Abstract
DNA polymerase gamma (Pol γ) is a nuclear encoded, mitochondrially located replicase that conducts all DNA synthesis in the organelle. Structurally, human Pol γ closely resembles bacteriophage T7 DNA polymerase. Perhaps due to this prokaryotic-like feature, Pol γ is highly susceptible to inhibition by drugs designed against HIV reverse transcriptase and HCV RNA polymerase. In this review, I summarize recent structural and biochemical studies towards understanding Pol γ-mediated antiviral drug toxicity.
Unlike the maintenance of nuclear DNA that involves multiple DNA polymerases, a single enzyme, Pol γ, performs all DNA synthesis in mitochondria [1]. Mitochondrial DNA (mtDNA) codes for tRNAs and rRNAs, necessary for protein synthesis in the organelle, but only encodes a subset of the genes for the oxidative phosphorylation electron transfer chain. If mitochondria did evolve from α-protobacteria, most of the bacterial genes, including genes for Pol γ and the other components of oxidative phosphorylation, have migrated to the nucleus. Mutations affecting Pol γ manifest as human diseases involving multisystems with a varied onset time (for reviews see [2-4]). This creates a challenge to correlate genotype with phenotype. A better understanding of the molecular basis for how Pol γ mutations translate to clinical disorders is urgently needed. Pol γ has also been linked to the toxicity of antiviral reagents designed to inhibit viral replication [5-8]. Most notably, these include nucleoside analogs designed to inhibit HIV reverse transcriptase (NRTIs) and hepatitis C virus RNA polymerase. Prolonged usage of NRTIs during AIDS treatment causes severe adverse reactions with clear indications of mitochondrial dysfunction [9].
Human Pol γ holoenzyme is comprised of a catalytic subunit Pol γA and an accessory dimeric subunit Pol γB. Pol γ contains three enzymatic activities, DNA polymerase (pol), 3′->5′ proofreading exonuclease (exo), and a 5′-deoxyribosephosphate (5′-dRP) lyase activity [10-12]. Despite all three active sites being located on Pol γA, Pol γB alters all activities upon formation of holoenzyme. In comparison to Pol γA alone, holoenzyme has elevated pol and 5′-dRP lyase activities but decreased exo activity [10,13-15]. Holoenzyme-catalyzed synthesis is also much more processive.
A crystal structure of human Pol γ holoenzyme has been recently determined [16]. Successful crystallization of Pol γ required modification of both subunits. An N-terminal poly-glutamine were deleted from Pol γA, and a non-essential four-helical bundle was removed from Pol γB. This deletion of Pol γB is critical for crystallization because a crystal contact is formed at this newly created surface. The holoenzyme crytals were initially assigned to P3221 space group, but later found to be twinned and belong to space group P32. The structure was solved using the detwinning function in the program cns [54].
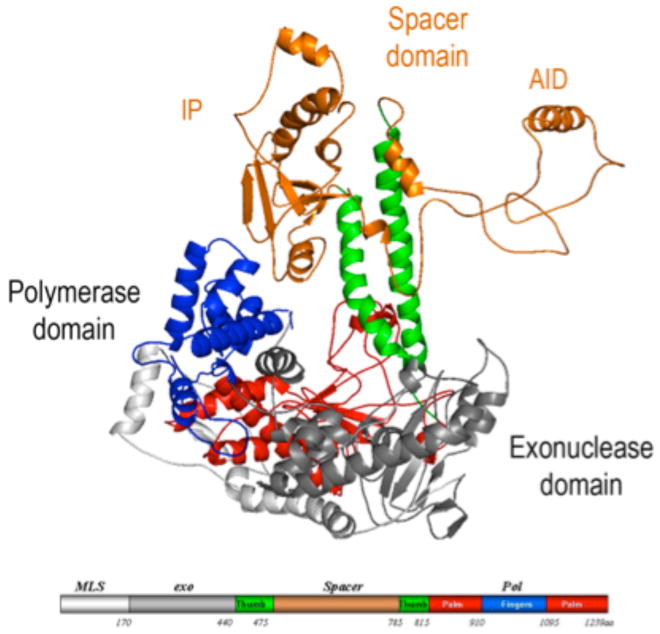
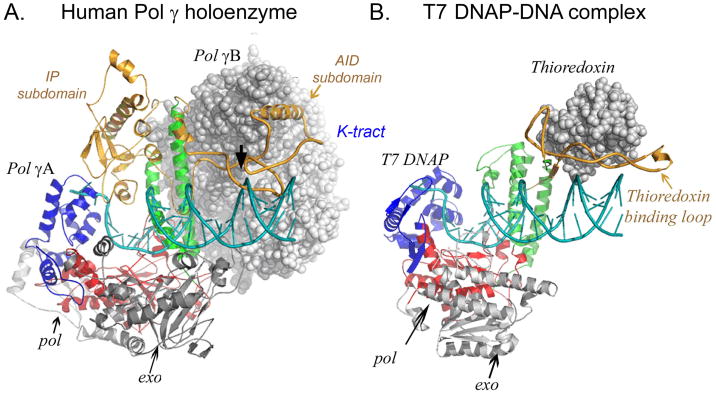
The PolγA exo domain is N-terminal located while the C-terminal polymerase (pol) domain is split by an important and unusually large spacer domain (Fig. 1). The pol domain presents a typical polymerase right-hand fold with palm, thumb and fingers subdomains. Despite the evidence for a bacterial origin of mitochondria, Pol γA closely resembles bacteriophage T7 DNAP structurally. The spacer domain can be subdivided into IP (intrinsic processivity) and AID (accessory interacting) subdomain. The IP subdomain presents a unique fold absent in other Pol A family enzymes, whereas the AID subdomain forms the major interacting surface with the accessory subunit Pol γB in a manner reminiscent of the way T7 gp5 binds to its accessory protein, thioredoxin, in forming T7 DNAP. The two catalytic palm domains superimpose so well (with a rmsd 2.3 Å) that a model of the Pol γ-DNA complex can be constructed by docking the primer-template DNA from the T7 DNAP-DNA structure [17] (Fig. 2).
Figure 1.
Structure of Pol γA. The pol domain shows a canonical ‘right-hand’ configuration with thumb (green), palm (red) and fingers (blue) subdomains, and the exo domain (grey). The spacer domain (orange) presents a unique structure and is divided into two subdomains. Domains are shown in a linear form where the N-terminal domain contains residues 1-170; exo: 171-440; spacer: 476-785; pol: 441-475 and 786-1239.
Figure 2.
Similarities between human Pol γ and T7 DNA polymerase. The catalytic subunits (ribbon diagram) of the two polymerases are oriented by superpositioning palm (red) subdomains. Pol g-DNA complex is constructed with a docked DNA from a T7 DNAP-DNA complex structure. The accessory subunits are presented in space-filling models.
Mammalian Pol γB is a structural homolog of certain class II aminoacyl-tRNA synthetases (aaRS), particularly of threonyl-tRNA and glycyl-tRNA synthetases [18], but no longer possesses aaRS activity. However, the tRNA-binding patch appears to be conserved, and Pol γB is predicted to bind to some tRNAs, although this has not been demonstrated directly. Interestingly, mammalian Pol γB is a homodimer, but its counterparts in insects (e.g., Drosophila and mosquito) are monomeric, and fungal Pol γ seems to have no requirement for an accessory subunit.
Processivity of Pol γ
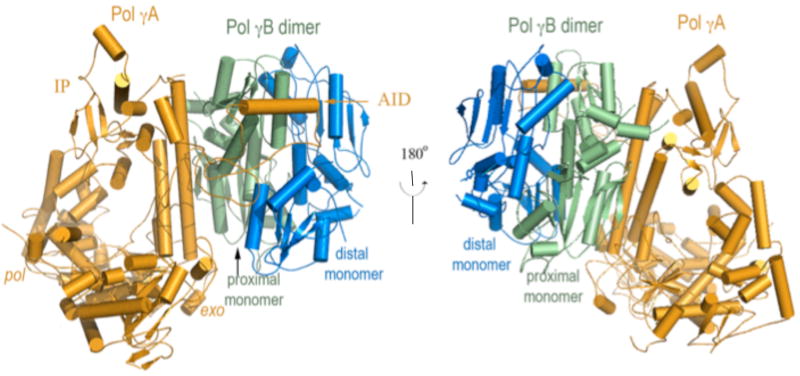
In humans, Pol γA interacts with the homodimeric Pol γB asymmetrically: an extensive interaction exists between Pol γA and the monomer of Pol γB proximal to the catalytic subunit, but makes very limited interactions with the distal monomer (Fig. 3). Such an arrangement suggests that a monomeric Pol γB should be functional. A monomeric human Pol γB was generated by changing the human protein so that it more closely resembles its insect counterpart. Deletion of a 4-helical bundle in Pol γB that constitutes 40% of the dimer interface but has no counterpart in Drosophila, and removal of two inter-subunit salt bridges, makes human Pol γB monomeric. This construct enabled comparative studies of the monomeric and dimeric forms.
Figure 3.
Structure of the heterotrimeric Pol γ holoenzyme containing one catalytic subunit Pol γA (orange) and the proximal (green) and distal (blue) monomers of Pol γB. Pol γA primarily interacts with the proximal monomer of the dimeric Pol γB.
Processivity is defined as the length of DNA synthesized per binding event, it is determined by at least two factors: the rate of polymerization (kpol), and the duration of DNA-binding (defined by the off rate, koff). Dimeric Pol γB increases the affinity of Pol γA for DNA and accelerates the polymerization rate. The two factors contribute synergistically to the increased processivity of holoenzyme and are neatly divided between the two Pol γB monomers: the proximal monomer enhances DNA affinity while the distal monomer accelerates the polymerization rate [19]. These results suggest that, in the absence of compensating changes in the catalytic subunit Pol γA, the mtDNAP of organisms with a monomeric Pol γB should exhibit lower processivity. This idea has not yet been tested under identical reaction conditions using mtDNAPs isolated from different sources. Of particular note is that human Pol γB represents a rare example of identical proteins having clearly distinct biological or biochemical functions.
The structure of holoenzyme also sheds light on the mechanism of processivity enhancement by Pol γB. The interaction between Pol γA and Pol γB in holoenzyme is largely maintained by the hydrophobic interaction between the Pol γA AID subdomain and the proximal Pol γB monomer. The AID subdomain is predicted to be disordered in the catalytic subunit, but association with Pol γB stabilizes the other surface of the amphipathic AID subdomain, which contains a high density of positively charged residues (496KQKKAKKVKK505, the K-tract) that can bind DNA [16]. The K-tract interaction doubles the DNA-contact length of Pol γA from 10 bp to ∼20 bp, clearly enhancing the affinity of holenzyme for DNA.
The holenzyme structure suggests that processivity enhancement by Pol γB is entirely indirect, even though the accessory protein possesses an intrinsic DNA-binding property. Pol γB DNA-binding site mutants still function as a processivity factor and there is little or no effect on holoenzyme activity on primed single-stranded DNA [20-22]. Nonetheless, the region is important in replication, as the same mutants inhibit synthesis by the intact replisome on duplex DNA [21]. This result suggests that Pol γB may stabilize the replisome by interacting with the mitochondrial helicase and/or SSB, or perhaps with forked DNA during replication. The Pol γA AID subdomain may also contribute to replisome formation as the corresponding subdomain in T7 gp5 directly interacts with T7 helicase [23].
Pol γB coordinates pol and exo activities
Intriguingly, Pol γB regulates pol and exo activities in opposite directions. While Pol γB increases the rate of DNA synthesis and the processivity of holoenzyme, it simultaneously reduces exonuclease activity. When an incorrect nucleotide is incorporated, it is shuttled to the exo site for excision. The two active sites, which are separated by 40 Å, alternate in function, as a primer strand can only be bound to one active site at any given time. Altering the binding equilibrium between the two sites therefore affects each activity in opposite directions.
The mechanism of simultaneous regulation by Pol γB of both polymerase and exonuclease activities of Pol γA activities was partially revealed during an investigation of certain Pol γA mutants. Substitutions of Pol γA R232 have been implicated in severe neurological and muscular disorders. The mutant Pol γA displays almost normal pol and exo activities – an expected result because R232 is located outside of either active site. However, the mutant holoenzyme shows almost no response to the presence of Pol γB even though the two subunits appear to interact normally [24]. The studies suggest that Pol γB biases the equilibrium of wild-type holoenzyme towards positioning the primer terminus in pol, thereby increasing the polymerization rate and reducing exonuclease activity. Substitution of Pol γA R232 appears to block the signal from Pol γB, possibly by preventing the necessary structural changes associated with shuttling the primer terminus between exo and pol. Mutant holoenzymes containing Pol γA R232 variants therefore exhibit elevated nucleolytic activity and reduced DNA synthesis. These data explain the low DNA content (3-5%) in patients harboring R232 mutants [25-28], and also suggests how the nucleolytic activity of Pol γA can be regulated by factors beyond the immediate vicinity of the exonuclease active site.
The third function of Pol γ, 5′-dRP lyase activity, is used during base-excision repair [29-31]. In mitochondria, the high concentration of reactive oxygen species generated in redox reactions is highly damaging to DNA, mtDNA experiences a 10-fold higher rate of oxidative damage than nuclear DNA [32,33]. Historically, DNA repair in mitochondria was thought to be minimal and damaged DNA was simply degraded. However, good evidence of robust repair machinery for base-excision repair, mismatch repair and double-strand break has now been found [10,34-36]. The 5′-dRP lyase activity of Pol γ positions it as an integral part of base-excision repair where, after removal of a damaged base by specific glycolyases, the resulting abasic site is eliminated by the lyase activity. Relative to the prototypical eukaryotic repair DNA polymerase, Pol β, Pol γ exhibits comparable 5′-dRP activity [10]. In a 5′-dRP lyase reaction, an electrophilic nitrogen, typically either an N-terminal group or an internal lysine, makes a nucleophilic attack on the C1′ of the 5′-dRP and forms a Schiff s base. Subsequently, through a β-elimination reaction, the 5′-dRP is excised. Evidence for the involvement of Pol γ in removal of 5′-dRP includes trapping a covalently linked Pol γ-DNA intermediate [13,34]. Upon 5′-dRP removal, the polymerase activity of Pol γ fills in the one nucleotide gap, allowing DNA ligase to restore the integrity of the DNA.
Like its regulatory roles in other Pol γA activities, Pol γB greatly enhances 5′-dRP lyase activity in holoenzyme [13]. Although the location of the lyase active site on Pol γA is currently unknown, regions of Pol γB affecting holoenzyme lyase activity have been identified. These contribute either to formation of Pol γB dimers or to its intrinsic DNA-binding activity. The nucleophile for Pol γA 5′-dRP activity is not known but its likely general area can be deduced from the function of Pol γB. Because Pol γB increases both pol and 5′-dRP lyase activities by biasing the primer terminus towards the pol site, the 5′-dRP lyase nucleophile should be located in the vicinity of pol.
Pol γ and antiviral drug toxicity
Successful AIDS treatment requires combinatory administration of drugs inhibiting HIV protease, reverse transcriptase and integrase (HAART therapy). The central components of HAART are inhibitors against HIV reverse transcriptase (RT), which is responsible for replicating the viral genome. There are two broad classes of inhibitors against HIV RT: non-nucleoside analogs that allosterically inhibit enzyme activity, and nucleoside analogs (NRTIs) that directly compete with incoming nucleotide triphosphates during synthesis.
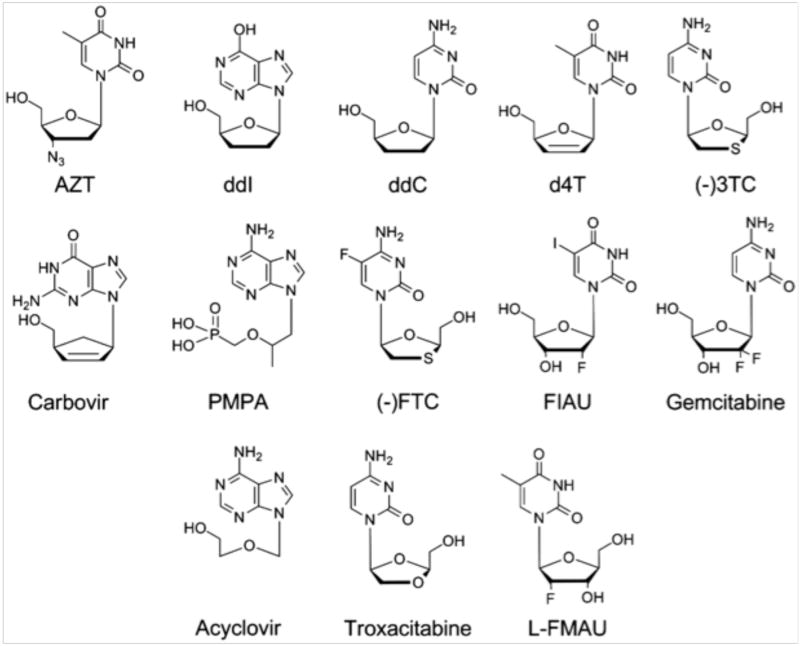
After phosphorylation by cellular enzymes, most NRTIs are dNTP analogs where the ribose moiety lacks a 3′-OH group (Fig. 4); they therefore serves as chain terminators. In principle, NRTIs can inhibit the activities of any human DNA polymerase causing drug toxicity, but studies have revealed that of the human DNA replicases, Pol γ, Pol δ and Pol ε, as well as the primase Pol α, Pol γ is inhibited 101-105-fold more effectively than other polymerases [37]. The high sensitivity of Pol γ to NRTIs inhibition explains, in part, the NRTIs mitochondrial toxicity. Other sources contribute to the mitochondrial adverse reactions are high drug concentrations as a consequence of active transporters and genetic predisposition [38,39]. However, since these inhibitors are effective against HIV-RT, human Pol γ must share some common structural features and/or mechanisms for inhibitor incorporation with the viral enzyme.
Figure 4.
Structures of selected nucleoside HIV reverse transcriptase analog inhibitors.
Selective inhibition of Pol γ indicates that it is either less discriminating between an NRTI and the substrate nucleotide or less efficient in excision of NRTI-containing primers. The rate of NRTI incorporation does not necessarily correlate with its rate of removal, suggesting that the pol and exo active sites exhibit independent discrimination criteria. For example, ddCTP, (-)3TC and (+)3TC are all cytidine derivatives (Fig. 4), but Pol γ incorporates ddCTP most effectively but removes it the least. Conversely, Pol γ incorporates (-)3TC least effectively but removes it the fastest [40,41]. Consequently, ddC is among the most toxic of inhibitors. These results indicate the most useful drugs should stimulate high nucleolytic activity and/or low incorporation for human Pol γ, but still be a good substrate for HIV RT.
Selective inhibition of Pol γ
To understand the basis for Pol γ's susceptibility to NRTIs, it is useful to compare the Pol γ structure to other DNA replicases to try and identify unique features of Pol γ that make it a target for NRTIs. It is also helpful to compare structures of Pol γ with HIV-RT to spot similarities and differences, and to try and exploit differences in the design of new antiviral drugs.
The high susceptibility of Pol γ to NRTIs may lie in structural features around the pol and exo domains. Pol γ belongs to the Pol A family, whereas other human replicases Pol α, Pol δ and Pol ε are Pol B family members. Regardless, the structures of both (and other) families are related: the palm subdomain is the most conserved, but large variations have been observed in the thumb and fingers subdomains [42-45], which form the DNA template and dNTP substrate-binding site, respectively.
Both A-family and B-family polymerases use a positively charged helix in the fingers domain, the ‘O-helix’, to bind the negatively charged phosphates of the incoming dNTP. To date, the most complete structural characterization of B-family polymerases is from bacteriophage RB69, where the structures of apo, and both synthesis and editing complexes have been determined [44,46,47]. Human Pol α and Pol δ share high sequence homology with RB69, especially around the exo and pol active sties. In an attempt to rationalize the varying toxicity of NRTIs towards human DNA polymerases, structures of RB69 DNAP and the composite Pol γ-DNA complex (Fig. 5-6) can be therefore used in an initial attempt to rationalize the varying toxicity of NRTIs towards human DNA polymerases.
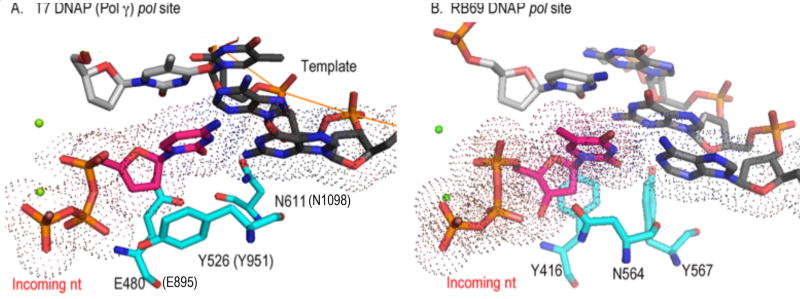
Figure 5.
Comparison of Pol A family and Pol B family in the pol site Pol A and Pol B members enforce W-C complementarity between the templating nucleotide and incoming nucleotide differently. In Pol γ (and T7 RNAP), they are accomplished by two sets of check-point residues in the minor groove: E895 and N1098 (E480 and N611) before translocation and R853 and Q1102 (R429 and Q615) after translocation, respectively. In RB69, both bps can be ensured by three tyrosines (Y416, Y564 and Y391) that are conserved in Pol α and Pol δ. The latter arrangement may be the more efficient and rigorous. Deoxyribose selection is conferred in part by Y951 on the O-helix of the fingers domain. The Y951F substitution in Pol γA reduces ddCTP incorporation by 5000 fold [53].
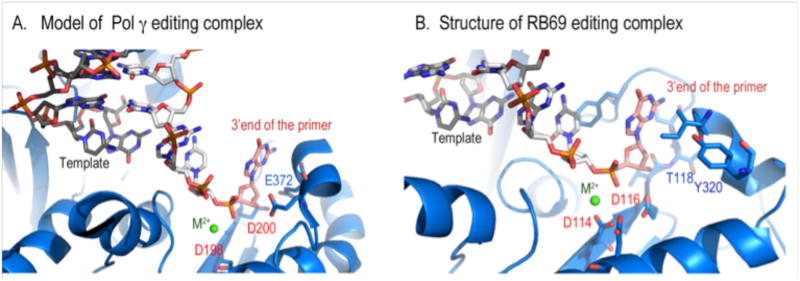
Figure 6.
Comparison of Pol A and Pol B editing complexes. A composed Pol g editing complex with a unwound primer-template from RB69 showing possible specific interaction between exo residue E374 with the 3′OH of the primer.
Several structural features may contribute to Pol γ's relatively low discrimination against NRTIs. In this context, discussions of polymerase fidelity should focus on Watson-Crick (W-C) base-pairing as well as the correct structure of the ribose, because Pol γ exhibits varied sensitivity to NRTIs containing different deoxyribose derivatives. Most replicating polymerases use conformational changes in the fingers domain after binding to dNTP to further increase the affinity of the correct nucleotide beyond W-C base-pairing interaction. The B-family RB69 DNAP apo enzyme undergoes a ∼60° rotation after binding to DNA and a dNTP, a value much larger than Pol γ can exhibit because its apo structure is more closed than other DNA polymerases. The extreme case is seen in the Y-family lesion-bypassing polymerases that conduct error-prone DNA synthesis, where the enzymes barely show any conformational changes in forming the equivalent ternary complex [48-50]. Pol A and Pol B members also use different amino acids to monitor W-C complementarity of an incoming nucleotide (Fig. 5), which allows for differences in the how the enzymes discriminate between the correct and incorrect substrate. A misincorporated nucleotide can be excised either before or after enzyme translocation. Again due to the structural difference in their fingers domain, Pol A and Pol B members accomplish mismatch discrimination using different residues.
To begin to understand Pol γ exo activity, we composed a Pol γ editing complex by superimposing the exo domain of RB69 DNAP editing complex to Pol γ and then docked the strand-separated primer-template onto Pol γ. Although obviously not rigorous, this exercise is not unreasonable because the exo domain does not undergo conformational changes upon DNA-binding for Pol A or Pol B polymerases [46,47,51], and the conformation of exo in apo Pol γ is likely very similar to that in its editing mode. The composite structure suggests that the Pol γ exo domain mainly interacts with the primer stand; significantly it predicts a specific interaction between an aspartate, D372, with the 3′-OH of the primer (Fig. 6), and thus a ddNMP-terminated primer may only bind weakly to the exo site. Weak binding leads to reduced hydrolysis [40,52].
The basis for the toxicity of NRTIs towards Pol γ holenzyme can be illustrated by comparing the structures of an NRTI bound to HIV RT with a modeled Pol γ –DNA complex. Similarities do exist between the two enzymes, including the arrangement of the palm catalytic subdomain, and these may indeed be the basis for toxicity. Significant differences are also found surrounding the incoming nucleotide-binding site (Fig. 7): Both human Pol γ and HIV RT utilize positively charged residues on the fingers domain to bind the negatively charged triphosphate of an incoming dNTP. However, the interaction of the two enzymes with the nucleoside moiety is different. In Pol γA, the fingers subdomain is α-helical whereas in HIV RT the fingers domain is composed of β sheets. Obviously, more explicitly useful comparisons have to wait for structures of Pol γ-DNA complexed to NRTIs. However, the predicted variations strongly suggest that significant differences exist between the substrate-binding sites of the two enzymes that could be exploited when designing higher potency antiviral drugs that exhibit lower toxicity. However, a series of structures of Pol γ, captured at all stages of inhibitor incorporation as well as in the editing mode, would be invaluable in understanding antiviral drug toxicity.
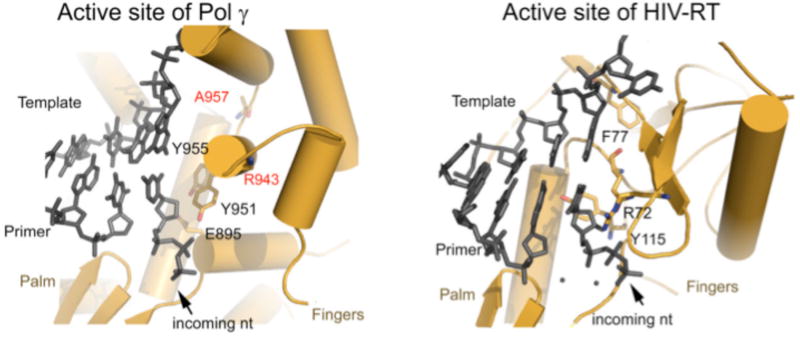
Figure 7.
Active sites of human Pol γ (left) and HIV-RT (right). The nucleotide binding site of the two enzymes are different: In Pol γ: it is likely bounded by E895, Y951, Y955, and Q1102, whereas in HIV RT the binding site is bounded by R72, F77, Y115, and Q151. The difference provides exploitable space for more potent and specific antiviral drug design.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp. 2001;235:247–263. doi: 10.1002/0470868694.ch20. discussion 263-246. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeviani M. Mitochondrial disorders. Suppl Clin Neurophysiol. 2004;57:304–312. [PubMed] [Google Scholar]
- 5.Lewis W, Dalakas MC. Mitochondrial toxicity of antiviral drugs. Nat Med. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- 6.Peters BS, Winer J, Landon DN, Stotter A, Pinching AJ. Mitochondrial myopathy associated with chronic zidovudine therapy in AIDS. Q J Med. 1993;86:5–15. [PubMed] [Google Scholar]
- 7.Arnaudo E, Dalakas M, Shanske S, Moraes CT, DiMauro S, Schon EA. Depletion of muscle mitochondrial DNA in AIDS patients with zidovudine-induced myopathy. Lancet. 1991;337:508–510. doi: 10.1016/0140-6736(91)91294-5. [DOI] [PubMed] [Google Scholar]
- 8.Dalakas MC, Illa I, Pezeshkpour GH, Laukaitis JP, Cohen B, Griffin JL. Mitochondrial myopathy caused by long-term zidovudine therapy. N Engl J Med. 1990;322:1098–1105. doi: 10.1056/NEJM199004193221602. [DOI] [PubMed] [Google Scholar]
- 9.Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- *10.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]; First characterization of 5′dRP DNA repair activity of human Pol γ.
- 11.Longley MJ, Ropp PA, Lim SE, Copeland WC. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 12.Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 13.Pinz KG, Bogenhagen DF. The influence of the DNA polymerase gamma accessory subunit on base excision repair by the catalytic subunit. DNA Repair (Amst) 2006;5:121–128. doi: 10.1016/j.dnarep.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J Biol Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- 15.Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- **16.Lee YS, Kennedy WD, Yin YW. Structural insights into human mitochondrial DNA replication and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first atomic structure of a eukaryotic DNA replicase holoenzyme
- 17.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. Embo J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell. 2001;7:43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 19.Lee YS, Lee S, Demeler B, Molineux IJ, Johnson KA, Yin YW. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J Biol Chem. 2010;285:1490–1499. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrodeguas JA, Pinz KG, Bogenhagen DF. DNA binding properties of human pol gammaB. J Biol Chem. 2002;277:50008–50014. doi: 10.1074/jbc.M207030200. [DOI] [PubMed] [Google Scholar]
- 21.Farge G, Pham XH, Holmlund T, Khorostov I, Falkenberg M. The accessory subunit B of DNA polymerase gamma is required for mitochondrial replisome function. Nucleic Acids Res. 2007;35:902–911. doi: 10.1093/nar/gkl1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrodeguas JA, Bogenhagen DF. Protein sequences conserved in prokaryotic aminoacyl-tRNA synthetases are important for the activity of the processivity factor of human mitochondrial DNA polymerase. Nucleic Acids Res. 2000;28:1237–1244. doi: 10.1093/nar/28.5.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdan SM, Marintcheva B, Cook T, Lee SJ, Tabor S, Richardson CC. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci U S A. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Lee YS, Johnson KA, Molineux IJ, Yin YW. A single mutation in human mitochondrial DNA polymerase Pol gammaA affects both polymerization and proofreading activities of only the holoenzyme. J Biol Chem. 2010;285:28105–28116. doi: 10.1074/jbc.M110.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of mutant Pol γ holoenzyme demonstrating how the accessory subunit functions to modulate both polymerase and proof-reading activities.
- 25.Ferrari G, Lamantea E, Donati A, Filosto M, Briem E, Carrara F, Parini R, Simonati A, Santer R, Zeviani M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain. 2005;128:723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- 26.Harrower T, Stewart JD, Hudson G, Houlden H, Warner G, O'Donovan DG, Findlay LJ, Taylor RW, De Silva R, Chinnery PF. POLG1 mutations manifesting as autosomal recessive axonal Charcot-Marie-Tooth disease. Arch Neurol. 2008;65:133–136. doi: 10.1001/archneurol.2007.4. [DOI] [PubMed] [Google Scholar]
- 27.Taanman JW, Daras M, Albrecht J, Davie CA, Mallam EA, Muddle JR, Weatherall M, Warner TT, Schapira AH, Ginsberg L. Characterization of a novel TYMP splice site mutation associated with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) Neuromuscul Disord. 2009;19:151–154. doi: 10.1016/j.nmd.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Kollberg G, Moslemi AR, Darin N, Nennesmo I, Bjarnadottir I, Uvebrant P, Holme E, Melberg A, Tulinius M, Oldfors A. POLG1 mutations associated with progressive encephalopathy in childhood. J Neuropathol Exp Neurol. 2006;65:758–768. doi: 10.1097/01.jnen.0000229987.17548.6e. [DOI] [PubMed] [Google Scholar]
- 29.Bogenhagen DF, Pinz KG, Perez-Jannotti RM. Enzymology of mitochondrial base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:257–271. doi: 10.1016/s0079-6603(01)68105-4. [DOI] [PubMed] [Google Scholar]
- 30.Copeland WC, Longley MJ. DNA polymerase gamma in mitochondrial DNA replication and repair. ScientificWorldJournal. 2003;3:34–44. doi: 10.1100/tsw.2003.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]; An excellent review of DNA repair processes in mitochondria
- 32.Nachman MW, Brown WM, Stoneking M, Aquadro CF. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 1996;142:953–963. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 34.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of Oxidative DNA Damage via FEN1-Dependent Long-Patch Base Excision Repair in Human Cell Mitochondria. Mol Cell Biol. 2008 doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinz KG, Bogenhagen DF. Characterization of a catalytically slow AP lyase activity in DNA polymerase gamma and other family A DNA polymerases. J Biol Chem. 2000;275:12509–12514. doi: 10.1074/jbc.275.17.12509. [DOI] [PubMed] [Google Scholar]
- 36.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37.Martin JL, Brown CE, Matthews-Davis N, Reardon JE. Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob Agents Chemother. 1994;38:2743–2749. doi: 10.1128/aac.38.12.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]; First illustration of the differential inhibition of human DNA polymerases by NRTIs
- 38.Yamanaka H, Gatanaga H, Kosalaraksa P, Matsuoka-Aizawa S, Takahashi T, Kimura S, Oka S. Novel Mutation of Human DNA Polymerase gamma Associated with Mitochondrial Toxicity Induced by Anti-HIV Treatment. J Infect Dis. 2007;195:1419–1425. doi: 10.1086/513872. [DOI] [PubMed] [Google Scholar]
- 39.Dolce V, Fiermonte G, Runswick MJ, Palmieri F, Walker JE. The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc Natl Acad Sci U S A. 2001;98:2284–2288. doi: 10.1073/pnas.031430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Feng JY, Johnson AA, Johnson KA, Anderson KS. Insights into the molecular mechanism of mitochondrial toxicity by AIDS drugs. J Biol Chem. 2001;276:23832–23837. doi: 10.1074/jbc.M101156200. [DOI] [PubMed] [Google Scholar]; Extension of the inhibitory studies of NRTIs to include both incorporation and excision by DNA Pol γ
- 41.Feng JY, Anderson KS. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry. 1999;38:9440–9448. doi: 10.1021/bi990709m. [DOI] [PubMed] [Google Scholar]
- 42.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 43.Kohlstaedt LA, Steitz TA. Reverse transcriptase of human immunodeficiency virus can use either human tRNA(3Lys) or Escherichia coli tRNA(2Gln) as a primer in an in vitro primer-utilization assay. Proc Natl Acad Sci U S A. 1992;89:9652–9656. doi: 10.1073/pnas.89.20.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 45.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 47.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 48.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 49.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 50.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat Struct Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 51.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. Embo J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng JY, Murakami E, Zorca SM, Johnson AA, Johnson KA, Schinazi RF, Furman PA, Anderson KS. Relationship between antiviral activity and host toxicity: comparison of the incorporation efficiencies of 2′,3′-dideoxy-5-fluoro-3′-thiacytidine-triphosphate analogs by human immunodeficiency virus type 1 reverse transcriptase and human mitochondrial DNA polymerase. Antimicrob Agents Chemother. 2004;48:1300–1306. doi: 10.1128/AAC.48.4.1300-1306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim SE, Ponamarev MV, Longley MJ, Copeland WC. Structural determinants in human DNA polymerase gamma account for mitochondrial toxicity from nucleoside analogs. J Mol Biol. 2003;329:45–57. doi: 10.1016/s0022-2836(03)00405-4. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]