A growing number of recent studies have highlighted bats as a reservoir for Leptospira bacteria, pointing out the potential role of bats in the epidemiology of the most widespread zoonotic disease in the world [1]. Because leptospirosis is a largely neglected disease, a number of unanswered questions remain about the ecology and evolution of Leptospira, especially those associated with bats. Here we summarize what has been recently learned about this emerging but enigmatic host–pathogen association. We show how this system can provide exciting new opportunities to obtain insights into the evolutionary ecology of bat-borne pathogens and propose future directions to disentangle the role of bats in human leptospirosis.
What Do We Know, Briefly, about Leptospirosis and Leptospira?
Leptospirosis is a bacterial disease of humans and animals caused by pathogenic spirochetes of the genus Leptospira. In humans, leptospirosis is an important (re-)emerging zoonosis of global public health concern [1], although tropical regions display the highest human incidence [2]. Over 500,000 human cases of severe leptospirosis are thought to occur each year worldwide, with a mortality rate of over 10%. Asymptomatic or subclinical human infections are common, making leptospirosis likely far more prevalent than currently diagnosed or recognized [3].
Leptospira are a complex of highly diversified bacteria comprising 22 species that include pathogenic (Leptospira interrogans, L. kirschneri, L. borgpetersenii, L. mayottensis, L. santarosai, L. noguchii, L. weilii, L. alexanderi, L. kmetyi, and L. alstonii), intermediate (i.e., species of unclear pathogenicity: L. broomii, L. fainei, L. inadai, L. licerasiae, L wolffii), and saprophytic (i.e., free-living and generally considered not to cause disease: L. biflexa, L. idonii, L. meyeri, L. terpstrae, L. vanthielli, L. wolbachii, L. yanagawae) species [4]. Alongside genetic characterization, serological classification (based on bacterial cell surface antigens) differentiates nearly 300 Leptospira serovars, of which more than 200 are considered pathogenic [5]. However, serovars are not indicative of the taxonomic relation among strains because one serovar may belong to more than one species (e.g., L. interrogans serovar Hardjo and L. borgpetersenii serovar Hardjo) and multiple serovars occur within the same species [6]. A wide variety of mammals can be infected, but rodents are recognized as significant reservoir hosts. Pathogenic and intermediate leptospires reside in the kidneys of infected animals and are spread through the excretion of urine into the environment [1]. Thus, contaminated soil or water as well as direct contact with infected animals are the main sources of leptospirosis.
To What Extent Are Bats Infected with Leptospira?
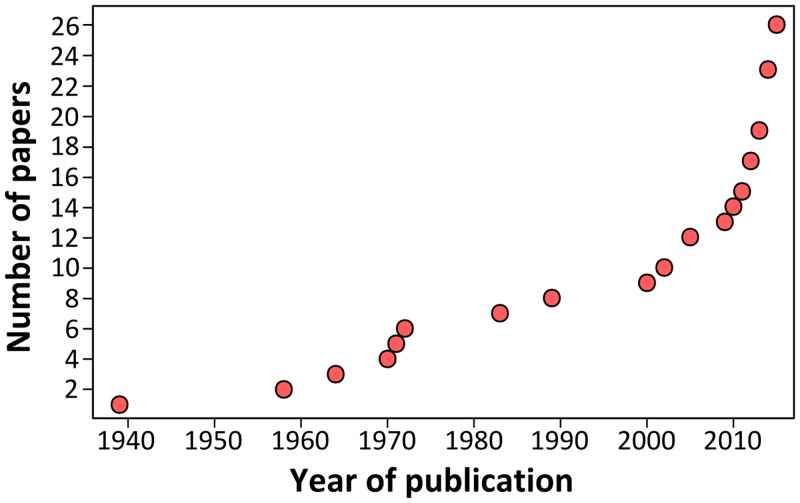
Growing scientific interest in bats as reservoirs of pathogens and the global importance of human leptospirosis have led to the emergence of investigations on the presence of Leptospira in wild bats during the last few years (Fig 1). Different techniques such as dark-field microscopy, serology by Microscopic Agglutination Test (MAT), PCR detection, and bacterial culture have been used. To date, Leptospira infection has been evidenced in over 50 bat species belonging to 8 of the 9 investigated bat families, encompassing various geographical regions in the tropics and subtropics [7–30] as well as Europe, although to a limited extent (Fig 2) [31,32]. Leptospira prevalence and seroprevalence in bat populations vary according to bat species and location. Given that bat sampling is often opportunistic, small sample sizes may account for the bias observed in the results. Moreover, a recent study revealed that the prevalence of Leptospira excretion in bat urine is highly variable over time, ranging from 6% to 45% within the same colony over a five-month period [20]. Thus, infection dynamics leading to variations in Leptospira shedding should be taken into account when bat populations are monitored for prevalence.
Fig 1. Cumulative number of publications investigating Leptospira infection in bats over the past 75 years.
Fig 2. Geographic distribution and diversity of Leptospira in bats.
Countries are highlighted in red when PCR and/or serology were found positive (corresponding to the following bat families: Phyllostomidae, Mollosidae, Vespertillionidae, Hipposideridae, Miniopteridae, Nycteridae, Mormoopidae, Pteropodidae) and in grey for negative results (Thyropteridae). When available, Leptospira diversity is shown in green when genetic data have been used for species identification and in blue when serological analyses have been performed for serovar determination.
Which Leptospira Infect Bats, and When?
There is increasing evidence that bats are infected by highly diverse leptospires, especially in tropical regions with high bat species richness [8,16,19]. Based on genetic identification, bats are infected by at least four species, i.e., L. interrogans, L. borgpetersenii, L. kirschneri, L. fainei, and likely yet-undescribed genetic clades (Fig 2) [8,19,28]. The use of multilocus sequence analysis has largely improved our view of Leptospira diversity in bats and has shown strong host specificity [19] as well as coinfection with multiple Leptospira [16,28]. The evolution of bat-borne Leptospira diversity and host specificity is probably linked to both cospeciation and host-switching events [33] but also to ecological features such as colony density, feeding behavior, and migration [34]. According to whole-genome [35] and field-based studies [36], which suggest that different Leptospira species have evolved towards different modes of transmission, bat species roosting in high-density colonies may be, for example, primarily infected by Leptospira dependent on host-to-host transmission, such as hypothesized for L. borgpetersenii [35].
Dynamics of Leptospira infection in bat populations remains largely overlooked. Bat roosting behavior is thought to favor Leptospira transmission via urine [20]. Indeed, the reproduction and aggregation behavior of bats within their roosts have been shown to be linked to active Leptospira transmission, leading to high rates of infection in maternity colonies [20]. As demonstrated for RNA viruses, increased prevalence during seasonal bat reproduction may thus be associated with higher risk of spillover [37–40]. However, based on current research, there is very little evidence to disentangle whether bat-borne Leptospira persist within the host and/or in the environment, as well as whether they are maintained in nature by perpetuation within and between bat colonies [38]. Field monitoring of Leptospira excretion in natural bat populations suggests that bats may develop an immune response after acute infection and then stop excreting Leptospira [20]. In contrast, observation of natural Leptospira-infected bats in Denmark showed that leptospires are able to colonize the renal tubules of bats followed by continuous excretion in urine up to five months. This would indicate that chronic infection may occur in bats [31], as already characterized in chronic asymptomatic animal carriers such as rats [41].
What Is the Public Health Risk of Bat-Borne Leptospira?
Because of their abundance and spatial distribution, bats may contribute to the global maintenance and dissemination of pathogenic leptospires. However, the role of bats as carriers of strains of leptospires associated with human leptospirosis remains uncertain. Direct transmission of bat-borne Leptospira to humans has already been suggested, but never evidenced, following a case of serologically confirmed human leptospirosis after bat exposure [42]. Contact with urine and contaminated water is the main form of disease transmission. Human encroachment into bat habitats as well as increasing urbanization, which facilitates bat roosting in artificial structures, are likely to increase the opportunity for bat-borne Leptospira spillover [34]. Indeed, evidence of leptospiral infection of kidneys has already been reported in bats roosting in schools and houses [10,16].
Indirect transmission of bat-borne Leptospira to humans may also occur through spillover between bat-borne Leptospira and other animal hosts, in particular ground-dwelling species such as rodents that reside or forage under bat roosts [8,13,15]. Such transmission between bats and rodents has already been suggested, as L. interrogans, a typical rodent-borne Leptospira species, has been evidenced in insectivorous and frugivorous bats [8,16]. Elucidating the ecological conditions that may favor bat-borne Leptospira transmission thus represents a major challenge for public health.
What Are Future Directions for Research into Bat-Borne Leptospira?
The widespread pattern and enigmatic features of Leptospira infection in bats represent a challenging opportunity to study the evolutionary ecology of bat-borne infectious agents of possible importance for public health. While other studies mostly focus on viruses, the study of transmission cycles involving bats and bacterial pathogens in particular will provide an original system to understand general patterns of bat-borne pathogen epidemiology.
As a model system, continued research on the ecology of host and bacteria is necessary. It has been recently shown that Leptospira excretion in bats can be highly dynamic [20], but ecological factors that drive spatial and temporal variations of infection remain uncertain. For example, what are the roles of environmental factors such as weather seasonal patterns in the transmission dynamics of Leptospira in bat populations? Is Leptospira infection in bats maintained through epidemic episodes during the bat reproductive season in maternity colonies, or does it persist endemically within any single local population? Do males play a particular role in dispersing Leptospira among colonies compared to phylopatric females? Some of these questions can be addressed using long-term data sets by monitoring bat population dynamics, Leptospira excretion, and immune response in bat colonies. Noninvasive urine sampling should be preferred, as it allows the collection of a high number of samples while limiting the disturbance of colonies [20]. This will require the validation of urine shedding as a good proxy of renal infection, as recently demonstrated in rats [43]. Parallel investigation of rodent populations in the vicinity of bat colonies would be necessary to assess potential exchanges between these two animal hosts, as already shown for other infectious agents such as paramyxoviruses [44,45].
Improvement of bacterial culture from noninvasive bat samples (such as urine) would be a crucial step for understanding Leptospira–bat associations. First of all, it would improve genetic characterization of bat-borne strains and thus provide a more comprehensive picture of Leptospira evolution in bats. Secondly, bacterial isolates would allow experimental studies to investigate chronic manifestations in bats, as already demonstrated for rodents [4], as well as the assessment of the survival of bat-borne Leptospira in soil and water, in order to determine the role of environment as a source of infection. Animal models would further enable assessment of host specificity and virulence of bat-borne strains and the potential for possible spillovers. Finally, the development of serological diagnostic tests, designed to express a narrow specificity towards bat-borne strains, will allow us to assess the potential exposure of rodent and domestic animal populations to bat-borne Leptospira and to determine the burden of acute and asymptomatic Leptospira infection in humans from bat origin.
Funding Statement
This work was made possible by funding from the National Research Foundation, South Africa (NRF – N00595) to MD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Adler B (2015) Leptospira and Leptospirosis. Curr Topics Microbiol 387: 1–293. [DOI] [PubMed] [Google Scholar]
- 2. Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N (2008) The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis 12: 351–357. [DOI] [PubMed] [Google Scholar]
- 3. Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, et al. (2010) Asymptomatic renal colonization of humans in the peruvian Amazon by Leptospira . PLoS Neglect Trop 4: e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ko AI, Goarant C, Picardeau M (2009) Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7: 736–747. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerqueira GM, Picardeau M (2009) A century of Leptospira strain typing. Infect Genet Evol 9: 760–768. 10.1016/j.meegid.2009.06.009 [DOI] [PubMed] [Google Scholar]
- 6. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. (2003) Leptospirosis : a zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 7. Bunnell JE, Hice CL, Watts DM, Montrueil V, Tesh RB, et al. (2000) Detection of pathogenic Leptospira spp. infections among mammals captured in the Peruvian Amazon basin region. Am J Trop Med Hyg 63: 255–258. [PubMed] [Google Scholar]
- 8. Matthias MA, Díaz MM, Campos KJ, Calderon M, Willig MR, et al. (2005) Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by bayesian phylogenetic analysis of 16S ribosomal DNA sequences. Am J Trop Med Hyg 73: 964–974. [PMC free article] [PubMed] [Google Scholar]
- 9. Zetun C, Hoffmann J, Silva R, Souza L, Langoni H (2009) Leptospira spp. and Toxoplasma gondii antibodies in Vampire bats (Desmodus rotundus) in Botucatu region, Brazil. J Venom Anim Toxins incl Trop Dis 15: 546–552. [Google Scholar]
- 10. Bessa TAF, Spichler A, Chapola EGB, Husch AC, de Almeida MF, et al. (2010) The contribution of bats to leptospirosis transmission in Sao Paulo City, Brazil. Am J Trop Med Hyg 82: 315–317. 10.4269/ajtmh.2010.09-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramirez NN, Alegre EA, De Biasio MB, Bastiani CE (2014) Detecciósn de leptospiras patógenas en tejido renal de murciélagos de Corrientes, Argentina. Rev Vet 25: 16–20. [Google Scholar]
- 12. Emanuel ML, Mackerras IM, Smith DJW (1964) The epidemiology of leptospirosis in North Queensland. J Hyg 62: 451–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smythe LD, Field HE, Barnett L, Smith CS, Sohnt MF, et al. (2002) Leptospiral antibodies in flying foxes in Australia. J Wildlife Dis 38: 182–186. [DOI] [PubMed] [Google Scholar]
- 14. Cox TE, Smythe LD, Leung LK-P (2005) Flying foxes as carriers of pathogenic Leptospira species. J Wildlife Dis 41: 753–757. [DOI] [PubMed] [Google Scholar]
- 15. Tulsiani SM, Cobbold RN, Graham GC, Dohnt MF, Burns M, et al. (2011) The role of fruit bats in the transmission of pathogenic leptospires in Australia. Ann Trop Med Parasitol 105: 71–84. 10.1179/136485911X12899838413501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lagadec E, Gomard Y, Guernier V, Dietrich M, Pascalis H, et al. (2012) Pathogenic Leptospira spp. in bats, Madagascar and Union of the Comoros. Emerg Infect Dis 18: 1696–1698. 10.3201/eid1810.111898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desvars A, Naze F, Benneveau A, Cardinale E, Michault A (2013) Endemicity of leptospirosis in domestic and wild animal species from Reunion Island (Indian Ocean). Epidemiol Infect 141: 1154–1165. 10.1017/S0950268812002075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desvars A, Naze F, Vourc’h G, Cardinale E, Picardeau M, et al. (2012) Similarities in Leptospira serogroup and species distribution in animals and humans in the Indian ocean island of Mayotte. Am J Trop Med Hyg 87: 134–140. 10.4269/ajtmh.2012.12-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dietrich M, Wilkinson DA, Soarimalala V, Goodman SM, Dellagi K, et al. (2014) Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol Ecol 23: 2783–2796. 10.1111/mec.12777 [DOI] [PubMed] [Google Scholar]
- 20. Dietrich M, Wilkinson DA, Benlali A, Lagadec E, Ramasindrazana B, et al. (2015) Leptospira and Paramyxovirus infection dynamics in a bat maternity enlightens pathogen maintenance in wildlife. Environ Microbiol: in press. [DOI] [PubMed] [Google Scholar]
- 21. Collier WA, Mochtar A (1939) Een serologisch afwijkende leptospira-stam uit der nier eener vleermuis. Gen Tschr Ned Ind 79: 226–231. [Google Scholar]
- 22. Alston JM, Broom JC (1958) Leptospirosis in Man and Animals. E. & S. Livingstone; Edinburgh, UK. [Google Scholar]
- 23. Van Peenen PFD, Light RH, Sulianti Saroso J (1971) Leptospirosis in wild mammals of Indonesia—Recent surveys. Se Asian J Trop Med 2: 496–502. [PubMed] [Google Scholar]
- 24. Thayaparan S, Robertson IAN, Amraan F, Ut LSU, Abdullah MT (2013) Serological Prevalence of Leptospiral Infection in Wildlife in Sarawak, Malaysia. Borneo J Res Sc Technol 2: 71–74. [Google Scholar]
- 25. Mgode GF, Mbugi HA, Mhamphi GG, Ndanga D, Nkwama EL (2014) Seroprevalence of Leptospira infection in bats roosting in human settlements in Morogoro municipality in Tanzania. Tanz J Health Res 16: 1–7. [DOI] [PubMed] [Google Scholar]
- 26. Everard CR, Fraser-Chanpong GM, Bhagwandin LJ, Race MW, James AC (1983) Leptospires in wildlife from Trinidad and Grenada. J Wildlife Dis 19: 192–199. [DOI] [PubMed] [Google Scholar]
- 27. Sebek Z, Sixl W, Reinthaler F, Valova M, Scneeweiss W, et al. (1989) Results of serological examination for leptospirosis of domestic and wild animals in the Upper Nile province (Sudan). J Hyg Epid Microb Im 33: 337–345. [PubMed] [Google Scholar]
- 28. Ogawa H, Koizumi N, Ohnuma A, Mutemwa A, Hang BM, et al. (2015) Molecular epidemiology of pathogenic Leptospira spp. in the straw-colored fruit bat (Eidolon helvum) migrating to Zambia from the Democratic Republic of Congo. Infect Genet Evol 32: 143–147. 10.1016/j.meegid.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harkin KR, Hays M, Davis R, Moore M (2014) Use of PCR to Identify Leptospira in Kidneys of Big Brown Bats (Eptesicus fuscus) in Kansas and Nebraska, USA. J Wildlife Dis 50: 651–654. [DOI] [PubMed] [Google Scholar]
- 30. Jobbins SE, Alexander KA (2015) Evidence of Leptospira sp. infection among a diversity of African wildlife species : beyond the usual suspects. Trans R Soc Trop Med Hyg: 349–351. 10.1093/trstmh/trv007 [DOI] [PubMed] [Google Scholar]
- 31. Fennestad KL, Borg-Petersen C (1972) Leptospirosis in danish wild mammals. J Wildlife Dis 8: 343–351. [DOI] [PubMed] [Google Scholar]
- 32. Tagi-Zade TA, Mardanly AS, Akhmedov IB, Alekperov FP, Gasanov SN (1970) Examination of bats for leptospirosis in the territory of Azerbaijan SSR. Zhurnal Mikrobiol Epidemiol i Immunobiol 9: 118–121. [PubMed] [Google Scholar]
- 33. Lei BR, Olival KJ (2014) Contrasting patterns in mammal-bacteria coevolution: Bartonella and Leptospira in bats and rodents. PLoS Neglect Trop 8: e2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hayman DTS, Bowen RA, Cryan PM, McCracken GF, O’Shea TJ, et al. (2013) Ecology of zoonotic infectious diseases in bats: current knowledge and future directions. Zoonoses Public Hlth 60: 2–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, et al. (2006) Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. P Natl Acad Sci USA 103: 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cosson J-F, Picardeau M, Mielcarek M, Tatard C, Chaval Y, et al. (2014) Epidemiology of Leptospira Transmitted by Rodents in Southeast Asia. PLoS Neglect Trop 8: e2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, et al. (2012) Seasonal Pulses of Marburg Virus Circulation in Juvenile Rousettus aegyptiacus Bats Coincide with Periods of Increased Risk of Human Infection. PLoS Pathog 8: e1002877 10.1371/journal.ppat.1002877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, et al. (2015) Ecological dynamics of emerging bat virus spillover. P Roy Soc B-Biol Sci 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Field H, Crameri G, Kung NY, Wang LF (2012) Ecological aspects of hendra virus. Curr Topics Microbiol 359: 11–23. [DOI] [PubMed] [Google Scholar]
- 40.Khan SU, Islam MA, Rahman MZ, Island A, Sazzad HMS, et al. (2013) Nipah virus shedding among Pteropus bats in the context of human outbreak in Bangladesh, 2012. ASTMH 62nd Annual Meeting, 13–17 November, Washington, DC.
- 41. Monahan AM, Callanan JJ, Nally JE (2009) Review paper: Host-pathogen interactions in the kidney during chronic leptospirosis. Vet Pathol 46: 792–799. 10.1354/vp.08-VP-0265-N-REV [DOI] [PubMed] [Google Scholar]
- 42. Vashi NA, Reddy P, Wayne DB, Sabin B (2010) Bat-associated leptospirosis. J Gen Intern Med 25: 162–164. 10.1007/s11606-009-1210-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Costa F, Wunder EA, Oliveira D De, Bisht V (2015) Patterns in Leptospira Shedding in Norway Rats (Rattus norvegicus) from Brazilian Slum Communities at High Risk of Disease Transmission. PLoS Neglect Trop 9: e0003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilkinson DA, Mélade J, Dietrich M, Ramasindrazana B, Soarimalala V, et al. (2014) Highly Diverse Morbillivirus-Related Paramyxoviruses in Wild Fauna of the Southwestern Indian Ocean Islands: Evidence of Exchange between Introduced and Endemic Small Mammals. J Virol 88: 8268–8277. 10.1128/JVI.01211-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, et al. (2012) Bats host major mammalian paramyxoviruses. Nat Comm 3: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]