Abstract
Large-scale genomic characterization of squamous cell lung cancers (SQCLC) has revealed several putative oncogenic drivers. There are, however, little data to suggest that these alterations have clinical relevance. We performed comprehensive genomic profiling of 79 stage IV SQCLCs (including next-generation sequencing) and analyzed differences in the clinical characteristics of two major SQCLC subtypes: FGFR1 amplified and PI3K aberrant. Patients with PI3K aberrant tumors had aggressive disease marked by worse survival (median OS 8.6 vs. 19.1 mo, p<0.001), higher metastatic burden (>3 organs 18% vs. 3%, p=0.025), and greater incidence of brain metastases (27% vs. 0% in others, p<0.001). We performed whole-exome and RNA sequencing on paired brain metastases and primary lung cancers to elucidate the metastatic process to brain. SQCLC primaries that gave rise to brain metastases exhibited truncal PTEN loss. SQCLC brain metastases exhibited a high degree of genetic heterogeneity and evidence of clonal differences between their primary sites.
Keywords: squamous cell lung cancer, FGFR1, PI3K, brain metastasis
Introduction
Squamous cell lung cancers (SQCLC) account for 20% of all lung cancers. This equates to 350,000 patients diagnosed with this disease worldwide every year. Half of these patients present with metastatic disease, and most will not be cured. These outcomes reflect the underlying aggressiveness of the disease and the paucity of new advances in the field, where the last substantial improvement in survival came with the introduction of cisplatin in the 1980s (1).
2004 marked a divergence in the management and conceptualization of the two most common histologic subtypes of lung cancer: SQCLCs and lung adenocarcinomas. The identification of EGFR tyrosine kinase inhibitor (TKI)-sensitizing mutations by three groups that year presaged the discovery of other druggable oncogenic events, primarily in adenocarcinomas, and including mutations in HER2 and BRAF and oncogenic rearrangements of ALK, RET, and ROS1. The breadth of these discoveries led to a rethinking of adenocarcinomas as a set of related but distinct oncogene-addicted cancers, each of which could be treated with a targeted therapy (2).
The majority of these events were found to not occur with any appreciable frequency in SQCLCs. Comprehensive molecular analyses of resected SQCLC tumors by The Cancer Genome Atlas (TCGA) Consortium did, however, identify somatic variants, copy number alterations, and gene expression changes that circumscribe genetic subsets of SQCLCs (3). Prior work showed that some of these key targets were both oncogenic and responsive to pharmacologic inhibitors, including FGFR1 amplification (4), DDR2 mutations (5), and hyperactivating PI3K pathway alterations (6, 7). In aggregate, these potential therapeutic targets are thought to occur in at least 50% of SQCLC cases (3, 8).
The TCGA analysis involved resection specimens from early stage (I-III) patients to ensure a precise histologic diagnosis and sufficient tumor content. The applicability of the findings to stage IV disease is, as a result, unknown. In addition, there were limited clinical data available for analysis in TCGA. As such, the clinical sequelae of biologic differences could not be assessed. These differences can provide necessary prognostic information to patients and the hypothetical groundwork to unravel the complex manifestations of these cancers.
In light of these gaps in our knowledge, we sought to identify differences in the clinical characteristics of patients with stage IV squamous cell lung cancers whose tumors were comprehensively tested for oncogenic drivers and to shed light on the biologic underpinnings of these differences. We focused, as the starting point for our study, on overall survival and patterns of metastasis in two distinct molecularly defined groups – FGFR1 and PI3K, both high-frequency pathways scored as significantly altered by TCGA and for which pre-clinical data supports potential roles as actionable drivers (4, 6, 7).
Results
Squamous cell lung cancer mutation analysis program (SQ-MAP) results
Seventy-nine patients with stage IV disease had sufficient archived tumor material for FISH for FGFR1 amplification, IHC for PTEN loss, Sequenom genotyping for hotspot mutations in eight oncogenes including PIK3CA, and exon capture/next-generation sequencing of 279 oncogenes and tumor suppressors by IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets, see Methods). For the purposes of this analysis, upstream aberrations in PI3K were defined as PIK3CA and PTEN mutations and PTEN loss, all relatively high frequency events in the TCGA analysis that have been shown to predict for response to pharmacologic inhibition in vitro or in vivo (7) and which are the exploratory predictive biomarkers used in a number of clinical trials (NCT01297491, NCT01655225).
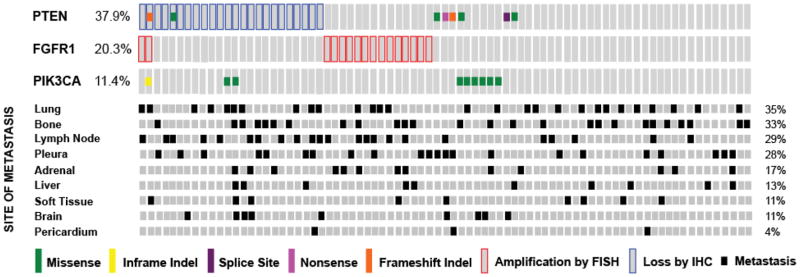
As shown in Figure 1, 20% of patients had FGFR1 amplification (95% CI: 12–31%), 29% had complete loss of PTEN (95% CI: 20–40%), 10% had PTEN mutations (95% CI: 5–19%), and 11% had PIK3CA mutations (95% CI: 6–20%). Thirteen percent (2/16) of FGFR1 amplified tumors had overlapping PTEN loss. Seventeen percent of tumors that had PTEN loss by IHC also had concurrent PTEN mutations, PTEN loss, or PIK3CA mutations. In total, 61% (95% CI: 49–71%) of patients had tumors that bore an aberration in either FGFR1 or the upstream PI3K pathway.
Figure 1.
Oncoprint depicting the frequency of FGFR1 amplifications and PI3K pathway aberrations (PIK3CA mutations, PTEN mutations, PTEN loss) in 79 stage IV SQCLC tumors. Percentages indicate the combined frequency of somatic mutations and copy number alterations for each listed gene. Sites of metastases for each case are shown below the oncoprint.
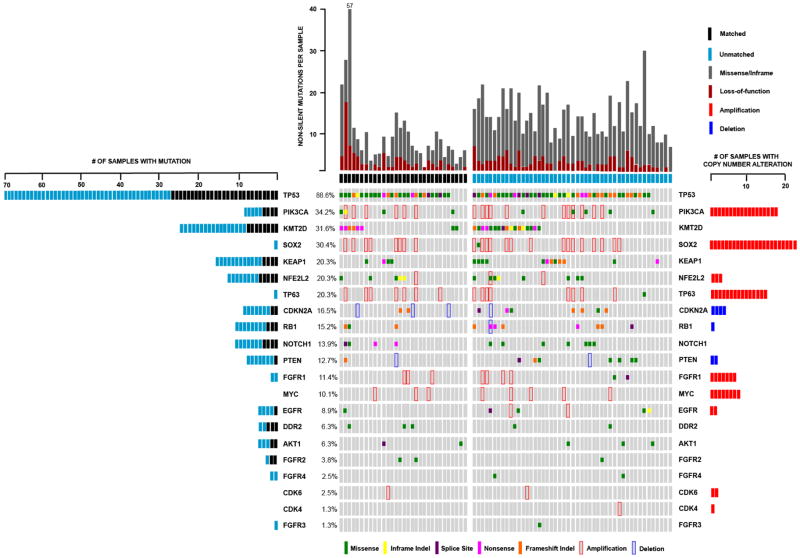
IMPACT results of selected genes, including those found significantly mutated by TCGA, are shown in Figure 2. Genes mutated in multiple patients included TP53 (89%), CDKN2A (11%), NFE2L2 (17%), and KEAP1 (20%). NFE2L2 and KEAP1 mutations appeared in non-overlapping cases, as expected (3). DDR2 mutations were found in 6% of patients (95% CI = 3–14%). Gene-specific copy number alterations were found at relatively high frequencies in chromosome 3 (SOX2, PIK3CA). A complete list of somatic mutations and copy number alterations detected by IMPACT is in Supplementary Table 1. A comparison of the frequency of selected genes found by SQ-MAP vs. TCGA is in Supplementary Table 2. A number of gene frequencies were significantly different by Fisher’s exact test but lost significance when adjusted for multiple testing effects.
Figure 2.
Summary of somatic mutations and copy number alterations in 79 stage IV SQCLC tumors tested by targeted exon sequencing through IMPACT.
Patient characteristics by genotype
Patient characteristics are shown in Table 1. Characteristics were by and large the same. There were no significant differences in age, sex, smoking history, Karnofsky performance status, or treatment with a platinum doublet by genotype. There were differences in the proportion of patients treated with genotype-directed targeted therapies. Thirty-one percent of patients with FGFR1 amplifications were treated with AZD4547, a specific and potent FGFR1-3 tyrosine kinase inhibitor. One patient with a PI3K alteration (PTEN loss) was treated with a PI3Kα inhibitor (buparlisib). No patient had a response (either partial or complete) to targeted therapy.
Table 1.
Clinical characteristics of 79 Stage IV SQCLC patients.
| PI3K (N=33) | FGFR1 (N=16) | Other (N=30) | p (PI3K vs. FGFR1) | p (PI3K vs. Other) | p (FGFR1 vs. Other) | |
|---|---|---|---|---|---|---|
| Age | ||||||
| Median (range) | 68 (47–85) | 73 (48–82) | 69 (45–87) | 0.10 | 0.36 | 0.42 |
| Sex | ||||||
| Female | 48% | 44% | 30% | >0.95 | 0.20 | 0.52 |
| Smoking history (pack years) | ||||||
| Median (range) | 40 (0–90) | 43 (1–100) | 27 (0–120) | 0.63 | 0.12 | 0.12 |
| Current or former | 94% | 100% | 80% | |||
| Never | 6% | 0% | 20% | >0.95 | 0.14 | 0.08 |
| KPS | ||||||
| ≤70% | 36% | 38% | 43% | >0.95 | 0.79 | 0.052 |
| Prior lines of therapy | ||||||
| Median (range) | 1 (0–4) | 2 (0–4) | 2 (0–4) | 0.19 | 0.002 | 0.15 |
| Treated with platinum doublet | ||||||
| Proportion (%) | 21/27 (78%) | 9/13 (69%) | 20/25 (80%) | 0.70 | >0.95 | 0.069 |
| Treated with targeted therapy | ||||||
| Proportion (%) | 1/27 (4%) | 4/13 (31%) | 0 | 0.03 | -- | -- |
| Number of metastatic sites | ||||||
| >3 | 6 (18%) | 1 (6%) | 0 | 0.40 | 0.025 | 0.35 |
Overall survival by genotype
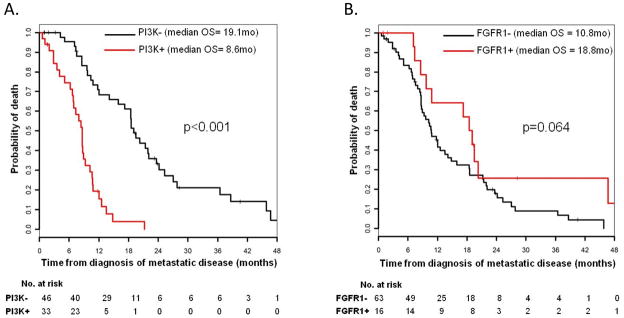
Median overall survivals (OS) were, on the other hand, significantly different among the genotypes: FGFR1 amplified = 18.8 mo (95% CI: 10.8-NR); PI3K aberrant = 8.6 mo (95% CI: 6.9–10.7); Others = 21.3 mo (95% CI: 15.9–27). Patients with PI3K aberrations had significantly worse OS than those without these aberrations (median OS 8.6 mo vs. 19.1 mo, p<0.001) (Figure 3A). OS was also worse compared to patients with FGFR1 amplification (median OS 8.6 mo vs. 18.8 mo, p<0.001). Patients with FGFR1 amplification had a trend towards improved OS vs. those without FGFR1 amplification (median OS 18.8 mo vs. 10.8 mo, p=0.064) (Figure 3B). Multivariate analysis using genotype, age, sex, performance status, and number of somatic changes as an additional genomic covariate (mutations + copy number) similarly demonstrated significantly worse OS for patients harboring a PI3K aberration (HR for death=5.6, 95% CI:2.8–11.2, p<0.0001). There was no significant association between OS and number of somatic changes as a continuous variable. Exclusion of the 2 patients who had overlapping FGFR1 amplification and PTEN loss or reassignment of them to the different genotype categories did not affect our results.
Figure 3.
Figure 3A: Kaplan Meier survival curves for OS in Stage IV SQCLC patients with and without PI3K pathway aberrations.
Figure 3B: Kaplan Meier survival curves for OS in Stage IV SQCLC patients with and without FGFR1 amplification.
Because PIK3CA amplification was found, post-hoc, in a substantial number of patients (23%), we also performed an overall survival analysis of this group of patients vs. those without PI3K pathway alterations. Median OS for those with PIK3CA amplified tumors vs. those without a PI3K pathway aberration was 10.8 mo (95% CI:8.6–19.6) vs. 20.2 mo (95% CI:14.1–24.2), p=0.15. There was, however, overlap with other PI3K aberrations: 40% of patients whose tumors bore amplified PIK3CA also had tumors that exhibited complete loss of PTEN.
Metastatic sites by genotype
Sites of metastatic disease are listed in Table 2. Brain metastases were the only site which varied significantly by genotype, occurring in 27% (N=9/33) of those with PI3K aberrations and 0% of all others (N=0/46) (p<0.001). Overall, brain metastases were relatively uncommon, occurring in 11% of the entire cohort. In addition, patients with PI3K aberrations exhibited a significantly higher burden of metastatic disease than other patients (>3 organ sites involved: 18% vs. 3%, p=0.025). Exclusion of the 2 patients who had overlapping FGFR1 amplification and PTEN loss or reassignment of them to the different genotype categories did not affect our results.
Table 2.
Sites of metastasis by genotype
| Site | PI3K (n=33) | % | p (vs. all others) | FGFR1 (n=16) | % | p (vs. all others) | Other (n=30) | % | Total (n=79) | % |
|---|---|---|---|---|---|---|---|---|---|---|
| Lung | 11 | 33 | 0.81 | 7 | 44 | 0.56 | 10 | 33 | 28 | 35 |
| Brain | 9 | 27 | <0.001 | 0 | 0 | 0.19 | 0 | 0 | 9 | 11 |
| Bone | 12 | 36 | 0.63 | 3 | 19 | 0.24 | 11 | 37 | 26 | 33 |
| Extrathoracic lymph node | 12 | 36 | 0.31 | 6 | 38 | 0.54 | 5 | 17 | 23 | 29 |
| Pleura | 6 | 18 | 0.13 | 5 | 31 | 0.76 | 11 | 37 | 22 | 28 |
| Liver | 4 | 12 | >0.95 | 1 | 6 | 0.68 | 5 | 17 | 10 | 13 |
| Adrenal | 7 | 21 | 0.37 | 3 | 19 | 0.72 | 3 | 10 | 13 | 16 |
| Soft tissue | 4 | 12 | >0.95 | 1 | 6 | 0.68 | 4 | 13 | 9 | 11 |
| Pericardium | 1 | 3 | >0.95 | 1 | 6 | 0.50 | 1 | 3 | 3 | 4 |
| Pancreas | 0 | 0 | >0.95 | 1 | 6 | 0.20 | 0 | 0 | 1 | 1 |
SQCLC brain metastases also exhibit PTEN loss
In an effort to better understand the biology underpinning the higher incidence of brain metastases in SQCLC patients harboring PI3K aberrations, we analyzed 6 surgically resected SQCLC brain metastases that had frozen tissue available for testing by whole exome sequencing, RNA sequencing, and IHC. Four of these patients had matched archived FFPE samples of their primary lung cancers. The clinical characteristics and treatment course of these patients is in Supplementary Table 3.
PTEN IHC and whole exome sequencing of both the primary lung tumors and brain metastases demonstrated complete loss of PTEN in 67% of cases (N=4/6). The other 2 cases exhibited weak PTEN staining (1+). There were no differences in PTEN loss between primary and metastasis pairs. A PIK3CA E542K mutation was present in 1 of the 6 patient brain metastasis samples (PP5), coincident with PTEN loss. A PIK3CG D521Y mutation was present in both the primary lung tumor and brain metastasis in 1 of 6 samples (PP4). Whole exome sequencing detected heterozygous loss of PTEN in all of the brain metastases. No other functionally validated mutations within the canonical PI3K pathway in either the primary lung tumors or brain metastases were found. No other common oncogenic drivers (e.g. EGFR, BRAF, KRAS, NRAS, HER2, MEK, DDR2, NFE2L2, KEAP1, FGFR1-4) were found. FISH demonstrated 8p11 amplification in PP4, but sequencing did not demonstrate focal amplification of FGFR1 in either the primary lung tumor or brain metastasis. The mutational burden and spectrum (missense, silent, non-coding, truncating) were similar in all cases save for the primary lung tumor from PP3 which showed a mutation burden roughly 10-fold that of the other samples. This was the only tumor biopsied after treatment with radiation (concurrent cisplatin + etoposide and radiation therapy).
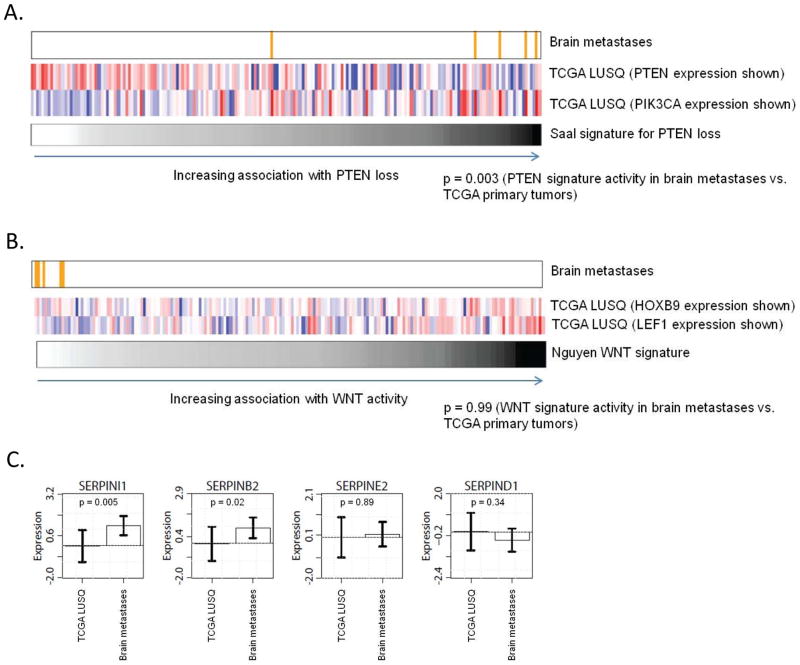
To further validate the IHC-based PTEN loss and heterozygous loss of PTEN signals, we performed RNA sequencing on each of the brain metastases and compared the gene expression results to an existing PTEN loss RNA signature derived by Saal et al (9). This analysis yielded PTEN signature scores in each brain metastasis commensurate with PTEN loss. These scores were significantly different from those derived from TCGA expression data for resected early stage primary lung cancers (p=0.003, Figure 4A).
Figure 4.
Figure 4A: PTEN loss signature score in resected SQCLC brain metastases. Values plotted to the right are more strongly correlated with PTEN loss. Expression of PIK3CA and PTEN from individual samples in the TCGA SQCLC analysis are plotted for reference, confirming a trend towards decreased PTEN expression and increased PIK3CA expression and signature scores consistent with PTEN loss. PTEN signature activities in the brain metastases were compared to the signatures in the TCGA primary tumors using a one-sided Wilcoxon rank-sum test.
Figure 4B: Pro-metastatic WNT gene signature score in resected SQCLC brain metatases. Values plotted to the right are more strongly correlated with WNT activity. Expression of HOXB9 and LEF1, key promoters of the brain metastatic process in lung adenocarcinomas, from individual samples in the TCGA SQCLC analysis are plotted as an SQCLC-specific reference. WNT signature activities in the brain metastases were compared to the signatures in the TCGA primary tumors using a one-sided Wilcoxon rank-sum test.
Figure 4C: Plasminogen activator (PA) inhibitor serpin gene expression levels from the TCGA SQCLC analysis and SQCLC brain metastases. There is no significant increase in PA serpin expression in the brain metastases vs. early stage tumors from TCGA.
SQCLC brain metastases are not marked by increased WNT signaling or serpin overexpression
Two mechanisms underlying the development of brain metastases in lung adenocarcinomas have been previously published, including activation of WNT signaling (with HOXB9 and LEF1 expression strongly correlated with metastatic potential) and upregulation of plasminogen activator (PA) inhibitory serpins (10, 11). Gene expression analysis of the SQCLC brain metastases did not show an association with increased WNT signaling (Figure 4B) or PA serpin overexpression (Figure 4C). We did confirm, as an internal control, that there was an association between HOXB9 and LEF1 expression and WNT signature activity in the TCGA cases.
SQCLC brain metastases and lung primaries are highly genetically divergent
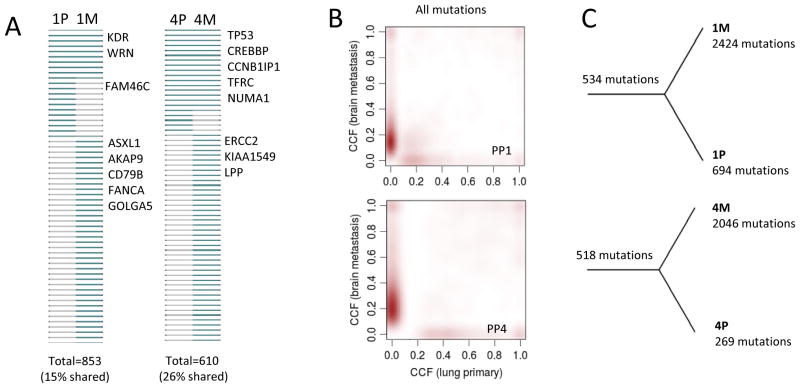
In order to better understand the degree and manner of divergence that might exist between brain metastases and their lung primaries, we performed whole-exome sequencing of paired lung primaries and brain metastases from four SQCLC patients (PP1-4) and whole-exome sequencing of an unpaired brain metastasis from one other (PP5). Tumor and normal DNA were sequenced at 100x coverage depth. In the PP1 paired set, 3,652 somatic mutations were detected of which 853 were non-silent. We classified each non-silent mutation as shared (present in both the lung primary and the brain metastasis) or unique (present in one, not the other). Of the 853 non-silent mutations in the PP1 pair, only 15% were shared between the lung primary and brain metastasis (Figure 5A). In the PP4 paired set, a total of 2,833 mutations were detected of which 610 were non-silent. Among these non-silent mutations, 26% were shared between the lung primary and brain metastasis. PP3 was excluded from paired analysis as a result of the hyper-mutated phenotype of the primary lung tumor; PP2 did not have a matched normal counterpart for the paired analysis.
Figure 5.
Patterns of shared and unique somatic mutations in pairs of lung cancer primary and brain metastasis tumors. A. Panel plots show non-silent mutations (present-blue, absent-gray) in the paired primary and brain metastasis from PP1 (1P, 1M) and PP4 (4P, 4M). Known cancer genes are annotated on the right side of each panel plot. The total number of non-silent mutations (percent shared mutations) is indicated at the bottom of each panel. B. Two-dimensional plots of cancer cell fraction (CCF) estimated from the variant allele frequencies of all mutations, adjusted for tumor purity and local copy number states. Increasing red intensity indicates higher mutation density. C. Phylogenetic trees derived from all mutations by neighbor-joining method.
To explore evidence of clonal subpopulations in the lung primaries and brain metastases, we estimated the cancer cell fraction for each mutation as described in the Methods section. The dominant clusters of mutations private to the lung primaries and brain metastases, respectively, showed clear evidence of subclonality, indicating a high degree of intra-tumor heterogeneity (Figure 5B, dense clouds along the x-axes and y-axes. Two independent methods of calculating cancer cell fractions (pyClone and FACETS) were used, and the results were highly concordant. These patterns in PP1 and PP4 were also seen when the analysis was restricted to mutations with ≥80x depth of coverage. Finally, construction of phylogenetic trees for all somatic mutations detected in the PP1 and PP4 paired sets yielded evidence of a branched evolutionary pattern (Figure 5C). Genes that were clonal in the brain metastases but subclonal in the lung primaries are shown in Supplementary Table 4.
Analysis of the copy number data from the paired sets also yielded evidence of great divergence between the lung primaries and brain metatases. In the PP1 paired set, only 24% of the copy number alterations were shared (Supplementary Figure 1). Major alterations observed in the primary tumor from PP1 included a gain in 7p and loss in 6q that were absent in the brain metastasis (Supplementary Figure 2A). A prominent gain of 5p and loss of 5q was observed in all of the brain metastases analyzed (Supplementary Figures 2B–2D). By contrast, not all of the primary tumors carried these aberrations; in PP1, the 5p gain was not present in the lung primary. FISH confirmed the presence of multiple copies of 5p in nearly all tumor cells analyzed from the brain metastases, with most harboring >6 copies (Supplementary Figure 3). This degree of aneuploidy was not seen in the PP1 primary lung tumor, which was the only primary tumor with sufficient material leftover for 5p assessment by FISH (disomy in 80% of cells).
Discussion
Our data demonstrate that somatic changes linked to activation of PI3K signaling in patients with stage IV SQCLCs are associated with a unique natural history of disease, reinforcing the preclinical data that show that these changes can engender a distinct biological program that can be targeted for therapeutic gain.
The frequency of PI3K and FGFR1 pathway changes in our stage IV dataset are similar to those reported by TCGA. TCGA found upstream changes in PI3K signaling (PIK3CA mutations, PTEN mutations) in 21% of the resected early stage cancers and FGFR1 amplification in 17% (3). We found PIK3CA and PTEN mutations in 21% of patients and FGFR1 amplification in 20%. In addition, we found PTEN loss by IHC in 29% of stage IV cases. A comparable frequency is unavailable from TCGA, as IHC was not performed, although Soria et al. found PTEN loss by IHC in 24% of early stage SQCLCs (12). The discrepancy between the frequencies of PTEN protein loss and genetic alterations such as copy number loss or PTEN mutations suggests that transcriptional or post-transcriptional mechanisms are likely at play, such as promoter hypermethylation (12).
While the mutation frequencies of some genes detected by IMPACT were significantly different from TCGA by Fisher’s exact test, none reached significance when adjusted for multiple testing. An example is AKT1, which was mutated in 6% of patients in our series and 0.6% in TCGA (Fisher’s exact test p=0.009, adjusted p=1). These mutations occurred in either the PH domain or kinase domain. Redistributing these patients into the PI3K aberrant group (only two of whom did not have any other upstream PI3K pathway alteration and so were not already in this category) did not alter our results. A larger sample size will be required to determine whether these differences in frequency are indeed significant, as some variation is expected from the greater depth of coverage associated with targeted exon sequencing. Interestingly, many of the genes with the greatest differences in frequencies are involved in transcriptional regulation rather than signal transduction. These are, to our knowledge, the first published next-generation sequencing results from patients with metastatic SQCLCs.
Several groups have reported survival differences associated with FGFR1 amplification in early stage SQCLCs with divergent results (13–16). While pre-clinical modeling has clearly shown that FGFR1 amplified cell lines and patient-derived xenografts do respond to FGFR1 inhibition (4, 17) and preliminary data from clinical trials has shown modest activity of selective FGFR1 inhibitors in patients with FGFR1 amplified SQCLCs (18, 19), responses have not been uniform and amplification does not translate to increased protein expression in many cases (20). We therefore note that within this genomic subgroup, heterogeneity in the activation of FGFR1 signaling could exist that FGFR1 amplification by itself might not adequately capture. This may be a confounder in our analysis of this genotype. As this is the first study to address FGFR1 amplified prognosis in metastatic disease, a direct comparison with prior clinical data, which is exclusively in early stage disease, is not possible.
To date, no group has yet reported clinical outcomes for patients with PI3K-aberrant SQCLCs. We showed, for the first time, that SQCLC patients with upstream PI3K aberrations have a significantly poorer survival compared to others. This holds true when taking into account other clinical and genomic variables on multivariate analysis. There were no differences in the number of lines of therapies between molecular subgroups. More patients received a targeted therapy in the FGFR1 amplified group, although the preliminary clinical activity of these agents is poor and this difference is unlikely to have favorably affected survival as a result (18, 19). The magnitude of the survival difference for patients with PI3K aberrations is substantial (HR for death=5.6) and exceeds any therapeutically-driven improvement in survival to date, arguing that stratification by this genotype category should be considered and PI3K status reported in future studies.
Consistent with this more aggressive phenotype, SQCLC patients with PI3K-abberant cancers had a surprisingly higher burden of metastatic disease in general and a higher incidence of brain metastases specifically. At 27%, this incidence approaches that seen in lung adenocarcinomas (21). This observation was reproduced in the resected SQCLC brain metastases we analyzed, where two-thirds exhibited complete loss of PTEN protein, all exhibited heterozygous loss of PTEN, and all had a pattern of gene expression consistent with PTEN loss.
With regard to functional implications, the retention of PTEN loss and PIK3CA mutations between the primary and metastatic sites suggests that PI3K signaling activation is necessary but not sufficient for the metastatic process to brain. As for evidence implicating PI3K signaling in the development of brain metastases, this is by and large correlative. Saal et al. demonstrated a correlation between PTEN loss and the development of distant metastases in breast cancer (9). Data from melanoma suggest that hyperactivation of AKT and PTEN loss are more common in brain metastases than extracranial metastases (22). Similar results correlating AKT and PTEN loss were found in an analysis of 52 breast cancer brain metastases (25% exhibited PTEN loss), though the lack of comparison to primary tumors that did not harbor brain metastases limit the interpretation of these data (23). Single-nucleotide polymorphism (SNP) analysis of AKT1, AKT2, PIK3CA, PTEN, and FRAP1 from predominantly lung adenocarcinoma (68%) primary tumors yielded an association between certain SNPs in AKT1, AKT2, and PIK3CA and risk of brain metastasis (24). The functional relevance of these SNPs is unknown, however. The pro-brain metastatic program triggered by PI3K pathway activation remains, as a result, undefined, although PTEN loss in particular is known to facilitate integrin-mediated cell spreading through impairment of cytoskeletal turnover (25). Other mechanisms that are important in the development of metastases in lung adenocarcinomas, including upregulated WNT signaling and overexpression of serpins, were not present in our series, suggesting that the mechanism that drives the metastatic process to brain differs by histology (10, 11). Potential pro-metastatic targets of interest from our study include γ and β clustered protocadherins on 5q31 (PCDHGA4, PCDHGA10, PCDHGB2, and PCDHB6) that are known to play a role in cell adhesion (26), which were clonal in the brain metastases and which were expressed at low levels relative to the TCGA primary lung dataset (Supplementary Figure 4).
Intriguingly, while there were no clonal differences in PTEN loss or PIK3CA mutations between the primary lung cancer and brain metastasis pairs, deeper genomic analysis did demonstrate evidence of clonal heterogeneity. The low degree of shared events (mutation and copy number) between the paired samples and the presence of multiple subclonal populations within each tumor sample indicates a high degree of genetic heterogeneity both within and between the lung primaries and the brain metastases analyzed. Thus, the originating brain metastatic clone may very well be different from the predominant primary lung cancer clone. Indeed, two recent studies of early stage NSCLCs reported a significant amount of mutational and copy number heterogeneity by multi-region sequencing in surgical samples of early stage patients (27, 28). Our data shows evidence of even greater heterogeneity in the metastatic setting, with as little as 15% of mutations shared between sites of disease, substantially less than the 76% of mutations shared across regions of primary lung tumors in early stage disease (27). Notwithstanding blood-brain barrier penetration by drugs, this offers an additional explanation- one which has to potential to be better elucidated- as to why metastatic brain disease is often difficult to treat.
In conclusion, our data provide evidence of a unique biological program associated with aberrations known to activate PI3K signaling in SQCLCs. This is the first such evidence of its kind in this disease, and simultaneously validates the analysis performed by TCGA in resection specimens and supports the current efforts to target this pathway pharmacologically. While it follows that targeting PI3K signaling may help to prevent or treat brain metastases in patients with SQCLCs, functional studies will be needed to determine the exact role of this pathway in the metastatic process. Our analysis of paired SQCLC primary lung and brain metastasis samples revealed the presence of copy number differences and subclonal somatic mutations between the lung primary and brain metastasis tumors as well as a low proportion of shared events between the two, indicating a clonally divergent evolutionary process and the requirement for multi-region sampling to track the origin of the metastatic subclone and to identify other alterations that contribute to the metastatic process to brain.
Materials and Methods
Study Design and Patients
Patients with pathologically-verified SQCLC with available tissue were approached for informed consent to an IRB/Privacy Board-approved protocol for testing of archived FFPE tumor specimens through a prospective testing platform termed Squamous Cell Lung Cancer Mutation Analysis Program (SQ-MAP). This study was conducted in accordance with the Declaration of Helsinki. There were no other eligibility criteria for this. While no stage restrictions were present for testing, the current analysis is limited to patients with stage IV disease. Seventy nine patients were identified for this study who had tumor sufficient for FISH for FGFR1 amplification, mass spectrometry-based genotyping (Sequenom™) for PIK3CA mutations (exons 9 and 20), immunohistochemistry (IHC) for PTEN loss, and targeted-exon sequencing for a large panel of mutations and copy number changes as described below.
Resected SQCLC brain lesions with formalin-fixed paraffin-embedded (FFPE) tumor blocks and frozen specimens as well as their paired primary lung tumor FFPE biopsies/resections were identified through an IRB/Privacy-Board Approved Biospecimen Utilization and Waiver of Authorization.
Pathologic verification
All pathologic samples were reviewed by thoracic pathologists at our institution using light microscopy and immunohistochemistry for TTF-1 and p63 (ΔN isoform) and selectively for Napsin A and mucicarmine as described previously (29).
FISH for FGFR1 amplification and chromosome 5p gain
FGFR1 amplification was determined through the use of an FGFR1/CEN8 dual color FISH probe from Zytovision (#Z-2072-200) in a CLIA laboratory. Four micron sections were generated from FFPE blocks for testing. Tumor specimens were deparaffinized in xylene and dehydrated in ethanol. FISH was performed according to manufacturer instructions with minor modifications. FISH analysis and signal capture was performed on fluorescence microscopes (AXIO, Zeiss) coupled with the ISIS FISH Imaging System (Metasystems). A minimum of fifty interphase nuclei from each tumor specimen was scored. An FGFR1/CEN8 ratio ≥ 2 in ≥ 10% tumor cells was used as the criterion to define FGFR1 amplification.
Gain of chromosome 5p was verified through the use of a 5p13 FISH probe, RP11-767K20, from Empire Genomics. The BAC clone RP11-767K20 was labeled by TAMRA (red signal). Four micron sections were generated from FFPE blocks for testing. FISH analysis and signal capture was performed on fluorescence microscopes (AXIO, Zeiss) coupled with the ISIS FISH Imaging System (Metasystems). A minimum of 100 interphase nuclei from each tumor specimen was scored. There was sufficient archived material to assess chromosome 5p status in the PP1, PP2, PP3, and PP4 brain metastases as well as the PP1 primary lung tumor.
PTEN loss
IHC for PTEN expression was performed using a mouse monoclonal antibody (Cell Signaling™ clone 138G6) on archival FFPE tissue sections in a CLIA laboratory. Heat induced epitope retrieval at 97–99C for 40 minutes was accomplished using Target Retrieval Solution pH 9.0 (DAKO) followed by incubation with primary PTEN antibody (dilution 1:50). Diaminobenzidin was used as the chromogen. Positive cytoplasmic and/or nuclear staining of blood vessel and stromal cells was used as an internal positive control. H-score was calculated according to the following formula: H-Score = % cells staining (0–100%) X intensity (range from 1 to 3), where an H-score of 0 corresponded to no staining and a score of 300 to maximum staining intensity in the entire tumor. Complete loss was defined as an absence of PTEN immunoreactivity in tumor cells with preserved reactivity in internal control cells (blood vessels and/or stromal cells).
Mutation analysis by Sequenom
Tumors were genotyped by the Sequenom Mass ARRAY™ system (Sequenom Inc., San Diego, CA). Briefly, samples were tested in duplicate using a series of multiplexed assays designed to interrogate hot-spot mutations in 8 oncogenes: EGFR, KRAS, BRAF, PIK3CA, NRAS, ERBB2/HER2, MAP2K1/MEK1 and AKT1. A total of 92 non-synonymous mutations were tested in 6 multiplex reactions. Genomic DNA amplification and single base pair extension steps were performed using specific primers designed with the Sequenom Assay Designer v3.1 software. The allele-specific single base extension products were then quantitatively analyzed using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) on the Sequenom MassArray Spectrometer. All automated mutation calls were confirmed by manual (visual) review of the spectra.
Next-generation sequencing through IMPACT
Genomic alterations in key cancer-associated genes were also profiled using exon capture by hybridization followed by next-generation sequencing (30). This assay, termed IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), encompasses all protein-coding exons and select introns of 279 cancer genes, and was the basis for a clinical version of this assay, termed MSK-IMPACT, for which methods and validation data are presented in detail (31). Genes were selected to include commonly implicated oncogenes, tumor suppressor genes, and components of pathways deemed actionable by current targeted therapies (Supplementary Table 5). We sheared DNA isolated from FFPE tissue for 6 minutes on a Covaris E220 instrument, prepared barcoded sequence libraries (New England Biolabs, Kapa Biosystems), and performed exon capture on barcoded pools by hybridization (Nimblegen SeqCap) using custom oligonucleotides. The amount of input DNA ranged from 112 to 500 ng (mean = 250 ng). Barcoded libraries were pooled at 100 ng per sample and used in a single exon capture reaction as previously described (32). To prevent off-target hybridization, we spiked in a pool of blocker oligonucleotides complementary to the full sequences of all barcoded adaptors to a final concentration of 10 micromolar. Hybridized DNA was subsequently sequenced on a single lane of an Illumina HiSeq to generate paired-end 75-base pair reads or 100bp reads. Data were demultiplexed using CASAVA, and reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Alignment tool (33). Local realignment and quality score recalibration were performed using the Genome Analysis Toolkit (GATK) according to GATK best practices (34). We obtained a mean unique on-target sequence depth of 355-fold (range = 60- to 1249-fold).
Sequence data were analyzed to identify three classes of DNA alterations: single-nucleotide variants, small insertions/deletion (indels), and copy number alterations. Single-nucleotide variants were called using muTect (35). For 31/79 tumors with patient-matched normal DNA, the somatic status could be inferred directly. For 48/79 tumors without matched normal DNA, we filtered out all silent variants and all additional variants in dbSNP but not in COSMIC (Catalog of Somatic Mutations in Cancer) (36). Indels were called using the SomaticIndelDetector tool in GATK. All candidate mutations and indels were reviewed manually using the Integrative Genomics Viewer. The mean sequence coverage was calculated using the DepthOfCoverage tool in GATK was used to compute copy number as described previously (32). For this analysis, all copy number alteration calls were normalized against the average ploidy of the tumor sample. Binomial exact confidence intervals for select gene alterations were calculated with the Clopper-Pearson method.
Whole exome sequencing somatic mutation analysis
Paired end whole exome sequencing (WES) was performed on tumor trios (primary tumor, brain metastasis, and matched normal) (dbGaP accession number phs000907.v1.p1). Somatic mutation detection was performed using standard algorithms by the Memorial Sloan Kettering Bioinformatics Core. Briefly, the raw reads were aligned to the hg19 b37 version of the human reference genome using the BWA algorithm (33). Pre-processing steps including removal of PCR duplicates using MarkDuplicates tool in Picard (37), realignment around indels, and quality scores recalibration using the GATK analysis toolkit (34). Somatic mutation calls were generated by the MuTect algorithm (35) and variant annotation was done using Oncotator (38). Each gene was scored and ranked by functional categories for the somatic mutation observed in the gene (truncation, deleterious missense, non-deleterious missense, silent) and the number of mutations observed across samples. Low expression genes as inferred from the RNA-sequencing data were removed. Gene set enrichment analysis was then performed to look for pathways and biological sets significantly enriched in the top-ranking mutated genes.
Whole exome sequencing copy number analysis
To identify copy number alterations from WES data, we applied seqDNAcopy (39) which extends the circular binary segmentation algorithm to next generation sequencing data. Similar to the log-ratio (logR) from copy number array, logR from WES is computed based on binned read counts between the tumor and the matched normal. Copy number abnormalities in the tumor sample cause changes in the logR values reflecting the true underlying copy number ratio. The logR measure was scaled by the ratio of total read counts in the tumor and normal. GC–bias observed in logR was corrected using loess regression. Off-target reads were removed to reduce noise. The variability of the logR from WES is expected to be inversely proportional to the average read count. Therefore, an inverse variance weighted version of CBS was used for copy number segmentation. seqDNAcopy was implemented in R. The Integrative Genomics Viewer (IGV) was then used to visualize genome-wide copy number profiles across samples.
Analysis of clonality
We developed a statistical and computational pipeline called FACETS that leverages rich information from germline polymorphic sites to estimate tumor purity, ploidy, and allele-specific copy number in DNA sequencing data of tumor-normal sample pairs (40). The output of FACETS also includes cellular fraction estimates to identify clonal versus subclonal copy number aberrations. Purity estimation by FACETS relative to pathological determination of tumor purity is shown in Supplementary Table 6. The purity, ploidy, and allele-specific copy number estimates were then used as PyClone (41) input to obtain estimates of cancer cell fraction for somatic mutations by correcting for tumor purity and local copy number states. Phylogenetic tree analysis was performed using Hamming distance computed using all somatic mutations and neighbor-joining method.
PTEN, WNT, and Serpin gene expression analyses
The PTEN loss signature genes and WNT lung cancer metastasis signature genes were obtained from Saal et al. (9) and Nguyen et al. (10), respectively. Log-transformed normalized counts were obtained from RNA-sequencing data and z-score transformed. RNA seq gene expression data from the TCGA study (primary lung tumors) were similarly normalized. The PTEN and WNT signature scores were then computed by aggregating signed expression values of the signature genes (signs obtained from the respective studies). Positive signs indicate positive correlation of signature gene expression with PTEN loss/WNT signaling, and negative signs indicate negative correlation of signature gene expression with PTEN loss/WNT signaling. Samples were then sorted by the PTEN and WNT signature scores. PTEN and WNT signature activities in the brain metastases were compared to the signatures in the TCGA primary tumors using a one-sided Wilcoxon rank-sum test.
Gene expression for key pro-metastatic serpins described in Valiente et al. (11) was assessed in the brain metastases and compared to expression in the TCGA primary tumors using a one-sided Wilcoxon rank-sum test.
Supplementary Material
Statement of Significance.
We performed next generation sequencing of metastatic squamous cell lung cancers and primary lung/brain metastasis pairs, identifying PI3K-aberrant tumors as an aggressive subset associated with brain metastases. We identified a genetic heterogeneity between lung primaries/brain metastases as well as clonal populations that may highlight alterations important in the metastatic process.
Acknowledgments
The MSKCC Sequenom facility was supported by the Anbinder Fund. The authors thank Dr. Laetitia Borsu for supervision of Sequenom assays, Edyta Brzostowski for her administrative supervision of SQ-MAP testing, the members of the MSKCC Bioinformatics Core, and the MSKCC Brain Tumor Center for provision of resected metastatic SQCLC specimens.
Grant support: Supported by the Geoffrey Beene Cancer Research Fund at Memorial Sloan Kettering and a Conquer Cancer Foundation of ASCO Career Development Award.
Footnotes
Conflicts of interest: PP- research funding from AstraZeneca and Novartis.
Author Contributions: PKP- writing, design, data collection, figures, data analysis, data interpretation, funding; RS- writing, data analysis, data interpretation, figures; NR- writing, analysis, interpretation; LW- data analysis, data interpretation; CS- data analysis, writing; AA- data analysis and writing; VS- data analysis and writing; HW- data analysis, data interpretation; MB- data analysis, data interpretation, design; ML- data analysis, data interpretation, design; MK- writing, design, funding.
References
- 1.Elliott J, Ahmedzai S, Hole D, Dorward A, Stevenson R, Kaye S, et al. Vindesine and cisplatin combination chemotherapy compared with vindesine as a single agent in the management of non-small cell lung cancer: a randomized study. Eur J Cancer Clin Oncol. 1984;20:1025–32. doi: 10.1016/0277-5379(84)90104-4. [DOI] [PubMed] [Google Scholar]
- 2.Rekhtman N, Paik P, Arcila M, Tafe L, Oxnard G, Moreira A, et al. Clarifying the spectrum of driver oncogene mutations in pure, biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clinical Cancer Research. 2011;18:1167–76. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and Focal FGFR1 Amplification Associates with Therapeutically Tractable FGFR1 Dependency in Squamous Cell Lung Cancer. Science Translational Medicine. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, et al. Mutations in the DDR2 Kinase Gene Identify a Novel Therapeutic Target in Squamous Cell Lung Cancer. Cancer Discovery. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu C, Fillmore Christine M, Koyama S, Wu H, Zhao Y, Chen Z, et al. Loss of Lkb1 and Pten Leads to Lung Squamous Cell Carcinoma with Elevated PD-L1 Expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spoerke JM, O’Brien C, Huw L, Koeppen H, Fridlyand J, Brachmann RK, et al. Phosphoinositide 3-Kinase (PI3K) Pathway Alterations Are Associated with Histologic Subtypes and Are Predictive of Sensitivity to PI3K Inhibitors in Lung Cancer Preclinical Models. Clinical Cancer Research. 2012;18:6771–83. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- 8.Drilon A, Rekhtman N, Ladanyi M, Paik P. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. The Lancet Oncology. 2012;13:e418–e26. doi: 10.1016/S1470-2045(12)70291-7. [DOI] [PubMed] [Google Scholar]
- 9.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She Q-B, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen DX, Chiang AC, Zhang XHF, Kim JY, Kris MG, Ladanyi M, et al. WNT/TCF Signaling through LEF1 and HOXB9 Mediates Lung Adenocarcinoma Metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valiente M, Obenauf Anna C, Jin X, Chen Q, Zhang Xiang HF, Lee Derek J, et al. Serpins Promote Cancer Cell Survival and Vascular Co-Option in Brain Metastasis. Cell. 2014;156:1002–16. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soria J-C, Lee H-Y, Lee JI, Wang L, Issa J-P, Kemp BL, et al. Lack of PTEN Expression in Non-Small Cell Lung Cancer Could Be Related to Promoter Methylation. Clinical Cancer Research. 2002;8:1178–84. [PubMed] [Google Scholar]
- 13.Craddock KJ, Ludkovski O, Sykes J, Shepherd FA, Tsao M-S. Prognostic Value of Fibroblast Growth Factor Receptor 1 Gene Locus Amplification in Resected Lung Squamous Cell Carcinoma. Journal of Thoracic Oncology. 2013;8:1371–7. doi: 10.097/JTO.0b013e3182a46fe9. [DOI] [PubMed] [Google Scholar]
- 14.Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, et al. Fibroblast Growth Factor Receptor 1 Gene Amplification Is Associated With Poor Survival and Cigarette Smoking Dosage in Patients With Resected Squamous Cell Lung Cancer. Journal of Clinical Oncology. 2013;31:731–7. doi: 10.1200/JCO.2012.43.8622. [DOI] [PubMed] [Google Scholar]
- 15.Tran TN, Selinger CI, Kohonen-Corish MRJ, McCaughan BC, Kennedy CW, O’Toole SA, et al. Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer. 2013;81:462–7. doi: 10.1016/j.lungcan.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Russell PA, Yu Y, Young RJ, Conron M, Wainer Z, Alam N, et al. Prevalence, morphology, and natural history of FGFR1-amplified lung cancer, including squamous cell carcinoma, detected by FISH and SISH. Mod Pathol. 2014;27:1621–31. doi: 10.1038/modpathol.2014.71. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Zhang L, Su X, Li M, Xie L, Malchers F, et al. Translating the Therapeutic Potential of AZD4547 in FGFR1-Amplified Non–Small Cell Lung Cancer through the Use of Patient-Derived Tumor Xenograft Models. Clinical Cancer Research. 2012;18:6658–67. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 18.Paik P, Shen R, Ferry D, Soria J-C, Mathewson A, Kilgour E, et al. A phase 1b open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers: Preliminary antitumor activity and pharmacodynamic data. J Clin Oncol. 2014;32(suppl):abstr 8035. doi: 10.1158/1078-0432.CCR-17-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogova L, Sequist L, Cassier P, Hidalgo M, Delord J-P, Schuler M, et al. Targeting FGFR1-amplified lung squamous cell carcinoma with the selective pan-FGFR inhibitor BGJ398. J Clin Oncol. 2014;32(suppl):abstr 8034. [Google Scholar]
- 20.Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, et al. FGFR1 mRNA and Protein Expression, not Gene Copy Number, Predict FGFR TKI Sensitivity across All Lung Cancer Histologies. Clinical Cancer Research. 2014;20:3299–309. doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenbygaard LE, Sorensen JB, Olsen JE. Metastatic pattern in adenocarcinoma of the lung: An autopsy study from a cohort of 137 consecutive patients with complete resection. The Journal of Thoracic and Cardiovascular Surgery. 1995;110:1130–5. doi: 10.1016/s0022-5223(05)80183-7. [DOI] [PubMed] [Google Scholar]
- 22.Niessner H, Forschner A, Klumpp B, Honegger JB, Witte M, Bornemann A, et al. Targeting hyperactivation of the AKT survival pathway to overcome therapy resistance of melanoma brain metastases. Cancer Medicine. 2013;2:76–85. doi: 10.1002/cam4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamo B, Deal A, Burrows E, Geradts J, Hamilton E, Blackwell K, et al. Phosphatidylinositol 3-kinase pathway activation in breast cancer brain metastases. Breast Cancer Research. 2011;13:R125. doi: 10.1186/bcr3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Yang J, Yu Q, Wu H, Liu B, Xiong H, et al. Associations between Single-Nucleotide Polymorphisms in the PI3K–PTEN–AKT–mTOR Pathway and Increased Risk of Brain Metastasis in Patients with Non–Small Cell Lung Cancer. Clinical Cancer Research. 2013;19:6252–60. doi: 10.1158/1078-0432.CCR-13-1093. [DOI] [PubMed] [Google Scholar]
- 25.Tamura M, Gu J, Matsumoto K, Aota S-i, Parsons R, Yamada KM. Inhibition of Cell Migration, Spreading, and Focal Adhesions by Tumor Suppressor PTEN. Science. 1998;280:1614–7. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 26.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, et al. Differential expression of individual gamma-protocadherins during mouse brain development. Molecular and Cellular Neuroscience. 2005;29:603–16. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–9. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–6. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rekhtman N, Ang D, Sima C, Travis W, Moreira A. Immunohistochemistry algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on a large series of whole-tissue sections with validation in small specimens. Mod Pathol. 2011;24:1348–59. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 30.Won H, Scott S, Brannon A, Shah R, Berger M. Detecting Somatic Genetic Alterations in Tumor Specimens by Exon Capture and Massively Parallel Sequencing. J Vis Exp. 2013;80:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng D, Mitchell T, Zehir A, Shah R, Benayed R, Syed A, et al. MSK-IMPACT: A hybridization capture-based next generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015 doi: 10.1016/j.jmoldx.2014.12.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagle N, Berger MF, Davis MJ, Blumenstiel B, DeFelice M, Pochanard P, et al. High-Throughput Detection of Actionable Genomic Alterations in Clinical Tumor Samples by Targeted, Massively Parallel Sequencing. Cancer Discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotech. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research. 2011;39:D945–D50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Institute B. Picard. 2015 Available from: http://broadinstitute.github.io/picard/
- 38.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: Cancer Variant Annotation Tool. Human Mutation. 2015 doi: 10.1002/humu.22771. n/a n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seshan V. Next Generation Sequencing Data Analysis. Springer; 2014. Detecting copy number changes and structural rearrangements using DNA sequencing. [Google Scholar]
- 40.Shen R, Seshan V. Working Paper 29. 2015. FACETS: Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing Memorial Sloan-Kettering Cancer Center, Dept of Epidemiology & Biostatistics Working Paper Series. [Google Scholar]
- 41.Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Meth. 2014;11:396–8. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.