Abstract
The bacterial envelope integrates essential stress-sensing and adaptive functions; thus, envelope-preserving functions are important for survival. In Gram-negative bacteria, envelope integrity during stress is maintained by the multi-gene Psp response. Mycobacterium tuberculosis was thought to lack the Psp system, since it encodes only pspA and no other psp ortholog. Intriguingly, pspA maps downstream from clgR, which encodes a transcription factor regulated by the MprAB-σE envelope-stress-signaling system. clgR inactivation lowered ATP concentration during stress and protonophore treatment-induced clgR-pspA expression, suggesting that these genes express Psp-like functions. We identified a four-gene set -- clgR, pspA (rv2744c), rv2743c, rv2742c – that is regulated by clgR and in turn regulates ClgR activity. Regulatory and protein-protein interactions within the set and a requirement of the four genes for functions associated with envelope integrity and surface-stress tolerance indicate that a Psp-like system has evolved in mycobacteria. Among Actinobacteria, the four-gene module occurred only in tuberculous mycobacteria and was required for intra-macrophage growth, suggesting links between its function and mycobacterial virulence. Additionally, the four-gene module was required for MprAB-σE stress-signaling activity. The positive feedback between envelope-stress-sensing and envelope-preserving functions allows sustained responses to multiple, envelope-perturbing signals during chronic infection, making the system uniquely suited to tuberculosis pathogenesis.
Keywords: clgR, pspA, rv2743c, rv2742c, actinobacteria
The intracellular pathogen Mycobacterium tuberculosis responds to host-generated stress by extensive metabolic remodeling that leads to growth arrest and reduced susceptibility to host defenses and most antimicrobial agents. The inability of the host immune response to eliminate the pathogen results in latent infection and billions of asymptomatically infected individuals (WHO, 2013). This huge reservoir of infected humans poses a serious public health threat, since weakening of host defenses leads to development of pulmonary disease and transmission of infection. Understanding mechanisms used by M. tuberculosis to sense and adapt to host-mediated stress is crucial for developing highly effective anti-tuberculosis strategies.
Stress-signal sensing typically occurs at the surface of pathogenic bacteria (Rowley et al., 2006). With M. tuberculosis, the cell surface serves both sensing and adaptive functions. For example, stress-sensing functions include two-component systems, anti-sigma factors, and membrane-bound serine/threonine protein kinases (Bretl et al., 2011, Rodrigue et al., 2006, Alber, 2009). Adaptive functions include structural remodeling of the bacterial envelope that alters signaling to macrophages and presumably leads to intracellular conditions favoring mycobacterial dormancy (Davidson et al., 1982, Cunningham & Spreadbury, 1998, Shui et al., 2007, Jain et al., 2007, Lavollay et al., 2008). Maintenance of envelope integrity may be essential for proper expression and integration of bacillary stress responses.
A cornerstone of the surface stress response of M. tuberculosis is the genetic network centered on the MprAB two-component system and the accessory sigma factor E (σE), two transcriptional regulators that activate each other (Manganelli & Provvedi, 2010, Tiwari et al., 2010). Sensing of stress occurs through MprAB, which may be activated by protein misfolding in the extracytoplasmic space (Bretl et al., 2014), and through σE, which is activated by stress-induced degradation of the phosphorylated form of its cognate anti-sigma factor (Barik et al., 2010). How sensing of envelope stress by the mprAB-sigE network translates into preservation of cell envelope integrity is unknown.
The mprAB-sigE regulon includes the transcription regulator clgR (rv2745c) and the adjacent rv2744c (Manganelli et al., 2001, Estorninho et al., 2010), a pspA homolog (Provvedi et al., 2009). The function of M. tuberculosis pspA is unknown, but in Gram-negative organisms, pspA is a member of the envelope-stress-responsive, multi-gene Phage shock protein (Psp) response system that mitigates envelope damage by preventing proton leakage across a damaged membrane, thereby maintaining proton motive force (Darwin, 2005, Darwin, 2013, Joly et al., 2010). In that system, PspA has a dual role: it is a stoichiometric inhibitor of the cognate PspF regulator, and it is an envelope-stabilizing effector as a component of an envelope-bound, multiprotein complex comprising PspA and the integral membrane proteins PspB and PspC (Darwin, 2005, Darwin, 2013, Joly et al., 2010). While the presence of a pspA homolog does not imply the presence of a complete Psp system, particularly in Gram-positive organisms (Joly et al., 2010), we were intrigued by the possibility that M. tuberculosis possesses a functional Psp system connected to the mprAB-sigE-clgR stress-signaling network. Such a connection would complete the logical need for stress signaling to help bacteria recover from envelope damage.
In the present work, we used gene expression profiling, mutant analyses, and protein-protein interaction studies to characterize operon structure and regulatory interactions among clgR, pspA and two downstream genes (rv2743c and rv2742c), and to assess the requirement for these genes in physiological processes typically associated with envelope integrity. We report that the four-gene module of M. tuberculosis is found only in tuberculous mycobacteria among Actinobacteria, and that it exhibits regulatory and functional characteristics similar to those seen with the Psp system of Gram-negative bacteria. Since three of four proteins in the M. tuberculosis module share no amino acid sequence homology with their Gram-negative counterparts, our work reveals a novel example of parallel or convergent evolution in bacteria of different phyla. Moreover, the four-gene module and the mprAB-sigE network are connected by a positive feedback circuit. Reciprocal positive regulation of surface-stress signaling and envelope maintenance functions could enable tubercle bacilli to respond to multiple surface-perturbing challenges from the macrophage during the course of infection, and therefore have an important role in mycobacterial virulence.
RESULTS
Relationship of clgR-pspA with energy generation and proton motive force
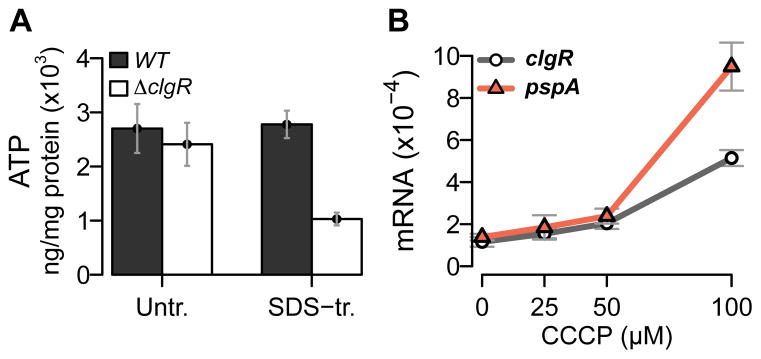
During envelope stress, the Psp regulon of Gram-negative bacteria affects cellular activities that depend on proton motive force and ATP level (Darwin, 2005, Darwin, 2013, Joly et al., 2010). To determine whether a putative clgR-pspA system has a similar role in M. tuberculosis, we measured levels of intracellular ATP in M. tuberculosis cultures stressed with bacteriostatic concentrations (0.03%) of sodium dodecyl sulfate (SDS) (see Methods). Two strains were used: a wild-type strain and a clgR deletion mutant, obtained by replacement of the clgR coding sequence with an antibiotic resistance cassette (Estorninho et al., 2010). While no difference was observed between the two strains in the absence of stress, the ATP levels in SDS-treated clgR mutant cultures were one-third those obtained with similarly stressed wild-type cells (Fig. 1A).
Figure 1.
(A) Effect of clgR deletion on intracellular ATP levels. Mid-log cultures of wild-type and clgR mutant strains were treated with 0.03% SDS for 24 hrs. Extracts from pre- and post-treatment cultures were then assayed for ATP levels and expressed as ng of ATP per mg of total protein. (B) Effect of treatment with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) on expression of clgR and pspA. Mid-log cultures of the wild-type strain were treated for 6 hrs with various concentrations of CCCP, as indicated, and harvested pre- and post-treatment. Total RNA was extracted, and amplicons were measured by quantitative RT-PCR using primers and gene-specific molecular beacons. Amplicon copy numbers were normalized to 16S rRNA. 0 μM = solvent-only control (corresponding to the highest solvent concentration used with the CCCP treatment). In this and subsequent figures, quantitative RT-PCR data are presented as mean values (+/− standard error of the mean) from triplicate experiments.
Energy generation correlates with maintaining proton motive force (PMF), and dissipation of PMF caused by treatment with proton ionophores, such as carbonyl cyanide m-chlorophenyl hydrazone (CCCP), induces expression of pspA and its product in E. coli and Salmonella typhimurium (Weiner & Model, 1994, Becker et al., 2005). When we examined the effect of CCCP treatment on the expression of clgR-pspA of M. tuberculosis, we observed CCCP-concentration-dependent induction of both genes (Fig. 1B). The lowered ATP concentration seen with the clgR mutant and the CCCP-mediated upregulation of clgR-pspA suggest that M. tuberculosis possesses an active Psp response.
Regulation of the clgR-pspA-rv2743c-rv2742c region by ClgR
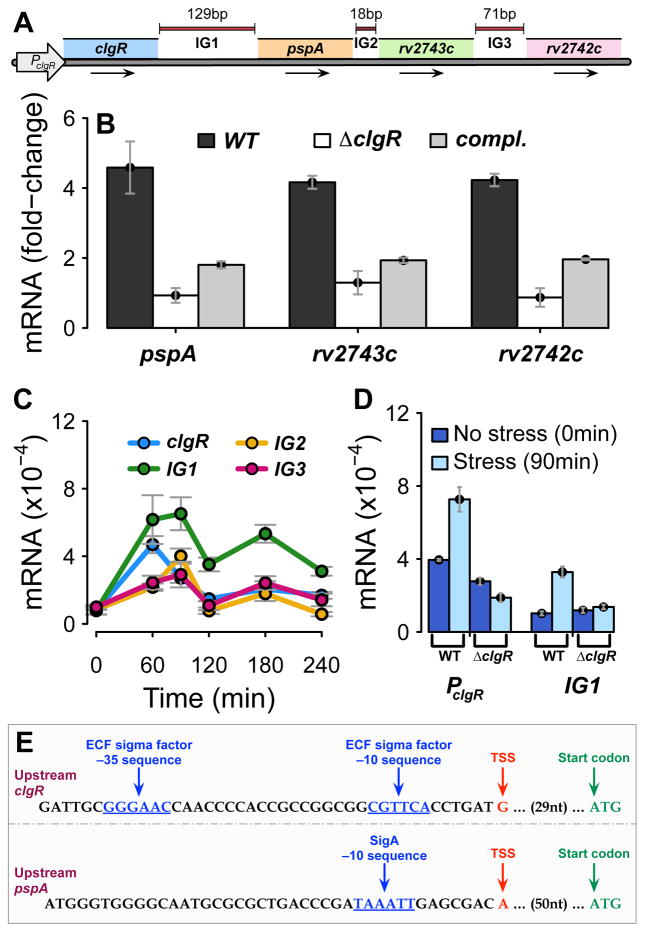
We next examined the regulatory structure of the genomic region encoding clgR-pspA. Downstream of pspA are two additional genes (rv2743c and rv2742c) that are transcribed in the same direction and that are separated from pspA and from each other by short (~20 to ~70 bp) intergenic regions (Fig. 2A). Since clgR, pspA, and rv2743c were previously reported to respond to surface stress in a sigE-dependent manner in microarray experiments (Manganelli et al., 2001), and the DNA upstream from clgR contains ClgR-binding sites (Estorninho et al., 2010), we first asked whether the stress response of this gene set was clgR-dependent. By using quantitative RT-PCR for transcript enumeration, we found that the surface-stress (0.03% SDS) response of pspA, rv2743c, and rv2742c was abrogated in the clgR deletion strain (Fig. 2B). Mutant complementation with clgR restored 35% to 40% of the downstream gene expression (Fig. 2B), indicating that expression of each of the three downstream genes required a functional clgR. Additionally, since all four genes exhibited similar stress response profiles (Fig. S1), and pair-wise co-transcription of clgR-pspA and pspA-rv2743c was previously reported with unstressed M. tuberculosis cultures (Roback et al., 2007), we tested the possibility of co-transcription within the four-gene set under surface stress. To do so, we used RT-PCR to determine whether RNA was generated from the intergenic regions (Fig. 2A) under the surface stress conditions applied in Fig. 2B. We found that not only were amplicons generated from the intergenic regions but also that these amplicons exhibited stress-response expression profiles similar to that of clgR (Fig. 2C) and of the downstream genes (Fig. S1). This result strongly suggests the presence of one or more multicistronic mRNA(s) encoded by the four-gene region. To further explore the regulatory structure of the region, we used promoter-probe technology and constructed lacZ reporter fusions with DNA sequences upstream from clgR and within the clgR-pspA intergenic region (IG1 in Fig. 2A). When reporter gene expression was measured in response to surface stress, lacZ expression was induced with both constructs (2- to 3-fold over the corresponding basal level) in wild-type but not in clgR deletion mutant cultures (Fig. 2D). The presence of two promoters explains why complementing the clgR deletion mutant restores only ~40% of wild-type levels of the downstream genes (Fig. 2B): in the complemented mutant, ClgR produced from an ectopic locus can induce the stress response of pspA-rv2743c-rv2742c only from the downstream promoter but not from the upstream promoter. To map the two promoters, we analyzed data obtained by genome-wide transcription start site mapping using RNA adapter ligation and next generation RNA sequencing (Shell et al., 2015) (see Methods). A transcription start site was identified 30 nucleotides upstream from the start of the clgR coding sequence (Fig. 2E). The corresponding −10 and −35 sequences are characteristic of a SigE promoter, which had been previously identified (Manganelli et al., 2001). An additional 5′ end associated with a strong SigA −10 motif was identified 51 nucleotides upstream of the pspA coding sequence (Fig. 2E) (no −35 site was identified, consistent with the reported absence of a consensus −35 site for M. tuberculosis SigA promoters (Cortes et al., 2013, Feklistov & Darst, 2011). The two transcriptional start sites identified here were also reported in a previous genome-wide mapping study employing a different methodology (Cortes et al., 2013). Overall, the results reported in this section strongly suggest that one or more multicistronic transcripts are expressed from two stress-responsive, clgR-dependent promoter sequences, one located upstream of clgR and another found in the clgR-pspA intergenic region. In agreement with the clgR dependence of both promoters assessed genetically, ClgR-binding sites were found by bioinformatics upstream of clgR (as also reported in (Estorninho et al., 2010)) and of pspA (Fig. S2).
Figure 2. Structure and regulation of genes in the clgR-pspA-rv2743c-rv2742c region.
(A) Schematic representation of the region. Arrows represent direction of transcription. The arrow PclgR indicates sequences upstream of clgR expressing promoter activity. IG = intergenic regions of indicated length, in bp. (B) Effect of clgR deletion on gene stress response. Mid-log cultures of wild-type, clgR deletion mutant, and complemented strains were treated with 0.03% SDS and harvested pre-treatment (time 0) and post-treatment (90 min). Amplicons were generated from total RNA by quantitative RT-PCR using primers and gene-specific molecular beacons from intragenic sequences for each gene, as indicated. Amplicon copy numbers, normalized to 16S rRNA, are presented as fold-change relative to time 0. Mean values (+/− standard error of the mean) were obtained from triplicate experiments. (C) Enumeration of intergenic-sequence-containing amplicons. Mid-log cultures of the wild-type strain were treated with SDS, and samples were harvested pre-treatment and at multiple times post treatment. Amplicons were generated from sequences internal to clgR (reference gene) and from intergenic sequences along the region, as indicated (see panel A). Data obtained from triplicate experiments were normalized to 16S rRNA. (Note that copy numbers of RT-PCR amplicons obtained with different primer sets and molecular beacons cannot be compared with each other due to variations in primer-associated efficiency of PCR amplification and molecular-beacon-dependent detection sensitivity). (D) Analysis of promoter probe constructs. Two promoter::lacZ fusions were generated with nucleotide sequences comprising either 382 bp upstream of clgR (PclgR, see panel A) or 250 bp upstream of pspA (IG1, see panel A) in wild-type and clgR mutant strains. lacZ transcripts were enumerated by quantitative RT-PCR prior to and 90 min following SDS treatment of mid-log cultures. Data obtained from triplicate experiments were normalized to 16S rRNA. (E) Promoter mapping upstream of clgR and pspA. The panel shows nucleotide sequences upstream of clgR and upstream of pspA (intergenic region IG1). Indicated in brown are the two transcription start sites (TSS) mapped in this work using the genome-wide mapping method described in Methods. The TSS in the top row had a ratio of converted to unconverted 5′ ends >1.74 (adjusted p ≤0.01 for probability of being a processed 5′ end). This TSS is preceded by −10 and −35 sequences (shown in blue) characteristic of the extracytoplasmic function (ECF) sigma factor SigE (Manganelli et al., 2001). The 5′ end in the bottom row had a converted/non-converted ratio of 1.58, and was confirmed to be a TSS based on the presence of a very strong SigA −10 motif (in blue) (Cortes et al., 2013, Feklistov & Darst, 2011). In both rows, the annotated first codon (ATG) is as reported in the M. tuberculosis genome sequence (http://tuberculist.epfl.ch/). nt = nucleotides.
Regulation of ClgR by downstream genes
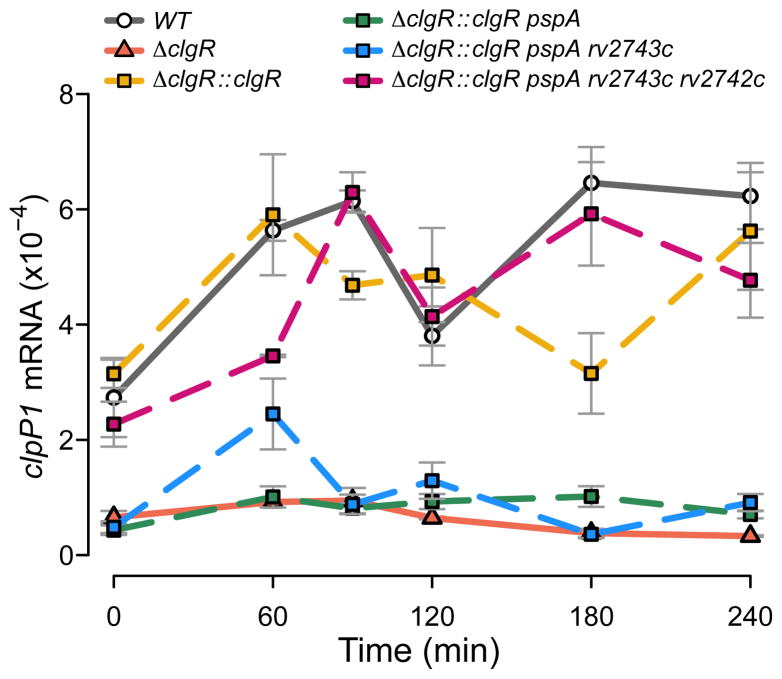
Since ClgR is the regulator of the clgR-pspA-rv2743c-rv2742c region, we characterized its own regulation. Induction of clgR by surface stress requires functional sigE ((Fig. S3) and (Manganelli et al., 2001)). To take into account potential post-transcriptional regulatory events, we assessed ClgR activity by using as read-out the expression of the target gene clpP1 (rv2461c), which is directly induced by ClgR (Sherrid et al., 2010). Stress (SDS)-mediated induction of clpP1 exhibited a biphasic temporal expression profile, with one peak at ~60–90 min post-stress and a second at ~180–240 min. The two peaks were separated by a trough at ~120 min post-stress (Fig. 3). Induction of clpP1 was absent in a clgR deletion mutant (Fig. 3). When we complemented the clgR mutant with clgR alone (by using an ectopic, integrated copy of the complementing DNA expressed from the native promoter), clpP1 induction was restored to wild-type levels at most time points (Fig. 2D). However, we were surprised to find that mutant complementation with clgR and pspA together abrogated clpP1 expression (Fig. 3): addition of pspA to the complementing DNA inhibited ClgR activity. The expression profile of clpP1 was fully restored only when both downstream genes, rv2743c and rv2742c, were added to the clgR-pspA complementing DNA (Fig. 3). Similar results were obtained when we measured expression of another ClgR target gene, hsp (rv0251c) (Estorninho et al., 2010) (Fig. S4A). Together, these genetic data show that pspA has an inhibitory effect on ClgR activity and that rv2743c and rv2742c reverse that effect. Functional data were consistent with the genetic results, as complementation of the clgR defect by the clgR-pspA-rv2743c-rv2742c DNA also restored wild-type levels of ATP in stressed cultures (Fig. S4B).
Figure 3. Effect on clpP1 stress response of clgR deletion and sequential complementation with clgR and downstream operon genes.
Expression of the sentinel target gene clpP1 was used as readout of ClgR activity (Sherrid et al., 2010). Complementation of the clgR deletion was performed sequentially by introducing clgR alone, clgR plus pspA, plus pspA-rv2743c, and plus pspA-rv2743c-rv2742c. Transcript copy numbers were generated and expressed as described in the legend to Fig. 2C. The reason why clgR alone complements the clpP1 expression defect of the clgR deletion mutant to similar levels as the clgR-pspA-rv2743c-rv2742c complementation can be explained by ectopically produced ClgR inducing pspA-rv2743c-rv2742c expression from the promoter located in the clgR-pspA intergenic region (Fig. 2D). Measurements of clpP1 transcripts performed using M. tuberculosis H37Rv and M. tuberculosis CDC1551 gave indistinguishable results (Fig. S8D), indicating that the clpP1 surface-stress response is conserved in different M. tuberculosis strains.
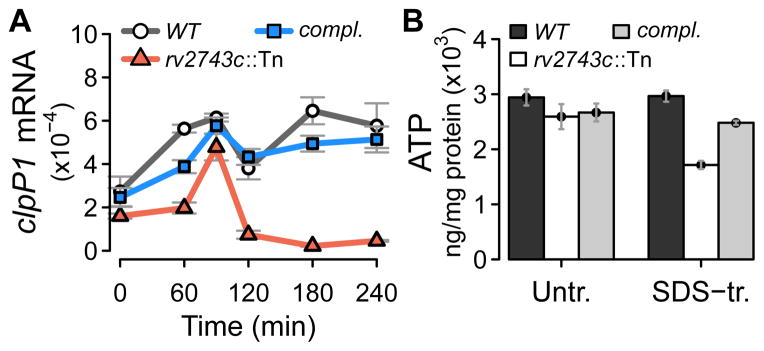
The effect of rv2743c-rv2742c on ClgR activity was further examined with a mutant of rv2743c carrying a transposon insertion that has a polar effect on the downstream gene rv2742c (data not shown). In this mutant, the second peak of the clpP1 profile disappeared (Fig. 4A) (the biphasic profile of the clpP1 response and the temporal effects of the rv2743c-rv2742c mutation are interpreted in the Discussion). In addition, the rv2743c-rv2742c mutation reduced intracellular ATP levels under surface-stress conditions to <60% of wild-type levels (Fig. 4B), thereby recapitulating functional effects observed with the clgR mutant (Fig. 1).
Figure 4. Effects of rv2743c inactivation on target gene expression and ATP levels.
(A) Effect on clpP1 stress response. ClgR activity was assessed as expression of the sentinel target gene clpP1 in an rv2743c transposon (Tn) insertion mutant. Complementation was performed with rv2743c-rv2742c to overcome the polar effect of the mutation on the downstream rv2742c (data not shown). Wild-type, mutant, and rv2743c-rv2742c complemented (compl.) strains were subjected to surface stress with SDS, and clpP1 transcripts were enumerated and presented as described in the legend to Fig. 2C. The reason why the rv2743c mutation did not phenocopy the clgR-pspA complementation of the clgR deletion mutant (Fig. 3) can be gene dosage effect due to the extra copy of pspA in the complementing DNA. (B) Effect on intracellular ATP levels. ATP levels were assayed in extracts from mid-log cultures of wild-type, mutant, and complemented strains pre- and post-treatment with 0.03% SDS for 24 hrs, and expressed as in the legend to Fig. 1A.
Effect of the pepD protease gene on ClgR activity
In light of the inhibitory effect of pspA on ClgR activity, described above with the clgR complementation studies (Fig. 3 and Fig. S4A), we reasoned that ClgR target gene expression profiles might be affected by mutations in accessory genes that alter PspA levels. One such gene is pepD (rv0983), which encodes a serine protease that targets PspA for proteolytic degradation (White et al., 2011). Indeed, in a pepD-deficient mutant the SDS-mediated stress response of the ClgR target gene clpP1 showed a striking decrease in the second peak of the clpP1 profile (Fig. S5). Thus, pepD has a temporal effect on ClgR activity similar to that described above for the rv2743c insertion mutant (Fig. 4A).
Protein-protein interactions
We next determined whether protein complexes form between ClgR, PspA and the Rv2743c and Rv2742c proteins. We constructed E. coli strains expressing IPTG-inducible, differentially tagged copies of each protein in all pairwise combinations, and then we assessed protein complex formation by immunoprecipitation, pull-down assays, and western blot analyses. We detected complex formation between ClgR and PspA (Fig. 5A, lane 3) but not between ClgR and either Rv2743c (Fig. 5B, lane 3) or Rv2742c (Fig. 5C, lane 3). Moreover, protein complexes formed between PspA and Rv2743c (Fig. 5D, lane 3) and between Rv2743c and Rv2742c (Fig. 5E, lane 3), but not between PspA and Rv2742c (Fig. 5F, lane 3). In summary, protein complexes formed only between ClgR and PspA, PspA and Rv2743c, and Rv2743c and Rv2742c.
Figure 5. Protein-protein interactions.
Pairwise protein-protein interactions were assessed between differentially tagged recombinant proteins expressed from IPTG-inducible promoters in E. coli. Whole cell lysates from IPTG-induced recombinant cultures expressing tagged proteins, alone or in pairs as indicated, were used for co-immunoprecipitation (IP; left column) or for pull-down of 6xHis tagged protein (PD; right column), followed by immunoblotting (IB) with the appropriate monoclonal antibody. Left column: IP was performed with anti-His (A and C) or with anti-S-tag antibody (B), followed by IB with anti-Myc antibody to detect ClgR. Lanes: 1) whole-cell lysate + IB (positive control); 2) IP (with cell extracts carrying ClgR-Myc alone) + IB; 3) IP (with cell extracts carrying ClgR-Myc plus a His-tagged or S-tagged protein) + IB. Right column: PD was performed with Ni-NTA agarose, followed by IB with anti S-tag antibody for detection of Rv2743c (D and E) or Rv2742c (F). Lanes: 1) whole-cell lysate + IB (positive control); 2) PD (with cell extracts carrying S-tag protein alone) + IB; 3) PD (with cell extracts carrying S-tag protein plus His-tag protein) + IB. The bottom row of each panel shows unidentified E. coli bands cross-reacting with anti-Myc monoclonal antibody (panels in the left column) or with the anti-S-tag monoclonal antibody (panels in the right column) that served as loading controls. Experiments in panels B, C, and F were repeated by switching tags in each corresponding protein pair to control for potential tag effects on the protein-protein interaction, with no change of result. S = S tag; M = Myc tag; H = His tag.
Preservation of envelope integrity
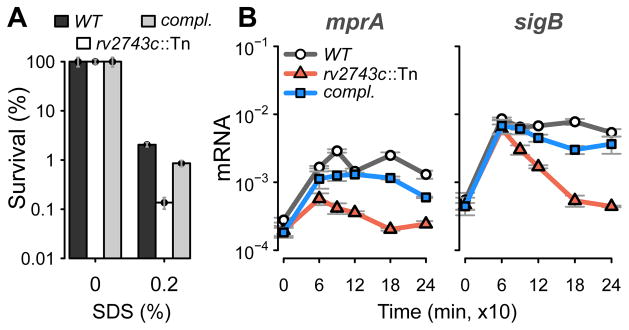
Since clgR and rv2743c mutants show reduction of cellular processes that are typically associated with envelope integrity, such as energy generation under stress (Figs. 4B and S4B), we asked whether the clgR-pspA-rv2743c-rv2742c region has an envelope-preserving activity. Since envelope integrity is expected to be linked to surface-stress tolerance, we measured survival of M. tuberculosis wild-type and rv2743c mutant cultures during treatment with bactericidal concentrations (0.2%) of SDS. Mutant survival was drastically lower (>90% reduction) than wild-type survival (Fig. 6A). A similar effect was observed with a clgR mutant (data not shown).
Figure 6. Effects of rv2743c inactivation.
(A) Tolerance to surface stress. Lethal activity of the detergent SDS was determined in wild-type, rv2743c mutant and rv2743c-rv2742c complemented (compl.) strains. Data are expressed as % survival relative to untreated cultures. Mean values (+/− standard error of the mean) from triplicate experiments are shown. (B) Surface-stress response of sentinel target genes of MprAB and σE. Transcripts for mprA and sigB were enumerated in wild-type, mutant, and complemented strains and presented as described in the legend to Fig. 2C. Tn = transposon.
Another measure of envelope integrity is proper functioning of the envelope-stress sensing system, which, as pointed above, centers on the MprAB-σE network. We examined the effects of genetic inactivation of rv2743c on the MprAB-σE network. As read-out for MprAB and σE activity, we measured expression of mprA (which contains regulatory MprA-binding sites (He et al., 2006)) and sigB (which is transcribed from a σE-dependent promoter (Song et al., 2008)). Inactivation of rv2743c reduced mprA and sigB expression to ~20% and ~10% of wild-type levels, respectively, after 90–120 min of treatment with SDS (Fig. 6B). This temporal effect was similar to that observed on ClgR activity with the same mutant (Fig. 4A). In addition, the rv2743c mutation reduced the mprA but not the sigB stress response profile in the first 90 min following SDS addition, suggesting that the effect is directly on MprA activity and indirectly (through the dampening of MprA) on σE activity. Together, the observed effects of the rv2743c mutation on surface stress tolerance and on the activity of the MprA-σE network establish a link between the clgR-pspA-rv2743c-rv2742c region and expression of envelope-stress sensing functions.
M. tuberculosis survival in macrophages
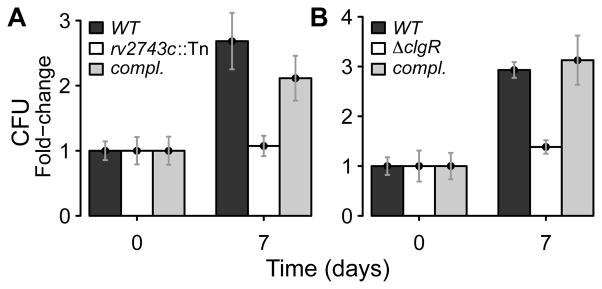
Given that the ClgR-PspA-Rv2743c-Rv2742c module maintains tolerance to surface stress in vitro, we tested whether the intact system is required for M. tuberculosis survival during macrophage infection. When rv2743c was inactivated, M. tuberculosis growth in primary human monocyte-derived macrophages was reduced 40% at seven days post-infection (Fig. 7A). Since mutant and wild-type cultures are indistinguishable when grown in rich medium, the rv2743c inactivation increases M. tuberculosis susceptibility to the intra-macrophage environment. Similar effects were observed with a clgR mutant (Fig. 7B). Thus, the clgR-pspA-rv2743c-rv2742c region facilitates intracellular growth of M. tuberculosis.
Figure 7. Effect of rv2743c and clgR inactivation for growth in macrophages.
Human monocyte-derived macrophages were infected with different M. tuberculosis strains. Bacterial counts (CFU) were normalized to 105 adherent cells per well. Data are expressed as fold-increase in CFU at day 7 relative to CFU enumerated at 4 hrs post-infection. Mean values (+/− standard error of the mean) from triplicate experiments are shown. (A) Wild-type, rv2743c mutant and rv2743c-rv2742c complemented (compl.) strains. Tn = transposon. (B) Wild-type, clgR mutant and complemented (compl.) strains. Complementation was with clgR-pspA-rv2743c-rv2742c DNA. Reduced growth of the clgR mutant in murine bone-marrow-derived macrophages was previously reported (Estorninho et al., 2010).
Comparison with the Psp proteins of Gram-negative bacteria
We next asked whether the proteins encoded by the clgR-pspA-rv2743c-rv2742c operon of M. tuberculosis share similarities with the proteins mediating the Psp response to envelope stress in Gram-negative Proteobacteria (reviewed in (Darwin, 2005, Darwin, 2013, Joly et al., 2010)). In that system, the “minimal” Psp module (Darwin, 2005) includes four adjacent genes, namely, the regulator pspF and the PspF-regulated pspABC. Thus, the gene organization in the Gram-negative module parallels that seen in M. tuberculosis. Except for PspA, which contains the PspA/IM30 domain characteristic of this protein family (Fig. 8), the other proteins encoded by the clgR-pspA-rv2743c-rv2742c region share no sequence homology with the Psp proteins of Gram-negative bacteria (Fig. 8). The transcriptional regulators ClgR and PspF are very different in length. Moreover, ClgR lacks the AAA+ ATPase domain characteristic of bacterial enhancer binding proteins such as PspF, and the respective helix-turn-helix DNA binding domains are located at opposite ends of the two proteins (Fig. 8). In addition, while PspB and PspC are both integral membrane proteins, only Rv2743c (but not Rv2742c) contains transmembrane domains (Fig. 8). Thus, the proteins encoded by clgR, rv2743c and rv2742c of M. tuberculosis are unrelated to the Psp system of Gram-negative organisms with respect to amino acid sequence.
Figure 8. M. tuberculosis (Mtb) and E. coli (Eco) Psp protein comparison.
(A) Mtb ClgR and Eco PspF. ClgR and PspF contain helix-turn-helix (HTH)-type DNA-binding domains (DBD). ClgR has an N-terminal HTH_XRE (SM00530, PF01381) DBD, while PspF has a C-terminal Fis-type HTH_8 DBD (PF02954, IPR002197). PspF contains AAA+ and sigma-54 interaction domains (IPR003593 and IPR002078, respectively). (B) Mtb PspA and Eco PspA. Both proteins were used to define PspA/IM30 (PF04012, IPR007157), a domain characterized by ~25% of conserved amino acid residues spanning the entire protein length. This domain is also predicted to contain coiled-coils. (C) Mtb Rv2743c and Eco PspB. Mtb Rv2743c and Eco PspB do not share significant sequence similarity; however, they are both predicted to be transmembrane (TM) proteins. Mtb Rv2743c is predicted to have two ~22-amino-acid TM helices connected by a short (4–6 amino acid) non-cytoplasmic loop. Mtb Rv2743c is also predicted to contain a C-terminal coiled-coil. Eco PspB is a single PspB domain (PF06667, IPR009554), which contains a predicted N-terminal TM helix (residues: 6–24) and C-terminal coiled-coil (residues: 36–64). (D) Mtb Rv2742c and Eco PspC. Mtb Rv2742c and Eco PspC share no significant sequence similarity. Sequence analysis of Rv2742c revealed only a single domain of unknown function (DUF3046, PF11248), which is conserved in actinobacteria. In contrast, Eco PspC has an N-terminal PspC domain (residues: 7–68, PF04024), a transmembrane helix (residues: 39–64), and a C-terminal coiled-coil (residues: 84–105).
Phylogenetic analysis
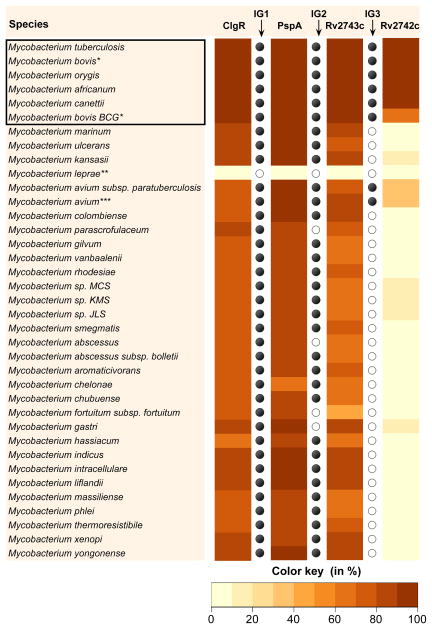
The distribution of the Psp system and its components in species encoding ClgR homologs was not investigated in previous phylogenetic analyses of the Psp system (for example (Joly et al., 2010)), presumably due to the absence of sequence homology between the regulators PspF and ClgR (Fig. 8). We performed protein sequence similarity searches in ~3,400 species of Actinobacteria and found that ~2,200 species carried a protein having at least 25% homology with M. tuberculosis ClgR (E-value < 1e-5). When we examined the ClgR-positive species for conservation of PspA-Rv2743c-Rv2742c, we found that the number of genes present correlated inversely with phylogenetic distance from Mycobacterium spp. (Fig. S6). Within Mycobacterium spp., only the M. tuberculosis complex contains the entire ClgR-PspA-Rv2743c-Rv2742c region: mycobacteria outside the M. tuberculosis complex showed <85% homology with Rv2743c and lacked Rv2742c (Fig. 9). Despite the lack of a fourth, contiguous gene in the module, the non-tuberculous species Mycobacterium smegmatis showed induction of clgR and the ClgR target clpP1 in response to surface stress (Fig. S7), and expression profiles resembled those seen in M. tuberculosis (Figs. 2C and 3). Thus, non-tuberculous mycobacteria encode three contiguous genes that appear to be sufficient for regulation of the module’s genes (we cannot exclude other, non-contiguous gene(s) participating to the Psp function). Overall, our analysis shows that the four-gene module is characteristic only of tuberculous mycobacteria.
Figure 9. Distribution of ClgR-PspA-Rv2743c-Rv2742c proteins in Mycobacterium spp.
Significant matches and similarity scores were obtained as described in Methods. The heat map represents the similarity score per target species or sub-species (or the highest score in the case of multiple matches), expressed as percent of protein sequence similarity for the longest contiguous match. Mycobacterial species were ordered on the basis of phylogenetic similarity ((Wattam et al., 2014) and http://www.patricbrc.org). The box contains the species in the M. tuberculosis complex. Intergenic distances (IG) between matches were calculated (in bp) in the target genomes. IG1, IG2 and IG3 denote distances between clgR-pspA, pspA-rv2743c and rv2743c-rv2742c, respectively. Filled black circles indicate intergenic distances that are similar to that of M. tuberculosis clgR-pspA-rv2743c-rv2742c operon, as shown in Fig. 2A. Open circles denote either absence of one gene in the pair, intergenic distances of >250 bp, or absence of genomic location information.
*Of 13 virulent M. bovis isolates analyzed, one (AF2122/97) contained a 23-bp insertion in pspC approximately two-thirds into the gene sequence, presumably giving rise to a truncated product. The same insertion was found in the genomes of 9 out of 14 substrains of the attenuated vaccine strain M. bovis Bacillus Calmette Guerin. The evolutionary significance of this polymorphism remains to be ascertained.
**Only Mycobacterium leprae was devoid of the entire ClgR-Psp module, as previously reported (Bretl et al., 2014), suggesting that loss of the Psp system is part of the genomic downsizing observed with this obligate intracellular pathogen (Cole et al., 2001).
***Only Mycobacterium avium among non-tuberculous mycobacteria showed a truncated form of Rv2742c (~1/3 the length of M. tuberculosis Rv2742c) at the appropriate distance from the preceding gene.
DISCUSSION
The present work connects envelope stress signaling with envelope maintenance in M. tuberculosis, thus completing the circle between sensing stress and recovering from damage. Our finding that the four-gene, clgR-pspA-rv2743c-rv2742c module is conserved only within the M. tuberculosis complex strongly suggests a link between this module and the ability of tuberculous mycobacteria to cause disease. Such linkage is consistent with the reduced intra-macrophage growth of M. tuberculosis we find associated with disruption of the clgR-pspA-rv2743c-rv2742c operon.
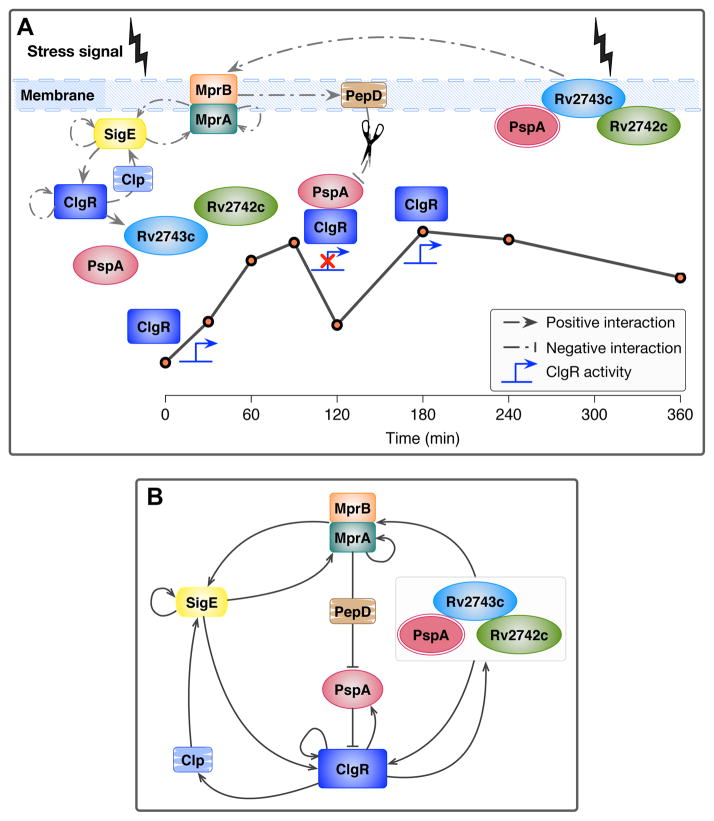
We found strong similarities between the clgR-pspA-rv2743c-rv2742c operon of M. tuberculosis and the Psp system of Gram-negative bacteria. For example, the minimal Psp module includes four adjacent genes, the regulator pspF and its three regulated genes, pspABC. In M. tuberculosis, the regulator is clgR, followed by pspA-rv2743c-rv2742c. With the Gram-negative system, the regulator’s transcriptional activity is normally blocked by formation of the PspF-PspA complex; envelope stress presumably alters PspA conformation and leads to formation of higher-order PspA multimers (Joly et al., 2010). The resulting multimeric structure results in (i) release of PspF, (ii) consequent PspF-dependent induction of the pspABC operon by σ54-containing RNA polymerase, and (iii) formation of a complex between PspA and the two integral membrane proteins PspB and PspC (Darwin, 2005, Joly et al., 2010). With M. tuberculosis, our data fit with a model (Fig. 10A) that parallels the Gram-negative scenario. We propose that surface-stress-dependent induction of the clgR-pspA-rv2743c-rv2742c operon by MprAB-σE results in formation of a complex between ClgR and PspA. This complex would block ClgR activity at ~90 min post-stress (Fig. 3). A second peak of ClgR activity (~120 min onwards) requires functional rv2743c-rv2742c (Fig. 4A), which we interpret as PspA forming a multiprotein complex with Rv2743c-Rv2742c and releasing ClgR from the inhibitory complex (this proposition implies that the pair-wise interactions observed with recombinant proteins (Fig. 5) reflect the ability to form a multiprotein complex in living cells). Thus, the ubiquitous protein PspA would exert parallel inhibitory function over the cognate regulator in M. tuberculosis (ClgR) and in Gram-negative organisms (PspF). Moreover, since PspA repurposing from inhibitor to effector activity in Gram-negative bacteria requires PspBC, these two proteins positively regulate PspF activity (Joly et al., 2010). Likewise, the M. tuberculosis rv2743c-rv2742c genes positively affect ClgR activity (Fig. 4A). Thus, despite the absence of amino acid sequence homology, Rv2743c and Rv2742c might serve as functional homologs of PspB and PspC of Gram-negative organisms. Indeed, the disappearance of the second peak of ClgR activity in a mutant deficient in pepD (Fig. S5), which normally targets PspA for degradation (White et al., 2011), supports our model: wild-type abundance of Rv2743c-Rv2742c may be insufficient to repurpose excess PspA found in the pepD mutant, leading to continuing inhibition of ClgR activity by PspA. These regulatory similarities between phylogenetically distant microorganisms emphasize the widespread importance of the four-gene Psp module.
Figure 10.
(A) Model of MprAB-σE-dependent maintenance of envelope integrity in response to surface stress. The figure shows interactions between the envelope-stress signaling MprAB-σE network and the envelope-preserving ClgR-PspA-Rv2743c-Rv2742c system. Interactions include the mprAB-sigE transcriptional network and the mprAB-dependent pepD, the downstream target clgR-pspA-rv2743c-rv2742c operon, and additional clgR-regulated genes (clpP1). The line plot represents the temporal, post-stress expression profile of clpP1, as a proxy of ClgR activity. The location of individual proteins, protein complexes, and interactions relative to the line plot depicts the proposed temporal course of events associated with propagation of signal (ClgR activity) through the Psp system, resulting membrane integrity, and MprAB stabilization. The scissors icon indicates proteolytic degradation. The red cross over the ClgR activity icon represents ClgR inactivation. (B) Feedback loops in the stress-response system of M. tuberculosis. The wiring diagram shows multiple regulatory feedback loops, transcriptional and post-transcriptional, operating between mprAB, sigE, clgR, the Psp system, and neighboring network genes and/or corresponding gene products under surface stress conditions. The abundance of feedback loops in the system does not allow distinguishing whether the observed functional effects of the pspA-rv2743c-rv2742c products are direct, or occur indirectly via the regulation of ClgR and its target gene products. Since clgR is connected to the mprAB/sigE sensing network through two positive feedbacks, links between envelope-stress-sensing and envelope-preserving functions exist regardless of whether pspA-rv2743c-rv2742c act directly or indirectly. Except for sigE autoregulation (Chauhan and Gennaro, unpublished), the regulatory interactions showed in this figure are all cited or discovered in the present manuscript. Arrowhead = positive regulation; Barhead = negative regulation.
Parallels also exist between the Gram-negative and the M. tuberculosis systems in terms of both function and sub-cellular localization. The membrane-bound PspABC complex of Gram-negative bacteria has envelope-stabilizing activities, with PspA exhibiting key effector functions (Darwin, 2005, Darwin, 2013, Joly et al., 2010). Similar effector functions are presumably exerted by the M. tuberculosis module, since clgR-pspA-rv2743c-rv2742c is induced when PMF is dissipated and is required to express phenotypes diagnostic of envelope integrity, such as ATP homeostasis and tolerance to surface stress. We note that the extensive network of positive feedback loops regulating this system (Fig. 10B) precludes using genetic analyses to assign these functional phenotypes to direct (envelope-preserving) effects or to indirect (ClgR activity modulating) effects of the pspA-rv2743c-rv2742c genes. However, the ability of pspA of M. tuberculosis to complement the alkaline pH-tolerance defect of a pspA mutant of E. coli (data not shown) implies that genes in the pspA-rv2743c-rv2742c module of M. tuberculosis can serve the same functions reported for Gram-negative bacteria. As with Gram-negative bacteria, the multiprotein complex between PspA, Rv2743c and Rv2742c may be targeted to the M. tuberculosis cell envelope, since Rv2743c is a membrane protein (Fig. 8). Intriguingly, subcellular fractionation analyses of M. tuberculosis have revealed both PspA- and Rv2742c-derived peptides in the cell membrane fraction, despite the absence of transmembrane domains or lipid anchoring motifs in these proteins (Mawuenyega et al., 2005, Rumschlag et al., 1990). These observations support the presence in envelope-stressed mycobacterial cells of an envelope-bound, multiprotein complex containing PspA, Rv2743c and Rv2742c proteins, parallel to the Gram-negative situation. The observation that clgR and its target genes are induced in response to surface stress in M. smegmatis (Fig. S7), which encodes only three of the four genes found in the Psp module of the M. tuberculosis complex (Fig. 9), provides an opportunity for parallel functional and structural analyses of the Psp module between tuberculous and non-tuberculous mycobacteria, and between mycobacteria and Gram-negative bacteria.
In conclusion, our work reveals parallel or convergent evolution of the Psp response in bacteria of different phyla in which conserved functions are expressed by unrelated proteins. The Gram-negative and M. tuberculosis systems have, however, evolved dynamic differences. In Gram-negative bacteria, pspF is constitutively expressed (Weiner et al., 1991), and the PspF-PspA complex is found in unstressed cells (Darwin, 2005, Joly et al., 2010). In M. tuberculosis, clgR is stress-induced, and our data point to formation of the ClgR-PspA complex in response to stress. The simpler, pre-stress/post-stress dichotomy seen in Gram-negative bacteria may allow a faster response. In contrast, post-stress formation of the inhibitory ClgR-PspA complex in M. tuberculosis, together with the positive feedback loops connecting the ClgR-PspA-Rv2743c-Rv2742c system with the MprAB-σE network (Fig. 6B), may be better suited for sustained responses to multiple, envelope-perturbing signals that are likely to occur over the long course of chronic infection. Such dissimilar dynamics point to how conserved functions can adapt to different bacterial life styles by varying regulatory modes.
EXPERIMENTAL PROCEDURES
Bacterial strains, reagents, and growth conditions
Escherichia coli XL1 blue (Agilent Technologies, Santa Clara, CA) was used for DNA cloning. E. coli BL21(DE3) (Novagen, La Jolla, CA) was used for recombinant protein expression. All E. coli strains were grown with aeration at 37°C in Luria-Bertani (LB) broth or agar (Thermo Fisher Scientific, Waltham, MA). Media were supplemented with 25 μg ml−1 chloramphenicol (Sigma, St. Louis, MO), 150 μg ml−1 hygromycin B (Roche Diagnostic, Mannheim, Germany), 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin sulfate (Thermo Fisher Scientific, Waltham, MA), as needed. M. tuberculosis knock-out mutants in sigE, clgR and pepD and corresponding complemented strains were previously reported (Manganelli et al., 2001, Estorninho et al., 2010, White et al., 2010). A transposon-insertion mutant in gene rv2743c was obtained from the BEI repository (NR18017; http://www.beiresources.org/Catalog). Complemented strains for clgR and rv2743c mutants were constructed in this work (described below in DNA manipulations). Gene numbering for M. tuberculosis is according to Cole et al (Cole et al., 1998). M. tuberculosis cultures were grown in Dubos Tween-albumin broth (Becton Dickinson, Sparks, MD) (liquid medium) or 7H10 (solid medium) (Difco, Franklin Lakes, NJ) supplemented with 0.05% Tween 80, 0.2% glycerol, and 10% ADN (2% glucose, 5% bovine serum albumin, 0.15 M NaCl). Liquid cultures of M. tuberculosis were grown in 25-ml tubes at 37°C with magnetic-bar stirring at 450 rpm. Plates were incubated at 37°C in sealed plastic bags. Both solid and liquid media were supplemented with 25 μg ml−1 kanamycin sulfate (ThermoFisher Scientific, Waltham, MA), 50 μg ml−1 hygromycin B, as needed. M. smegmatis cultures were grown in 7H9 broth (Difco, Franklin Lakes, NJ) supplemented with 0.05% Tween 80 and 0.2% glycerol.
DNA manipulations
Construction of complementing plasmids
A clgR knock-out mutant was complemented with DNA comprising an increasing number of open reading frames. These fragments were obtained by using the same forward primer (mapping 382 bp upstream of the clgR coding sequence) paired with a different reverse primer mapping at the 3′end of clgR, pspA, rv2743c or rv2742c to amplify the DNA sequences encompassing clgR, clgR-pspA, clgR-pspA-rv2743c or clgR-pspA-rv2743c-rv2742c, respectively. Amplified fragments were cloned in an integrative E. coli-mycobacteria shuttle vector pMV306kan (Braunstein et al., 2001, Stover et al., 1991). To take into account the polar effect of the rv2743c mutation on rv2742c, a DNA fragment encompassing clgR-pspA-rv2743c-rv2742c was generated for complementation of the rv2743c mutant with rv2743c-rv2742c expressed from the native promoters, and cloned in the integrative plasmid pMV306Hyg. To inactivate clgR and pspA in this complementing DNA, an in-frame deletion was introduced in pspA, and an in-frame stop codon was introduced in clgR, in the following way. Genomic DNA was used as template to amplify two DNA fragments: (i) a 895-bp fragment spanning from 382 bp upstream of clgR to 45 bp downstream of pspA, and (ii) a 1,848-bp fragment spanning from 771 bp downstream of pspA to 69bp downstream of the rv2742c ORF. The first fragment was further used as template for site-directed mutagenesis (performed by overlap extension PCR (Datta et al., 2002)) to introduce an in-frame stop codon (TAA) 54 bp downstream of the start codon of the clgR open reading frame. The fragment containing the in-frame mutation of clgR was cloned at the KpnI-XbaI sites while the second fragment was cloned at the XbaI-HindIII sites in plasmid pMV306-Hyg. Cloning these two fragments next to each other resulted in a 723-bp in-frame deletion in pspA. Thus, the resulting fragment contained the entire clgR-pspA-rv2743c-rv2742c region in which both clgR and pspA were non-functional. For complementation of the hsp mutant, a DNA fragment spanning the hsp coding sequence and the upstream 400 bp region was amplified by PCR. The amplified fragments were cloned in the integrative plasmid pMV306-Hyg. In all cases, the nucleotide sequence of the complementing DNA fragment was verified by DNA sequence analysis. Nucleotide sequences of PCR primers are listed in Table S1. Constructs were introduced in the corresponding mutant strains by electroporation. Transformants were selected on plates supplemented with appropriate antibiotics and verified by PCR analysis.
Construction of lacZ fusions
For construction of promoter::lacZ fusions, DNA fragments containing sequences upstream of clgR (382 bp) and upstream of pspA (250 bp) plus the first eight codons of the corresponding open reading frame were amplified. Amplified fragments were cloned in frame with lacZ in the promoter probe E. coli-mycobacteria shuttle plasmid pJEM13 (Timm et al., 1994). Nucleotide sequences of PCR primers are listed in Table S1. The resulting plasmids were introduced by electroporation in M. tuberculosis wild-type and clgR mutant. Transformants were selected on 7H10 agar plates supplemented with kanamycin.
SDS treatment
For gene expression analyses, mid-log cultures of M. tuberculosis were treated with SDS to select a bacteriostatic concentration of the detergent (0.03%) (Fig. S8A) that induced sigE (Fig. S8B), and had no lethal effect in a 6-hr treatment on any of the mutants used (Fig. S8C). Following treatment with 0.03% SDS, 1-ml culture aliquots were collected at various time intervals up to 4 hrs and used for RNA extraction. For assessment of relative susceptibility to bactericidal SDS concentrations, wild-type and mutant M. tuberculosis cultures were grown to mid-log phase, and aliquots of approximately 105 cells were treated for 24 hrs with a bactericidal concentration of the detergent (0.2%) (Fig. 6A). After incubation, samples were diluted and plated on stressor-free medium to determine the fraction of surviving cells relative to samples taken at the time of SDS treatment.
Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) treatment
Mid-log-phase cultures of M. tuberculosis were treated with CCCP at increasing concentrations (up to 100 μM). Since CCCP is dissolved in DMSO, a solvent-only control (corresponding to the highest DMSO concentration used with the CCCP treatment) was also included. Following 6 hrs of treatment, 1-ml culture aliquots were collected and used for RNA extraction.
RNA extraction and enumeration of bacterial transcripts
Bacterial cell pellets were resuspended in 1 ml TRI reagent (Molecular Research Center, Cincinnati, OH) and 0.8 ml zirconia beads (0.1-mm diameter, BioSpec Products, Inc., Bartlesville, OK). Cells were disrupted in a bead-beater (Mini-Beadbeater-16, BioSpec Products, Bartlesville, OK) by three 45-sec pulses, each separated by 10 min incubation on ice. Cells were lysed by adding 100 μl BCP Reagent (Molecular Research Center, Cincinnati, OH) and vigorous mixing for 10 minutes. After 5-min incubation at room temperature, the tubes were centrifuged for 30 min at 12,000 × g at 4°C. The aqueous phase was transferred to fresh tubes containing 500 μl isopropanol for overnight precipitation. After four cycles of overnight precipitation, samples were washed with 75% ethanol, air-dried, and resuspended in diethyl pyrocarbonate (DEPC)-treated H2O for storage at −80°C. Reverse transcription was performed with random hexameric primers and ThermoScript™ Reverse Transcriptase (Invitrogen, Carlsbad, CA). Enumeration of mRNAs was carried out by qPCR using gene-specific primers, molecular beacons, and AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA) in a Stratagene Mx4000 thermal cycler (Agilent Technologies, La Jolla, CA).
Nucleotide sequences of PCR primers and molecular beacons are listed in Table S1. The M. tuberculosis 16s rRNA copy number was used as a normalization factor to enumerate bacterial transcripts per cell, as previously described (Shi et al., 2003).
E. coli constructs for protein expression
The coding regions for clgR, pspA, rv2743c, and rv2742c of M. tuberculosis were each amplified using gene-specific primers (Table S1) and cloned into either of the compatible plasmids pETDUET and pACYCDUET (EMD Chemicals, San Diego, CA) under an IPTG-inducible T7 promoter. A fragment carrying the clgR coding sequence was cloned at the MCS1 site of plasmid pACYCDUET to express a recombinant product carrying a C-terminal Myc tag. The pspA coding sequence was cloned at the MCS1 of pETDUET to obtain a N-terminally 6xHis-tagged protein. The rv2743c coding sequence was cloned at the MCS2 site in pETDUET or in the pspA-pETDUET to obtain a C-terminally S-tagged protein. The rv2742c coding sequence was cloned at the MCS1 site of either pETDUET or rv2743c-pETDUET to obtain an N-terminally 6xHis-tagged protein, and it was also cloned at the MCS2 of pspA-pETDUET to be expressed as a C-terminally S-tagged protein. The resulting DNA was transformed into E. coli XL1 Blue. Transformants were selected on antibiotic-containing LB agar plates and analysed by restriction endonuclease mapping and DNA sequencing. For expression of recombinant proteins, plasmids expressing differentially tagged protein pairs or each protein alone were electroporated into E. coli BL21(DE3) cells. Transformants expressing protein or proteins pair were selected on plates supplemented with the appropriate antibiotics.
Transcription start site (TSS) mapping
Replicate cultures of M. tuberculosis H37Rv were grown to late-log phase (optical density ~1) in 7H9 Middlebrook broth supplemented with 10% OADC, 0.05% Tween-80, and 0.2% glycerol. Cell pellets were disrupted in Trizol (Life Technologies, Carlsbad, CA) in Lysing Matrix B tubes in a FastPrep-24 instrument (MP Biomedicals, Santa Ana, CA), and RNA was extracted according to the manufacturer’s instructions. RNA was subsequently treated with DNase Turbo (Life Technologies, Carlsbad, CA) and purified by RNeasy (Qiagen, Hilden, Germany) before depletion of ribosomal RNA with a MICROBExpress kit (Life Technologies, Carlsbad, CA). Transcription start sites were mapped as described (Shell et al., 2015). Briefly, two parallel Illumina sequencing libraries were made from each RNA preparation: a “converted” library, which captured RNA 5′ ends endogenously bearing 5′ triphosphates or 5′ monophosphates, and a “non-converted” library, which captured only RNA 5′ ends endogenously bearing 5′ monophosphates. For the “converted” library, RNA was pre-treated with polyphosphatase (Epicentre, Madison, WI). Samples were purified by RNeasy (Qiagen, Hilden, Germany) and ligated to an adapter oligo SSS392 (TCCCTACACGACGCTCTTCCGAUCU; normal font indicates deoxyribonucleotides and italics indicate ribonucleotides) with T4 RNA ligase I (New England Biolabs, Ipswitch, MA). Ligation products were purified and sheared in a Covaris sonicator. cDNA was synthesized with oligo SSS397 (CTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNN, where “N” represents a degenerate base) using Superscript III (Life Technologies, Carlsbad, CA). Purified cDNA was subsequently amplified by eight PCR cycles using primers containing additional Illumina adapter sequences (oligo SSS398, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTC, and a reverse oligo CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGTGCT, where “XXXXXX” is a 6-nucleotide Illumina index sequence). PCR products were size-selected by agarose gel electrophoresis to a range of 150–500 nucleotides, purified with a Qiagen gel extraction kit followed by Ampure XP beads (Beckman Coulter, Pasadena, CA), and subjected to an additional four PCR cycles with oligo SSS401 (AATGATACGGCGACCACCGAGATC) and oligo SSS402 (CAAGCAGAAGACGGCATACGAGAT). PCR products were purified with Ampure XP beads and sequenced on an Illumina Genome Analyzer. 56-nucleotide, paired-end reads were mapped using the sequence alignment algorithm Ssaha2. Peaks in 5′-end coverage were identified and filtered based on the absolute read count values in the converted libraries as well as the ratio of these read counts to coverage in RNAseq expression libraries. For each filtered peak, the ratio of “1st nucleotide” coverage in the converted/non-converted libraries was determined after pooling coverage from replicate libraries. Gaussian mixture modeling of bimodally distributed ratios was used to estimate means and standard deviations for two skewed normal distributions (processed and unprocessed 5′ ends). RNA 5′ ends with ratios >1.74 had a cumulative probability of ≤0.01 of belonging to the processed 5′ end population after Benjamini-Hochberg adjustment, and were designated transcription start sites.
Recombinant protein expression and immunoprecipitation
To test protein-protein interactions, mid-log E. coli BL21(DE3) cultures expressing two differentially tagged proteins singly or together were treated with 200 μM IPTG for 2 hrs. 50 ml cultures were harvested and subjected to lysis in 1 ml B-PER reagent as per manufacturer’s protocols (Thermo Scientific, Rockford, IL). The whole-cell lysate was clarified by centrifugation. Total protein content of the cleared lysates was measured with the Bradford assay. Cleared lysates containing fixed amounts of total protein were incubated with monoclonal anti-His or anti S-tag antibody following manufacturer’s protocols (EMD Chemicals, San Diego, CA). Immune complexes were pulled down with 20 μl protein A/G agarose (EMD Millipore, Billerca, MA). Agarose-bound complexes were washed twice with 200 μl B-PER reagent, and the resulting samples were boiled in 20 μl of 2× Laemmli sample buffer for 15 min. Proteins were separated by SDS-PAGE, transferred onto PVDF membranes for Western blot analysis with anti-Myc antibody (Life technology, Grand Island, NY, USA).
Protein-protein interactions were also analyzed by pull-down assays. Cell lysates expressing recombinant 6xHis tagged protein, alone or together with the potential interacting partner, were incubated with Ni-NTA agarose (EMD Chemicals, San Diego, CA). Slurry was washed twice with 500 μl wash buffer (EMD Chemicals, San Diego, CA), boiled with 2x Laemmli buffer and subjected to SDS-PAGE. Western blot analysis was performed with anti-S tag antibody, as above. Detection was carried out by incubation with anti-mouse IgG-HRP-conjugate (EMD Chemicals, San Diego, CA) and enhanced chemiluminescence (Cell Signaling Technology, Danvers, MA).
Measurement of ATP
Exponentially growing cultures were treated with 0.03% SDS for 24 hrs. One-ml aliquots of untreated and SDS-treated cultures were harvested for total ATP measurement, as described (Wayne & Hayes, 1996). Briefly, cell pellets were resuspended in 0.025M HEPES, pH 7.75 (supplemented with 0.02% Tween-80). Aliquots of 0.05 ml of cell suspensions were diluted in equal volume of 0.025M HEPES, and 0.04 ml of chloroform was added to the each tube. The resulting sample was heated at 80°C for 20 min, followed by addition of 4.9 ml of 0.025M HEPES. A 0.05 ml aliquot of the sample was mixed with an equal volume of the luciferin-luciferase reagent, and light unit was recorded in a luminometer (Glomax-R, Promega Corporation, Madison, WI). Light units were converted to concentrations of ATP by use of a standard curve of light units measured with various freshly prepared dilutions of a 100 ng ml−1 ATP standard solution. Concentration of ATP was normalized to total protein content.
Culture and infection of human monocyte-derived macrophages (MDM)
Human buffy coats were obtained from the New York Blood Center (Long Island City, NY) and peripheral blood mononuclear cells (PBMC) were prepared by Ficoll density gradient centrifugation (Ficoll-Paque™, GE Healthcare, Uppsala, Sweden) as described (Davies & Gordon, 2005). Isolated PBMC were washed and resuspended in RPMI-1640 medium (Corning, Manassas, VA) supplemented with 10% fetal bovine serum (Seradigm, Radnor, PA) and 4 mM L-glutamine (Corning, Manassas, VA), plated at a density of 5×106 cells per well in 24-well plates, and incubated overnight in humidified atmospheric air containing 5% CO2 at 37°C to let monocytes adhere, following standard protocols (for example, (Sharma et al., 2009)). Non-adherent cells were removed by washing three times with 1x PBS; adherent cells were maintained in supplemented RPMI-1640 medium and allowed to differentiate for 4 days in a 5% CO2 incubator at 37°C. At day 5, medium was removed and MDM were counted and infected with M. tuberculosis strains. Bacterial inoculum for infection was prepared by diluting a frozen bacterial stock in supplemented RPMI-1640 medium to obtain an MOI of 0.1 (CFU per cell). Bacterial clumps were disrupted by vortexing with sterile 3-mm diameter glass beads for 2 min, and this suspension was used for MDM infection. After 4 hrs of infection, extracellular bacteria were removed by washing three times with 1x PBS, and fresh supplemented RPMI-1640 medium was added to cells. Medium was replenished also at days 2 and 5 post-infection. At 4 hrs (to determine infecting dose) and at day 7 post-infection, bacterial CFU were enumerated by lysing infected MDM with 0.05% SDS and plating serial dilutions of cell lysates on agar plates. Prior to lysis, an aliquot of cells was used to calculate the number of adherent cells in each well, and CFU were normalized to 105 adherent cells.
In silico nucleotide and amino acid sequence analyses
To identify putative ClgR binding sites, ClgR binding sequences were obtained from [16]. MEME software [43] was used to identify all binding sites and to generate the corresponding motifs and the sequence logo. The tool MAST [44] was used to calculate the p-values associated with the sequence matches.
Analysis of primary amino acid sequence similarity was performed using the Blosum62 matrix in Geneious Pro (Biomatters Ltd., Auckland, New Zealand). Comparative modeling and consensus secondary structure predictions were carried out using Phyre (Kelley & Sternberg, 2009), which incorporates the results of calculations made using Psi-Pred (McGuffin et al., 2000), SSPro (Pollastri et al., 2002), and Jpred (Cole et al., 2008). Additional protein and domain classification was carried out using InterPro (Hunter et al., 2012), InterProScan (Jones et al., 2014), Pfam (Mistry et al., 2007, Sonnhammer et al., 1997, Finn et al., 2011) and SMART (Schultz et al., 1998, Letunic et al., 2014). Coiled-coil predictions were done using Coils (Lupas et al., 1991). Transmembrane regions were predicted using Phobius (Kall et al., 2004), TMHMM (Sonnhammer et al., 1998), HMMTop (Tusnady & Simon, 1998, Tusnady & Simon, 2001) and TMPred (Hofmann & Stoffel, 1993).
For the phylogenetic analyses, protein sequence similarity searches were carried out using NCBI BLAST (Altschul et al., 1990, Altschul et al., 1997) with each of M. tuberculosis clgR-pspABC proteins as query against a set of 3,436 genomic sequences from Actinobacteria (taxid: 201174) obtained from the PATRIC database ((Wattam et al., 2014) and http://www.patricbrc.org). Significant matches (E-value < 1e-5) from each genome were obtained, and similarity scores for each significant match was calculated as percent of the number of positive (i.e., identical plus conserved substitutions) amino acid residues divided by the length of the query protein.
Supplementary Material
Acknowledgments
We are grateful to Graham Stewart for the clgR mutant and complemented strain and Thomas Zahrt for the pepD mutant and complemented strain. We also thank Karl Drlica, David Perlin, Richard Pine, and Xilin Zhao for critical comments on the manuscript. The work was supported by NIH grant HL106788 to MLG.
Footnotes
The authors have no conflict of interest to declare.
References
- Alber T. Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr Opin Struct Biol. 2009;19:650–657. doi: 10.1016/j.sbi.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S, Sureka K, Mukherjee P, Basu J, Kundu M. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol Microbiol. 2010;75:592–606. doi: 10.1111/j.1365-2958.2009.07008.x. [DOI] [PubMed] [Google Scholar]
- Becker LA, Bang IS, Crouch ML, Fang FC. Compensatory role of PspA, a member of the phage shock protein operon, in rpoE mutant Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;56:1004–1016. doi: 10.1111/j.1365-2958.2005.04604.x. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Brown AM, Kurtz S, Jacobs WR., Jr Two nonredundant SecA homologues function in mycobacteria. J Bacteriol. 2001;183:6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretl DJ, Bigley TM, Terhune SS, Zahrt TC. The MprB extracytoplasmic domain negatively regulates activation of the Mycobacterium tuberculosis MprAB two-component system. J Bacteriol. 2014;196:391–406. doi: 10.1128/JB.01064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretl DJ, Demetriadou C, Zahrt TC. Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol Mol Biol Rev. 2011;75:566–582. doi: 10.1128/MMBR.05004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin AJ. The phage-shock-protein response. Mol Microbiol. 2005;57:621–628. doi: 10.1111/j.1365-2958.2005.04694.x. [DOI] [PubMed] [Google Scholar]
- Darwin AJ. Stress relief during host infection: The phage shock protein response supports bacterial virulence in various ways. PLoS Pathog. 2013;9:e1003388. doi: 10.1371/journal.ppat.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta P, Dasgupta A, Bhakta S, Basu J. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J Biol Chem. 2002;277:24983–24987. doi: 10.1074/jbc.M203847200. [DOI] [PubMed] [Google Scholar]
- Davidson LA, Draper P, Minnikin DE. Studies on the mycolic acids from the walls of Mycobacterium microti. J Gen Microbiol. 1982;128:823–828. doi: 10.1099/00221287-128-4-823. [DOI] [PubMed] [Google Scholar]
- Davies JQ, Gordon S. Isolation and culture of human macrophages. Methods Mol Biol. 2005;290:105–116. doi: 10.1385/1-59259-838-2:105. [DOI] [PubMed] [Google Scholar]
- Estorninho M, Smith H, Thole J, Harders-Westerveen J, Kierzek A, Butler RE, Neyrolles O, Stewart GR. ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology. 2010;156:3445–3455. doi: 10.1099/mic.0.042275-0. [DOI] [PubMed] [Google Scholar]
- Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Hovey R, Kane J, Singh V, Zahrt TC. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol. 2006;188:2134–2143. doi: 10.1128/JB.188.6.2134-2143.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler. 1993;347:166. [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Petzold CJ, Schelle MW, Leavell MD, Mougous JD, Bertozzi CR, Leary JA, Cox JS. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci U S A. 2007;104:5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MP, Buck M. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Provvedi R. An integrated regulatory network including two positive feedback loops to modulate the activity of sigma(E) in mycobacteria. Mol Microbiol. 2010;75:538–542. doi: 10.1111/j.1365-2958.2009.07009.x. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Voskuil MI, Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor sigma E: role in global gene expression and survival in macrophages. Mol Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- Mawuenyega KG, Forst CV, Dobos KM, Belisle JT, Chen J, Bradbury EM, Bradbury AR, Chen X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell. 2005;16:396–404. doi: 10.1091/mbc.E04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007;8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- Provvedi R, Boldrin F, Falciani F, Palu G, Manganelli R. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology. 2009;155:1093–1102. doi: 10.1099/mic.0.024802-0. [DOI] [PubMed] [Google Scholar]
- Roback P, Beard J, Baumann D, Gille C, Henry K, Krohn S, Wiste H, Voskuil MI, Rainville C, Rutherford R. A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res. 2007;35:5085–5095. doi: 10.1093/nar/gkm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- Rumschlag HS, Yakrus MA, Cohen ML, Glickman SE, Good RC. Immunologic characterization of a 35-kilodalton recombinant antigen of Mycobacterium tuberculosis. J Clin Microbiol. 1990;28:591–595. doi: 10.1128/jcm.28.3.591-595.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sharma M, Bose M. Mycobacterium tuberculosis infection of human monocyte-derived macrophages leads to apoptosis of T cells. Immunol Cell Biol. 2009;87:226–234. doi: 10.1038/icb.2008.97. [DOI] [PubMed] [Google Scholar]
- Shell SS, Chase MR, Ioerger TR, Fortune SM. RNA sequencing for transcript 5′-end mapping in mycobacteria. In: Parish T, Roberts DM, editors. Mycobacteria Protocols. 3. New York, NY: Springer Science+Business Media; 2015. [Google Scholar]
- Sherrid AM, Rustad TR, Cangelosi GA, Sherman DR. Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS ONE. 2010;5:e11622. doi: 10.1371/journal.pone.0011622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Jung YJ, Tyagi S, Gennaro ML, North RJ. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc Natl Acad Sci U S A. 2003;100:241–246. doi: 10.1073/pnas.0136863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. Sensitive profiling of chemically diverse bioactive lipids. J Lipid Res. 2007;48:1976–1984. doi: 10.1194/jlr.M700060-JLR200. [DOI] [PubMed] [Google Scholar]
- Song T, Song SE, Raman S, Anaya M, Husson RN. Critical role of a single position in the −35 element for promoter recognition by Mycobacterium tuberculosis SigE and SigH. J Bacteriol. 2008;190:2227–2230. doi: 10.1128/JB.01642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Timm J, Lim EM, Gicquel B. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J Bacteriol. 1994;176:6749–6753. doi: 10.1128/jb.176.21.6749-6753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Balazsi G, Gennaro ML, Igoshin OA. The interplay of multiple feedback loops with post-translational kinetics results in bistability of mycobacterial stress response. Phys Biol. 2010;7:036005. doi: 10.1088/1478-3975/7/3/036005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner L, Brissette JL, Model P. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 1991;5:1912–1923. doi: 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- Weiner L, Model P. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc Natl Acad Sci U S A. 1994;91:2191–2195. doi: 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, He H, Penoske RM, Twining SS, Zahrt TC. PepD participates in the mycobacterial stress response mediated through MprAB and SigE. J Bacteriol. 2010;192:1498–1510. doi: 10.1128/JB.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Savaryn JP, Bretl DJ, He H, Penoske RM, Terhune SS, Zahrt TC. The HtrA-like serine protease PepD interacts with and modulates the Mycobacterium tuberculosis 35-kDa antigen outer envelope protein. PLoS ONE. 2011;6:e18175. doi: 10.1371/journal.pone.0018175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Global tuberculosis report. Geneva, Switzerland: World Health Organization; 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.