
| γ Stalk (+LC1) | 0.053 | 19 |
| LC1 | 0.014 | — |
The values of Bmax/Kd were determined from the slopes of linear fits to Figure 6A. The Kd values were estimated from the values of Bmax/Kd for the γ stalk and γ stalk–LC1 complex, assuming that Bmax = 1.
DISCUSSION
LC1 locates at the γ MTBD region
With regard to the location of LC1 within the γ HC, LC1 was proposed to bind to the AAA+ ring of the γ HC in a previous, less-well-supported model (Patel-King and King, 2009; Figure 7A). By EM observation of wild-type and mutant OADs, we found that the γ head has a large stalk tip compared with the α and β heads (Figure 2). This feature is consistent with previous rotary-shadowed EM images (Goodenough and Heuser, 1984) but has not been explicitly reported. Specificity of the binding of LC1 to the γ stalk was consistent with the results of the pull-down assays (Figure 4). Ni-NTA-nanogold labeling of His-tagged LC1 revealed that LC1 was located at the large stalk tip (Figures 1A and 3C). Averaged images of the stalk tip suggested that an additional structure, most likely corresponding to LC1, was associated at the MTBD region (Figure 3B). Association of LC1 with the γ MTBD region was also verified by pull-down assays using chimeric stalk constructs and an MTBD fragment (Figure 5 and Supplemental Figure S6B). Previous cross-linking experiments followed by vanadate-mediated ultraviolet photocleavage showed that LC1 is associated with the C-terminal region of the γ HC distal to the P1 site of AAA1 (Benashski et al., 1999). Because this region contains the stalk (including the MTBD), these results are consistent with our conclusion. A new model of LC1 localization to the γ head domain is shown in Figure 7B.
FIGURE 7:
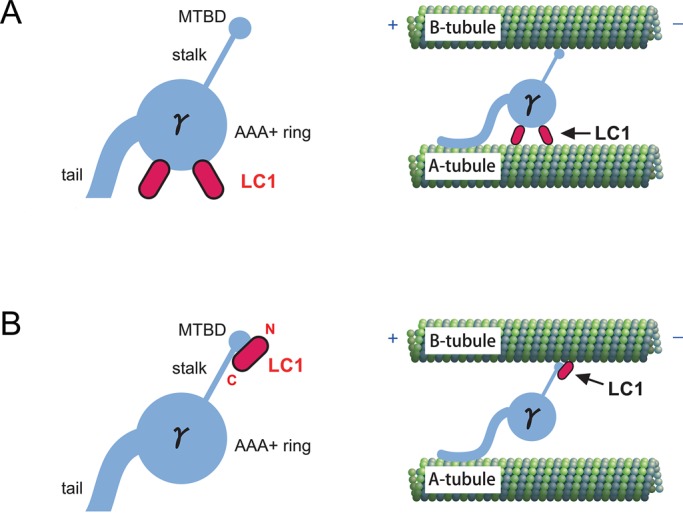
Model of the LC1 localization within the γ head domain and the axoneme. (A) Previous model of LC1 localization. Two copies of LC1 were assumed to bind to the AAA+ ring of the γ head domain. Inside the axoneme, LC1 was presumed to tether the AAA+ ring to the A-tubule of DMT. The model was adopted from Patel-King and King (2009). (B) A new model of LC1 localization. One copy of LC1 is believed to locate on the γ MTBD region with its N-terminus oriented toward the tip and C-terminus toward the base. In this model, LC1 interacts with the B-tubule of the DMT together with the γ MTBD inside the axoneme. LC1 is likely to regulate OAD activity by modulating the affinity of the γ MTBD for the B-tubule. Note that the LC1-binding site within the MTBD region was not determined in this study.
Sequence comparison between the MTBD regions in Chlamydomonas α, β, and γ HCs indicates that their overall sequences are similar to each other, except that the γ MTBD has a significantly longer insert sequence between helix 2 (H2) and helix 3 (H3; Supplemental Figure S7B). The numbers of amino acid residues in the inserts compared with the cytoplasmic dynein MTBD are as follows: α HC, 2 aa; β HC, 4 aa; and γ HC, 15 aa. This feature was also present in the Tetrahymena and the human HCs homologous to the Chlamydomonas γ HC (Supplemental Figure S7B). A recent nuclear magnetic resonance study on the MTBD of Chlamydomonas inner arm dynein-c showed that a 13-aa insert sequence between the H2 and H3 forms an extended structure called the “flap” (Kato et al., 2014). By homology modeling, we performed a structural prediction on the MTBD of α, β, and γ HCs and found that only the γ MTBD forms the “flap” structure (Supplemental Figure S9). Although we were not able to dissect the LC1-binding site within the γ MTBD region, this flap structure is specific to the γ MTBD among the three HCs of OAD complex, and therefore it is a plausible candidate for the LC1 binding site. High-resolution structural studies using cocrystallization and mutational analysis will clarify this point.
Orientation of LC1 on the γ MTBD
The labeling of His-tagged LC1 with Ni-NTA-nanogold at either the N- or the C-terminus (Figure 3C) suggested that LC1 is located on the γ MTBD region with its N-terminus oriented toward the tip and its C-terminus toward the base (Figure 7B). Previous mutational analysis showed that several basic residues in the N-terminal region are important for LC1's binding to the DMT (King and Patel-King, 2012). Based on the LC1's orientation in our model, the N-terminal region should locate close to a microtubule (Figure 7B), and this model is consistent with the results of a previous mutation study. Because the LC1–γ MTBD association persists in high-salt conditions, it might be mediated by the β-sheet face of leucine-rich repeat structure of LC1, where a hydrophobic patch is present (Patel-King and King, 2009).
Previously LC1 was presumed to associate with the A-tubule of the DMT (Figure 7A). This model was based on the results of in situ immunogold EM as well as on the assumption that LC1 was too short (∼7 nm long at most) to bridge the gap between the AAA+ ring and the B-tubule (>10 nm; Patel-King and King, 2009). However, considering that LC1 locates to the γ MTBD region, LC1 is likely to interact with the B-tubule of the adjacent DMT (Figure 7B).
Number of LC1 molecules associated with the γ head
In the previous model, two copies of LC1 were presumed to bind to the γ head domain (Patel-King and King, 2009; Figure 7A). From our results on the stoichiometry of binding, the number of LC1 molecules bound to the γ MTBD was estimated to be one (Figure 4C). Although our data do not totally rule out the possibility of two copies of LC1 associating with the γ head domain, this possibility does not appear very high. When the OAD complex was affinity purified by taking advantage of the protein tag on the recombinant LC1, the purified OAD complex did not contain endogenous LC1 (Supplemental Figure S1, Aiii and Cii). Because endogenous LC1 was present in both Chlamydomonas and Tetrahymena transformants, the affinity-purified OAD should contain endogenous LC1 in addition to recombinant LC1 if two copies of LC1 were attached to a single OAD complex. This result suggests that either there is no LC1 present other than the one at the MTBD or the association of the other copy of LC1 to the γ head domain is very weak (Figure 7B). This model is compatible with a previous biochemical result showing that LC1 binds to OAD complex at a 1:1 molar ratio (Barkalow et al., 1994).
Implications for LC1's regulatory mechanism of the OAD complex
Although LC1 itself had a weak microtubule-binding ability, binding of LC1 decreased γ stalk's affinity for the microtubule by about half in the MT copelleting assay (Figure 6). There are alternative possible explanations for this mechanism. First, LC1 might cause a steric hindrance to the γ stalk's microtubule binding. Second, LC1's binding could induce a structural change in the γ MTBD region; for example, a low-affinity structure of the γ MTBD for microtubules might be stabilized. High-resolution structural information would be needed to fully understand the mechanism. Note that there are differences in tubulin modifications between in vitro–assembled microtubules and the B-tubule of DMT (Westermann and Weber, 2003), and the values of affinities in Figure 6 and Table 1 might not accurately reflect the strength of the interaction between the γ stalk-MTBD and B-tubule.
Previously LC1 was proposed to work as a tether between the γ HC and the DMT (Patel-King and King, 2009). However, our results do not support this possibility, since LC1 decreased the γ stalk's affinity for the microtubule. One possible LC1 function may be to regulate the affinity of the γ MTBD for the B-tubule so that the γ HC can interact with the B-tubule only when it is necessary. Previously LC1 was found to be important for the transition from effective stroke to recovery stroke in Chlamydomonas flagellar beating (Patel-King and King, 2009). In Paramecium, a 29-kDa light chain of the OAD complex (p29), possibly corresponding to Paramecium LC1, undergoes phosphorylation, which enhances the OAD activity (Hamasaki et al., 1991; Barkalow et al., 1994; Kutomi et al., 2012). Future studies focusing on the relationship of LC1 and force generation of the OAD complex, such as in vitro motility studies using the OAD complex with or without LC1, will answer the question.
Conclusions
In this study, we found a novel feature of OAD: the stalk tip of the γ HC is larger than the tips in the other two HCs, and this is due to the presence of LC1 at the MTBD region of the γ HC. Further, our results suggest that LC1 functions to modulate the binding affinity of the γ head to microtubules. It is unexpected that dynein LC subunit localizes at the MTBD, which directly affects the interaction between dynein and microtubules. These findings provide the basis for understanding the regulatory function of LC1 in ciliary and flagellar motility and offer new insight into diverse and complicated regulatory mechanisms of the dynein complex.
MATERIALS AND METHODS
The strains of Chlamydomonas and Tetrahymena used in this study, the construction and purification of the proteins, and the computational analysis are described in the Supplemental Materials and Methods.
Electron microscopy
Purified OAD-complex samples were applied to carbon grids prehydrophilized as in Sakato et al. (2007), rinsed with MME buffer (30 mM 3-(N-morpholino)propanesulfonic acid, 5 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, pH 7.4), and negatively stained with 1.5% (wt/vol) uranyl acetate. The grids were observed in an H-7500 electron microscope (Hitachi, Tokyo, Japan) at 40,000× magnification operating at 80 kV. The EM images were recorded on either a 1024 × 1024–pixel charge-coupled device camera (FastScan-F114; Tietz Video and Image Processing Systems, Gauting, Germany) or film (Electron Microscope FG Film; Fujifilm, Tokyo, Japan).
Single-particle analysis
Single-particle analysis was performed using the SPIDER program (Frank et al., 1996) as in Torisawa et al. (2014). For single-particle analysis, images recorded on film were digitized using a Super CoolScan 9000 ED scanner (Nikon, Tokyo, Japan). To average Tetrahymena DYH3 head fragments, 1486 molecules with the large stalk tip were selected by visual inspection and boxed manually using X3d (Conway et al., 1999). The images were aligned according to the single-reference algorithm, followed by a multireference algorithm with iterative process. The major group obtained was subclassified by K-means clustering. The images were further aligned with reference to the stalk region by applying a mask to most of the AAA+ ring region. For the averaging of the stalk tip region, only the stalk-tip region was boxed and processed similarly. For the oda11 (βγ two-headed) OAD complex, each of the two heads (with or without a large stalk tip) was processed separately.
Ni-NTA-nanogold labeling
Ni-NTA-nanogold particles were prepared according to Kitai et al. (2011). Purified OAD complex solution (∼20 μl) was mixed with Ni-NTA-nanogold particle solution (1–2 μl) so that the molar ratio of the OAD complex and nanogold would be 1:1 and incubated on ice for 30 min. Afterward, the samples were negatively stained as described. For quantification, the center distances between the AAA+ rings and the gold particles bound to the stalk tip were measured manually using ImageJ software (National Institutes of Health, Bethesda, MD). Although the gold particles were sometimes found on or within the AAA+ ring, this apparent binding might reflect an incidental overlap between the stalk and the AAA+ ring because of some interactions with the carbon grid in negative-staining EM. Therefore such gold particles were excluded from the quantification.
Pull-down assays
For pull-down assays using the His tag added to the stalk constructs, Dynabeads His-Tag Isolation and Pulldown (Life Technologies, Carlsbad, CA) was used. Stalk constructs (2.1 μM) and LC1 (4.3 μM) were incubated in 50 μl of His-tag binding buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.01% Tween-20) for 10 min at 25°C. Four microliters of Dynabeads solution (50% slurry) was added to the reaction solution and incubated for 10 min at 25°C. Dynabeads were isolated from solution (unbound fraction) using a magnet. Dynabeads were washed once with 800 μl of wash buffer (50 mM Tris-HCl, pH 8.0, 90 mM NaCl, 20 mM imidazole, 0.01% Tween-20), and bound protein was eluted from the beads by boiling in sample buffer.
For GST pull-down assays, stalk constructs or chimeric stalk constructs (229 nM) and GST-LC1 (316 nM) were incubated in 450 μl of phosphate-buffered saline (PBS) containing 1 mM dithiothreitol (DTT) for 30 min at 25°C. Then the proteins were incubated with 10 μl of Novagen GST•Bind Resin (Merck KGaA, Darmstadt, Germany) for 30 min at 4°C with rotation. GST•Bind Resin was collected by centrifugation, and the unbound protein fraction was subjected to trichloroacetic acid (TCA) precipitation. After the GST•Bind Resin was washed once with 450 μl of PBS containing 1 mM DTT, bound protein was eluted from the resin with 30 μl of GST elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM glutathione) for 5 min at 25°C. Quantitative GST pull-down assays were performed similarly, except that 158 nM GST-LC1 was incubated with 52.6–631 nM His-γ stalk. After TCA precipitation and SDS–PAGE, the amount of His-γ stalk in the unbound fraction was determined by comparing the intensities of the bands with those from known concentrations of His-γ stalk. The molar ratio of His-γ stalk to GST-LC1 was determined from the ratio of the two bands in the bound fraction.
Microtubule copelleting assay
Tubulin was purified from porcine brain as in Vallee et al. (1986), polymerized into microtubule, and stabilized with 40 μM paclitaxel. Microtubule, 5 μM, was incubated with an increasing concentration of His-γ stalk, γ stalk–LC1 complex, or LC1 in 45 μl of NAP5 buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl) containing 15 μM paclitaxel for 30 min at 25°C. The solution was subjected to ultracentrifugation (120.2 rotor, 75,000 rpm, 10 min, 27°C; Optima TLX ultracentrifuge; Beckman Coulter, Brea, CA). The supernatant and precipitated fraction were examined by SDS–PAGE, and images of the gels were digitized. The ratios of the band intensities in supernatant and precipitation were determined, and the protein concentration in each fraction was calculated from the input protein concentrations.
Supplementary Material
Acknowledgments
We thank T. Kobayashi for helpful discussion. This work was supported by Japanese Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant 25113504 to Y.Y.T. and Grant-in-Aid for Japan Society for Promotion of Science Fellowship to M.I.
Abbreviations used:
- LC1
light chain-1
- MT
microtubule
- OAD
outer arm dynein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0289) on September 23, 2015.
REFERENCES
- Acar S, Carlson DB, Budamagunta MS, Yarov-Yarovoy V, Correia JJ, Niñonuevo MR, Jia W, Tao L, Leary JA, Voss JC, et al. The bipolar assembly domain of the mitotic motor kinesin-5. Nat Commun. 2013;4:1343. doi: 10.1038/ncomms2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkalow K, Hamasaki T, Satir P. Regulation of 22S dynein by a 29-kD light chain. J Cell Biol. 1994;126:727–735. doi: 10.1083/jcb.126.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Kabututu ZP, Hill KL. Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J Cell Sci. 2007;120:1513–1520. doi: 10.1242/jcs.004846. [DOI] [PubMed] [Google Scholar]
- Benashski SE, Patel-King RS, King SM. Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the γ heavy chain. Biochemistry. 1999;38:7253–7264. doi: 10.1021/bi990466y. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. Control of flagellar bending: a new agenda based on dynein diversity. Cell Motil Cytoskeleton. 1994;28:199–204. doi: 10.1002/cm.970280303. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 2008;183:923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Steven AC. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- DiBella LM, Benashski SE, Tedford HW, Harrison A, Patel-King RS, King SM. The Tctex1/Tctex2 class of dynein light chains dimerization, differential expression, and interaction with the LC8 protein family. J Biol Chem. 2001;276:14366–14373. doi: 10.1074/jbc.M011456200. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Furuta A, Yagi T, Yanagisawa HA, Higuchi H, Kamiya R. Systematic comparison of in vitro motile properties between Chlamydomonas wild-type and mutant outer arm dyneins each lacking one of the three heavy chains. J Biol Chem. 2009;284:5927–5935. doi: 10.1074/jbc.M807830200. [DOI] [PubMed] [Google Scholar]
- Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heuser J. Structural comparison of purified dynein proteins with in situ dynein arms. J Mol Biol. 1984;180:1083–1118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- Hamasaki T, Barkalow K, Richmond J, Satir P. cAMP-stimulated phosphorylation of an axonemal polypeptide that copurifies with the 22S dynein arm regulates microtubule translocation velocity and swimming speed in Paramecium. Proc Natl Acad Sci USA. 1991;88:7918–7922. doi: 10.1073/pnas.88.18.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth J, Fliegauf M, Olbrich H, Kispert A, King SM, Mitchison H, Zariwala MA, Knowles MR, Sudbrak R, Fekete G, et al. Identification and analysis of axonemal dynein light chain 1 in primary ciliary dyskinesia patients. Am J Respir Cell Mol Biol. 2005;33:41–47. doi: 10.1165/rcmb.2004-0335OC. [DOI] [PubMed] [Google Scholar]
- Kato YS, Yagi T, Harris SA, Ohki S, Yura K, Shimizu Y, Honda S, Kamiya R, Burgess SA, Tanokura M. Structure of the microtubule-binding domain of flagellar dynein. Structure. 2014;22:1628–1638. doi: 10.1016/j.str.2014.08.021. [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS. Functional architecture of the outer arm dynein conformational switch. J Biol Chem. 2012;287:3108–3122. doi: 10.1074/jbc.M111.286211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitai T, Watanabe Y, Toyoshima YY, Kobayashi T, Murayama T, Sakaue H, Suzuki H, Takahagi T. Simple method of synthesizing nickel-nitrilotriacetic acid gold nanoparticles with a narrow size distribution for protein labeling. Jpn J Appl Phys. 2011;50:095002. [Google Scholar]
- Kobayashi T, Morone N, Kashiyama T, Oyamada H, Kurebayashi N, Murayama T. Engineering a novel multifunctional green fluorescent protein tag for a wide variety of protein research. PLoS One. 2008;3:e3822. doi: 10.1371/journal.pone.0003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- Koonce MP. Identification of a microtubule-binding domain in a cytoplasmic dynein heavy chain. J Biol Chem. 1997;272:19714–19718. doi: 10.1074/jbc.272.32.19714. [DOI] [PubMed] [Google Scholar]
- Kurkowiak M, Zie¸tkiewicz E, Witt M. Recent advances in primary ciliary dyskinesia genetics. J Med Genet. 2015;52:1–9. doi: 10.1136/jmedgenet-2014-102755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutomi O, Hori M, Ishida M, Tominaga T, Kamachi H, Koll F, Cohen J, Yamada N, Noguchi M. Outer dynein arm light chain 1 is essential for controlling the ciliary response to cyclic AMP in Paramecium tetraurelia. Eukaryotic Cell. 2012;11:645–653. doi: 10.1128/EC.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. Structural mechanism of the dynein power stroke. Nat Cell Biol. 2014;16:479–485. doi: 10.1038/ncb2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazor M, Alkrinawi S, Chalifa-Caspi V, Manor E, Sheffield VC, Aviram M, Parvari R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am J Hum Genet. 2011;88:599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton L, Tikhonenko I, Banavali NK, LeMaster DM, Koonce MP. A low affinity ground state conformation for the dynein microtubule binding domain. J Biol Chem. 2010;285:15994–16002. doi: 10.1074/jbc.M109.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Toba S, Edamatsu M, Watai-Nishii J, Hirokawa N, Toyoshima YY, Kikkawa M. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J. 2004;23:2459–2467. doi: 10.1038/sj.emboj.7600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat Struct Mol Biol. 2010;17:761–767. doi: 10.1038/nsmb.1832. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, King SM. An outer arm dynein light chain acts in a conformational switch for flagellar motility. J Cell Biol. 2009;186:283–295. doi: 10.1083/jcb.200905083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol. 2012;178:199–206. doi: 10.1016/j.jsb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Redwine WB, Hernández-López R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, Burgess SA. AAA+ ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Patel-King RS, King SM. An outer arm Dynein conformational switch is required for metachronal synchrony of motile cilia in planaria. Mol Biol Cell. 2010;21:3669–3679. doi: 10.1091/mbc.E10-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakato M, Sakakibara H, King SM. Chlamydomonas outer arm dynein alters conformation in response to Ca2+ Mol Biol Cell. 2007;18:3620–3634. doi: 10.1091/mbc.E06-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Sleigh MA. Ciliary activity of cultured rabbit tracheal epithelium: beat pattern and metachrony. J Cell Sci. 1981;47:331–347. doi: 10.1242/jcs.47.1.331. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, Spassky N, Murcia NS, Garcia-Verdugo JM, Marin O, Rubenstein JLR, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat Struct Mol Biol. 2012;19:492–497. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Kato Y, Morii H, Edamatsu M, Toyoshima YY, Tanokura M. The dynein stalk head, the microtubule binding-domain of dynein: NMR assignment and ligand binding. J Biomol NMR. 2008;41:89–96. doi: 10.1007/s10858-008-9242-6. [DOI] [PubMed] [Google Scholar]
- Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, Kojima H, Toyoshima YY, Furuta KY. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol. 2014;16:1118–1124. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- Toyoshima YY. Chymotryptic digestion of Tetrahymena 22S dynein. I. Decomposition of three-headed 22S dynein to one-and two-headed particles. J Cell Biol. 1987;105:887–895. doi: 10.1083/jcb.105.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB. Reversible assembly purification of microtubules without assembly-promoting agents and further purification of tubulin, microtubule-associated proteins, and MAP fragments. Methods Enzymol. 1986;134:89–104. doi: 10.1016/0076-6879(86)34078-3. [DOI] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–948. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Wu H, Maciejewski MW, Marintchev A, Benashski SE, Mullen GP, King SM. Solution structure of a dynein motor domain associated light chain. Nat Struct Mol Biol. 2000;7:575–579. doi: 10.1038/76804. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Saito K, Sutoh M, Nishizaka T, Toyoshima YY, Yajima J. Torque generation by axonemal outer-arm dynein. Biophys J. 2015;108:872–879. doi: 10.1016/j.bpj.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.