Abstract
Background: MicroRNA (miRNA) have been shown to regulate gene expression in many cancers. MiR-182 has recently been found to be prognostic for patients treated with radical prostatectomy for prostate cancer. We sought to assess miR-182 as a prognostic marker and understand its role in prostate cancer progression and metastasis.
Methods: We analysed miR-182 expression among 147 men treated for prostate cancer using biochemical recurrence and metastasis as the endpoints. We examined miR-182 expression in prostate cancer cells and created cell lines that overexpressed miR-182 for functional assays. Finally, we examined pathways through which miR-182 may function using prediction algorithms and confirmed by Western blotting and knock-down assays.
Results: We found that miR-182 was not associated with biochemical recurrence (p=0.1111) or metastasis (p=0.9268) following radical prostatectomy. However, in mechanistic assays, we found that miR-182 expression was higher among aggressive prostate cancer cells and that ectopic miR-182 expression resulted in increased proliferation, migration and invasion in vitro. We identified FOXO1 as regulated by miR-182 in prostate cancer cells, confirmed that ectopic miR-182 expression resulted in diminished FOXO1 levels, and showed that miR-182 inhibition results in increased FOXO1 levels. Expression of FOXO1 (p=0.0014) in tumors from patients who developed biochemical recurrence compared to tumors from patients who were recurrence-free five years after their radical prostatectomy.
Conclusions: Our findings suggest that miR-182 may act to increase prostate cancer proliferation, migration and invasion through suppression of FOXO1. This may be valuable in the development of further therapeutic interventions.
Keywords: MicroRNA, prostatic neoplasms, prostatectomy.
Introduction
Patients with prostate cancer may experience a phenotypically wide range of behaviours ranging from indolent disease that does not require treatment to a highly aggressive and fatal disease which progresses quickly. Thus far, we have limited tools to risk stratify these patients - tumor grade, stage and prostate specific antigen (PSA) as a tumor marker. These clinical parameters have been used to create a number of risk stratification schemes and guidelines. Recent work has demonstrated that patients with the same Gleason score1 or risk category2 may experience vastly different clinical outcomes. Therefore, there is an ongoing need for the development and validation of new biomarkers.
Further, compared with other sites such as breast and colon, targeted oncologic treatments are lacking in prostate cancer. Elucidation of the metastatic pathways in prostate cancer may facilitate the development of novel, targeted therapies.
MicroRNAs (miRNAs) are involved in critical steps of metastatic progression, such as migration, invasion, and angiogenesis, and their aberrant expressions have been reported in numerous cancer subtypes—including prostate cancer3. MiRNAs are endogenously expressed, ~18-25 nucleotide, single-stranded RNA molecules. They regulate gene expression at the post-transcriptional level primarily by binding to the 3'-untranslated region (UTR) of a target messenger RNA (mRNA). MiRNAs can regulate gene expression by inhibiting protein translation of a target mRNA4, and also by degrading their target mRNA by affecting its stability, thereby inhibiting protein synthesis indirectly5,6. MiRNAs that are overexpressed in tumors often target tumor suppressors associated with various steps of the tumor progression; thus, identifying these miRNAs may be vital in the management of disease.
MiR-182, a member of the miR-96/-182/-183 cluster7, has been shown to promote invasion and migration of melanoma cells, and to enhance their metastatic ability by directly repressing Forkhead Box O3 (FOXO3) and Microphthalmia-associated Transcription Factor-M (MITF-M)8. In breast cancer cells, miR-182 appears to regulate cell proliferation and survival by directly targeting Forkhead box O1 (FOXO1)9. Recent work has shown that miR-182 may be involved in prostate cancer progression10 though the authors did not assess its mechanism in prostate cancer.
For these reasons, we investigated the possible role of miR-182 in prostate cancer progression, with the hope of gaining more insights into the mechanisms of prostate cancer pathogenesis. We analyzed the expression levels of miR-182 in a large cohort of primary prostate tumors in various clinical stages, normal prostate tissues, as well as several prostate cancer cell lines.
Materials and Methods
Prostate Sample Collection
In order to assess the role of miR-182 in prostate cancer recurrence and metastasis, we examined a cohort of men treated with radical prostatectomy for clinically localized prostate cancer at Sunnybrook Health Sciences Centre (Toronto, Canada) between 1998 and 2006. Following radical prostatectomy, the specimen was oriented using anatomic landmarks and the prostate and seminal vesicles were painted right side green and left side blue11. A random transverse midsection of the prostate was taken perpendicular to the rectal surface, frozen in liquid nitrogen, and stored at -80°C. Banked slices of specimens were photocopied, oriented (anterior, posterior, right and left), quadrisected, and cut on the cryostat. Sections were stained with H&E and reviewed by a pathologist to delineate cancerous and non-cancerous regions on the stained slides and a corresponding image. The marked areas were used to guide extraction of the tumor tissue from which RNA was extracted. All research was conducted with the approval of Sunnybrook Health Sciences Centre research ethics board.
Quantitative Real-Time PCR
Total RNA was extracted from prostate tissues using TRIzol Reagent (Invitrogen) according to the manufacturers' instructions. 1 µg of total RNA was reversed transcribed using QuantiTect Reverse Transcription Kit (Qiagen). Quantitative Real-Time PCR analysis of miR-182, was performed in triplicate using miScript SYBR Green PCR Kit (Qiagen) on LightCycler Real-time PCR system (Roche applied science). The miRNA level was normalized by a reference gene, RNU6B. Primers for each miRNA as well as for RNU6B were commercially purchased (miScript Primer Assays, Qiagen). For genes other than miRNAs, two primer sets were used. The Primers used are as follows: FOXO1 (Forward: AAGAGCGTGCCCTACTTCAA, Reverse: CTGTTGTTGTCCATGGATGC), FOXO3 (Forward: TCAATCAGAACTTGCTCCACC, Reverse: GGACTCACTCAAGCCCATGTT), Beta-actin (Forward: GGAGAATGGCCCAGTCCTC, Reverse: GGGCACGAAGGCTCATCAT). The relative amount of genes in each sample was calculated based on the crossing-point analysis (RelQuant, version 1.01). For semiquantitative end point PCR analysis, cDNA was subjected to 40 rounds of amplification, and PCR products were electrophoresed and visualized in ethidium bromide-stained 1.5% agarose gels.
Clinical records
The medical records and of patients were thoroughly reviewed using standardised data entry forms by trained data abstractors and stored within a prostate cancer-specific database. Biochemical recurrence was defined as a PSA increase of at least 0.2 ng/mL on at least two separate consecutive measurements that are at least 3 months apart. Metastasis was defined as lesions within the bone identified on radionuclide bone scan and lymphadenopathy or visceral lesions identified by computed tomography imaging of the abdomen, pelvis and chest. These examinations were undertaken at the discretion of the treating urologist. From a total of 147 patients for whom miR-182 expression levels were available, 17 were excluded for missing Gleason score or pre-operative PSA levels. Of the remainder, adequate follow-up information was available on 100 patients for recurrence status and 74 patients for metastasis status. There were no statistically significant differences between patients with and without complete follow-up data with respect to age at treatment, pre-operative PSA, Gleason score, margin status or seminal vesicle invasion.
Statistical Analysis
We first examined baseline characteristics of the study population using univariate statistics: Wilcoxon rank sum test for continuous data and Fisher's exact test for categorical data. We then divided each study population into high and low miR-182 groups based on the median miR-182 for that group. Univarate statistics using Wilcoxon rank sum test and Fisher's exact test were used to assess the relationship between miR-182 group and accepted prognostic factors.
We then performed univariate and multivariate logistic regression to examine the odds ratio for recurrence and metastasis by miR-182 risk group. Multivariate analyses were adjusted for age, pre-operative PSA level, Gleason score, seminal vesicle invasion and margin positivity. Nonlinear regression (curve fit) and Pearson product-moment correlation coefficient were used for correlation analysis of miR-182 and FOXO1. Analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, USA) and GraphPad Prism v4.0 (GraphPad Software Inc, La Jolla, CA, USA).
Cell Lines
The non-tumorigenic human prostatic epithelial cell line, RWPE-1, and human prostate cancer PC-3, Du145, LNCaP, and VCaP cell lines were obtained from the American Type Culture Collection. LNCaP cells were stably transduced with the pLemiR lentiviral vector (Open Biosystems) using standard techniques. The base medium for RWPE-1 cells was provided by Invitrogen as Keratinocyte Serum Free Medium (K-SFM; Gibco). This kit is supplied with the two additives required to grow this cell line: bovine pituitary extract and human recombinant EGF. PC-3 cells are maintained in F-12K Medium, DU145 in Eagle's Minimum Essential Medium, VCaP in Dulbecco's Modified Eagle's Medium, and LNCaP and its transduced derivative cells in RPMI-1640 Medium, all supplemented with 10% fetal bovine serum (FBS) (Life Technologies-Invitrogen Corp.). Cells were incubated at 37ºC and 5% CO2 in a humidified incubator.
Soft Agar Assay
The bottom layer was obtained by covering 6-well dishes with 1 ml of 0.8% agarose in Dulbecco's Modified Eagle Medium 2X (DMEM 2X) containing 10% Fetal bovine serum (FBS). After an hour, 1.2x104 stably transduced LNCaP cells with either empty vector or with miR-182-expressing vector were seeded in triplicate in 1 ml DMEM 2X containing 0.5% agarose and 10% FBS. After two weeks, colonies were photographed and scored.
Wound Healing Assay
LNCaP cells were seeded in six-well culture plates and grown to confluence forming a monolayer covering the surface of the entire well. After cells were serum-starved in serum-free RPMI for 18 h, the wound was created in the center of the cell monolayer by the gentle removal of the attached cells with a 10µl pipette tip. Debris was removed by PBS wash, and the cells received fresh RPMI with 10% FBS and 10 mmol/L of hydroxyurea. Cells were photographed with a Zeiss Axiovert 200M inverted microscope at the intervals of 0, 24, and 72 hours.
Transwell Migration Assay
A suspension of 1x105 stably transduced LNCaP cells with either empty vector or with miR-182-expressing vector in 0.5% FBS medium was added to BD cell culture inserts with 8-μm porous membrane coated with 20% FBS (BD Biosciences, Bedford, MA). Cells were incubated for 2-5 days at 37°C in a 5% CO2 incubator. To quantify migrating cells, cells remaining on the top-side of the membrane were removed using a cotton-tipped swab, and cells that had migrated to the underside were visualized with a Zeiss Axiovert 200M inverted microscope and photographed.
Invasion Assay
A suspension of 1x105 cells in RPMI media (600 µL) containing 1% serum was added to transwell Boyden chamber containing 8.0 µm pore size filters coated with matrigel (BD Biosciences, Bedford, MA). The lower chamber was filled with 1.5 mL of media with 10% FBS as a chemo-attractant. Cells were incubated for 5 days under standard culture conditions. Cancer cells remaining on the top-side of the membrane were removed and cells that had migrated to the underside were visualized under a Zeiss phase contrast microscope, and photographed.
Western Blotting
Total cell extracts were prepared from sub-confluent cells using Nonidet P-40 lysis buffer (20 mM HEPES-KOH [pH 7.6], 100 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 0.25% Nonidet P-40, 2 μg/ml leupeptin, 2 μg/ml pepstatin, and 102 μg/ml cycloheximide). Lysates were mixed with 2X SDS-loading buffer containing 10% β-mercaptoethanol, boiled for 5 min, and loaded onto 12% SDS-polyacrylamide gel for electrophoretic separation. The proteins were transferred onto a nitrocellulose membrane (Bio-Rad Laboratories, CA) at 300 milliamps for 40 minutes at room temperature (RT). Membranes were then blocked for nonspecific binding with 5% non-fat dry milk in TBST (TBS with 0.1% Tween 20; pH 7.4) for 1 hour, and incubated with the primary antibodies overnight at 4ºC. The following monoclonal and polyclonal antibodies were used: anti-FOXO1 and anti-FOXO3a (Cell Signalling, USA); anti-MiTF and anti-Smad1 (abcam, USA): anti-β-Tubulin (Sigma). After washing with TBST, membranes were incubated with species-specific secondary antibody conjugated to horseradish peroxidase enzyme for 1 hour at RT, and washed three times with TBST. Proteins were detected using an enhanced chemiluminescence detection reagent kit (Amersham Biosciences Inc.) and Kodak X-OMAT-AR film for autoradiography.
Transfection of siRNA and miRNA inhibitors
The miRNA inhibitor of miR-182, and FOXO1 siRNA used in our study were designed by and purchased from Qiagen. 1.5 x 105 cells were seeded per well in a 24-well plate. 5 nM of siRNA (control and FOXO1 siRNA) and 50 nM of miRNA inhibitor (control and anti-miR-182) were diluted into 100 μl of serum free medium. Subsequently, 3 μl of HiPerFect Transfection Reagent (Qiagen) was added to the mixture and incubated at room temperature for 10 min. The transfection complexes formed were then added drop-wise onto cells and incubated under normal growth conditions. Cells were harvested 48 hours after the transfections.
Results
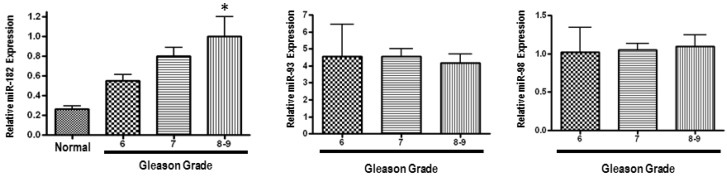
We first analysed MiR-182 expression in 147 primary prostate cancer specimens, obtained after radical prostatectomy, and in 5 normal prostate tissue samples using quantitative Real-Time PCR (q-PCR). MiR-182 mRNA expression was progressively upregulated with increasing Gleason grade, and was significantly higher in tumors with Gleason 8 or 9 as compared with normal prostate tissue (Figure 1). Expression levels of two control miRNAs (miR-93 and miR-98) did not show any difference between the groups.
Figure 1.
miR-182 is over-expressed in human prostate tumors, and its up-regulation is associated with clinical progression. MiR-182 expression was measured using qRT-PCR in various prostate tumor samples representing different Gleason grades compared with normal prostate tissue. Sample categories were distributed as follows: Normal (n=5), Gleason 5 or 6 (n=32), Gleason 7 (n=101), Gleason 8 or 9 (n=14). All samples were normalized to RNU6B expression. Expression levels of miR-93 and miR-98 were also assessed, but did not show a significant difference. (* p < 0.05).
In order to assess the role of miR-182 in prostate cancer recurrence, we examined a cohort of 100 patients treated with radical prostatectomy for whom complete follow-up data were available. Baseline characteristics of the study cohort, stratified by recurrence status, are presented in Table 1. Gleason score and margin positivity were significantly associated with recurrence (p=0.02 and <0.0001, respectively). Median follow-up was 5.0 years (IQR 3.0-7.0 years). The median miR-182 level was 0.540 in patients without recurrence and 0.636 in those with recurrence (p=0.2898). We then stratified the patients into low and high miR-182 groups based on the overall median miR-182 level (0.595). MiR-182 expression level was associated with age but not the other baseline tumor characteristics (Table 2).
Table 1.
Baseline characteristics of study cohort for patients with complete follow-up data on recurrence.
| No recurrence | Recurrence | p-value | |
|---|---|---|---|
| Age (median, IQR) | 61.5 (56.0-67.0) | 64.0 (58.0-68.0) | 0.1155 |
| PSA (ng/mL) | 0.6833 | ||
| ≤4 | 7 (14%) | 4 (8%) | |
| 4 - 10 | 32 (64%) | 34 (68%) | |
| >10 | 11 (22%) | 12 (24%) | |
| Grade (Gleason score) | 0.0249 | ||
| 6 | 13 (26%) | 3 (6%) | |
| 7 | 33 (66%) | 41 (82%) | |
| 8-10 | 4 (8%) | 6 (12%) | |
| Seminal vesicle invasion | 0.2336 | ||
| Present | 4 (8%) | 9 (18%) | |
| Absent | 46 (92%) | 41 (82%) | |
| Margin status | <0.0001 | ||
| Positive | 21 (42%) | 41 (82%) | |
| Negative | 28 (56%) | 6 (16%) | |
| Missing | 1 (2%) | 1 (2%) |
Table 2.
Association between miR-182 levels and known prognostic factors for patients with complete follow-up data on recurrence.
| Low MiR-182 | High MiR-182 | p-value | |
|---|---|---|---|
| Age (median, IQR) | 60.0 (56.0-67.0) | 64.5 (60.0-68.0) | 0.0428 |
| PSA (ng/mL) | 0.1551 | ||
| ≤4 | 8 (16%) | 3 (6%) | |
| 4 - 10 | 29 (58%) | 37 (74%) | |
| >10 | 13 (26%) | 10 (20%) | |
| Grade (Gleason score) | 0.4783 | ||
| 6 | 8 (16%) | 8 (16%) | |
| 7 | 39 (78%) | 36 (70%) | |
| 8-10 | 3 (6%) | 4 (14%) | |
| Seminal vesicle invasion | 1.000 | ||
| Present | 44 (88%) | 43 (86%) | |
| Absent | 6 (12%) | 7 (14%) | |
| Margin status | 0.2710 | ||
| Positive | 2 (4%) | 0 (0%) | |
| Negative | 20 (40%) | 16 (32%) | |
| Missing | 28 (56%) | 34 (68%) |
We examined the role of miR-182 in prostate cancer metastasis in a cohort of 74 patients treated with radical prostatectomy. The baseline characteristics were not significantly different between patients experiencing metastasis and those who did not (Table 3). Again, median follow-up was 5.0 years (IQR 3.0-7.0 years). The median miR-182 level was 0.590 in patients without recurrence and 0.580 in those with recurrence (p=0.7880). We then stratified the patients into low and high miR-182 groups based on the overall median miR-182 level (0.590). MiR-182 expression level was not associated with any of the baseline characteristics (Table 4) in this cohort.
Table 3.
Baseline characteristics of study cohort for patients with complete follow-up data on metastasis.
| No metastasis | Metastasis | p-value | |
|---|---|---|---|
| Age (median, IQR) | 61.0 (57.0-66.0) | 61.5 (58.0-67.0) | 0.6921 |
| PSA (ng/mL) | 0.7644 | ||
| ≤4 | 8 (13%) | 0 (0%) | |
| 4 - 10 | 40 (63%) | 7 (70%) | |
| >10 | 16 (25%) | 3 (30%) | |
| Grade (Gleason score) | 0.0993 | ||
| 6 | 14 (22%) | 0 (0%) | |
| 7 | 45 (70%) | 8 (80%) | |
| 8-10 | 5 (8%) | 2 (20%) | |
| Seminal vesicle invasion | 0.0703 | ||
| Present | 5 (8%) | 3 (30%) | |
| Absent | 59 (92%) | 7 (70%) | |
| Margin status | 0.0691 | ||
| Positive | 32 (50%) | 9 (90%) | |
| Negative | 31 (48%) | 1 (10%) | |
| Missing | 1 (2%) | 0 (0%) |
Table 4.
Association between miR-182 levels and known prognostic factors for patients with complete follow-up data on metastasis.
| Low MiR-182 | High MiR-182 | p-value | |
|---|---|---|---|
| Age (median, IQR) | 59.5 (56.0-67.0) | 62.5 (58.0-66.0) | 0.1774 |
| PSA (ng/mL) | 0.2491 | ||
| ≤4 | 6 (16%) | 2 (6%) | |
| 4 - 10 | 21 (55%) | 26 (72%) | |
| >10 | 11 (29%) | 8 (22%) | |
| Grade (Gleason score) | |||
| 6 | 8(21%) | 6 (17%) | |
| 7 | 27 (71%) | 26 (72%) | 0.8674 |
| 8-10 | 3 (8%) | 4 (11%) | |
| Seminal vesicle invasion | 0.2627 | ||
| Present | 6 (16%) | 2 (6%) | |
| Absent | 32 (84%) | 34 (94%) | |
| Margin status | 0.4832 | ||
| Positive | 19 (50%) | 22 (61%) | |
| Negative | 18 (47%) | 14 (39%) | |
| Missing | 1 (3%) | 0 (0%) |
MiR-182 score (high vs. low) was not significantly associated with either recurrence or metastasis in univariable logistic regression. There were insufficient events in the metastasis cohort to allow for multivariable regression though we observed no association between miR-182 score and recurrence on multivariable analysis (Table 5).
Table 5.
Univariable and Multivariable logistic regression model examining factors predicting recurrence following prostate cancer treatment.
| Prognostic factor | Univariable Odds Ratio (95% CI, p-value) | Mulitvariable Adjusted Odds Ratio (95% CI, p-value) |
|---|---|---|
| RECURRENCE | ||
| MiR-182 Score | ||
| Low | Referent | Referent |
| High | 1.91 (0.86-4.22, 0.1111) | 1.39 (0.51-3.79, 0.5263) |
| Age | 1.05 (0.99-1.12, 0.1067) | 1.04 (0.96-1.13, 0.2962) |
| PSA (ng/mL) | ||
| ≤4 | Referent | Referent |
| 4 - 10 | 1.86 (0.497-6.96, 0.3571) | 4.98 (0.95-26.18, 0.0583) |
| >10 | 1.909 (0.44-8.35, 0.3905) | 2.37 (0.37-15.04, 0.3599) |
| Grade (Gleason score) | ||
| 6 | Referent | Referent |
| 7 | 5.38 (1.42-20.49, 0.0136) | 8.31 (1.72-40.19, 0.0084) |
| 8-10 | 6.50 (1.09-38.63, 0.0396) | 5.86 (0.65-53.44, 0.1160) |
| Seminal vesicle invasion | ||
| Present | 2.52 (0.72-8.81, 0.1469) | 0.93 (0.17-5.15, 0.9298) |
| Absent | Referent | Referent |
| Margin status | ||
| Positive | 6.83 (2.66-17.59, <0.0001) | 11.22 (3.45-26.46, <0.0001) |
| Negative | Referent | Referent |
| METASTASIS | ||
| MiR-182 Score | N/A | |
| Low | Referent | |
| High | 1.07 (0.28-4.04, 0.9268) | |
| Age | 1.02 (0.92-1.14, 0.6834) | N/A |
| PSA (ng/mL) | N/A | |
| ≤10 | Referent | |
| >10 | 1.29 (0.30-5.57, 0.7368) | |
| Grade (Gleason score) | N/A | |
| 6-7 | Referent | |
| 8-10 | 2.95 (0.49-17.82, 0.2384) | |
| Seminal vesicle invasion | N/A | |
| Present | 5.06 (0.99-25.86, 0.0516) | |
| Absent | Referent | |
| Margin status | N/A | |
| Positive | 8.72 (1.04-72.94, 0.0457) | |
| Negative | Referent | |
Note: Multivariable logistic regression was not possible for the metastasis cohort due to a lack of events.
Though we were unable to demonstrate that miR-182 expression was predictive of prostate cancer prognosis in our cohort, we moved forward with mechanistic studies based on previous work which has demonstrated that miR-182 may be prognostically significant in prostate cancer patients treated with radical prostatectomy10.
MiR-182 over-expression enhances the tumorigenic properties of LNCaP cells
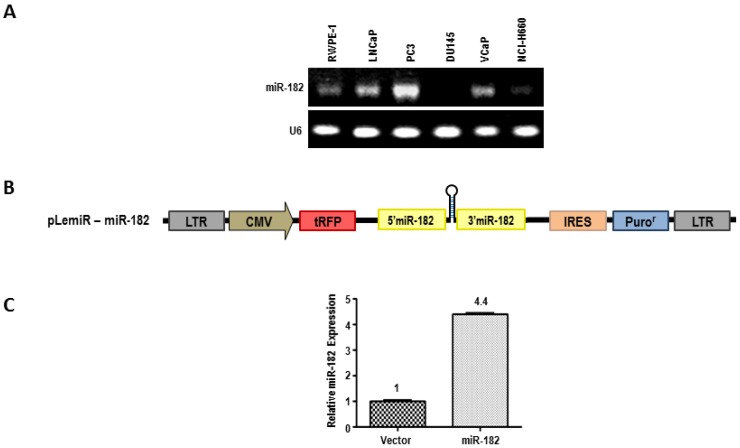
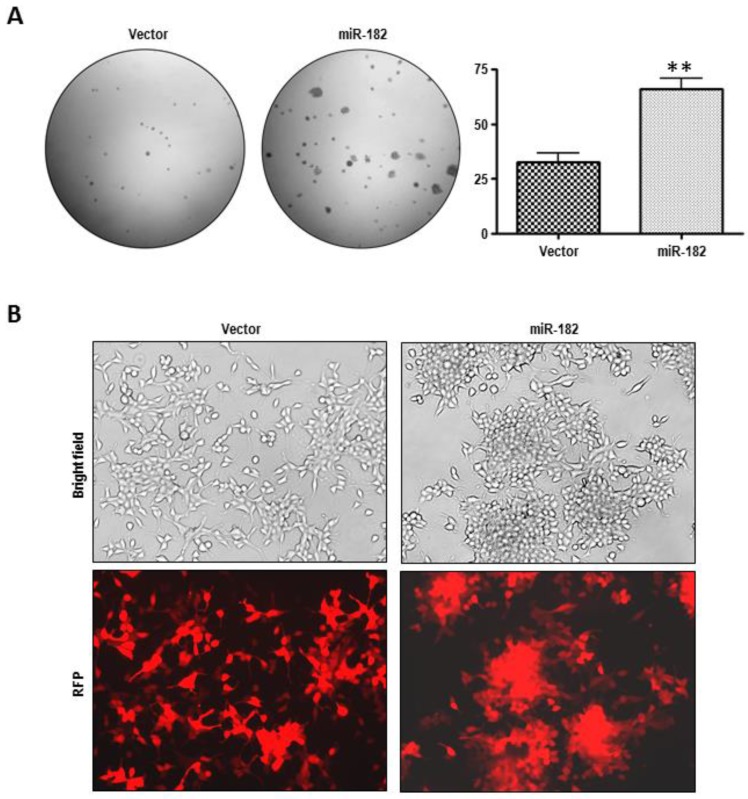
We next assessed the levels of miR-182 expression by semi-quantitative end point PCR analysis in a series of prostate cell lines (Fig. 2A). The expression of miR-182 appears to be highest in highly aggressive PC3 prostate carcinoma cells, compared to less invasive cell lines, such as DU-145 and LNCaP. To assess the biological effects of miR-182, we used lentiviral pLemiR vectors (Figure 2B) to increase miR-182 expression in LNCaP cells. LNCaP cells stably transduced with miR-182, hereafter referred to simply as miR-182, were found to express more than 4-fold higher levels of miR-182 than the empty vector control, abbreviated as “vector” (Fig. 2C). To analyze tumorigenic potential of these cells, we tested their anchorage-independent growth in soft agar as a marker of tumorigenicity. As shown in figure 3A, miR-182 cells developed more colonies in soft agar compared to vector control (p = 0.0007), indicating that miR-182 supports anchorage-independent growth of LNCaP cells. Importantly, miR-182 cells exhibited sharp morphological differences in culture (Fig. 3B). They appear to lose contact inhibition and form aggregates. The appearance of these irregular spherical structures is dense and individual cells are difficult to visualize. In contrast, vector control LNCaP cells exhibit a typical LNCaP morphology in bright field and fluorescent microscopy, growing as loosely adherent cells that do not form a uniform monolayer. Therefore, miR-182 overexpression enhances the tumorigenic properties of LNCaP cells.
Figure 2.
MiR-182 over-expression in LNCaP cells. (A), Expression levels of miR-182 was analyzed by semi-quantitative end point PCR in a series of Human Prostate Cell Lines. (B), Lentiviral pLemiR construct used to deliver miR-182 expression in LNCaP cell line. (C) Relative expression of miR-182 in stably transduced LNCaP cells with empty vector control (Vector) or with miR-182-expressing vector (miR-182).
Figure 3.
MiR-182 over-expression promotes proliferation and morphologic change in LNCaP cells. (A), Growth in soft agar of vector and miR-182 LNCaP cells. Graph is representative of 3 independent experiments (n=3). (B) Morphological transformation in culture of LNCaP cells with empty or miR-182-expressing vector. RFP: Red Fluorescent Protein.
miR-182 enhances the migratory and invasive abilities of LNCaP cells
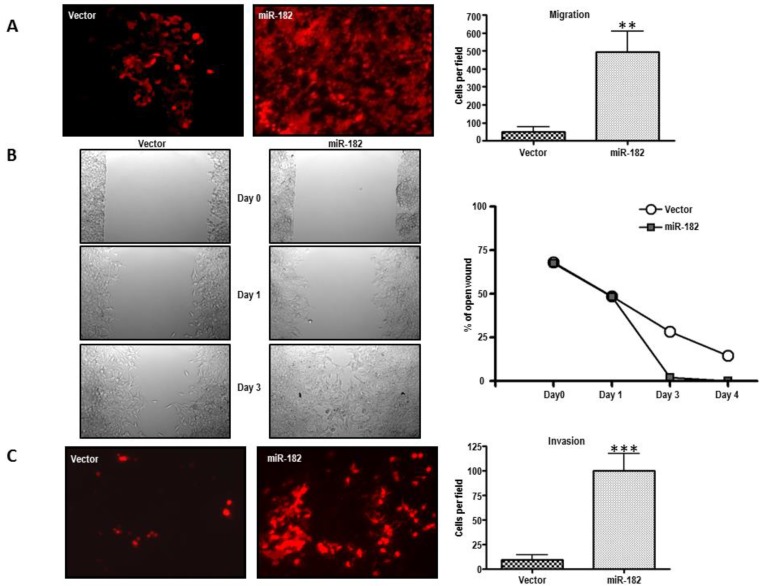
We then examined whether miR-182 might be involved in metastasis. We therefore investigated the role of miR-182 in cell migration of stably transduced LNCaP cells. When these cells were analyzed in a trans-well migration assay, the results shown in figure 4A indicate that miR-182 increased the migration of LNCaP cells by more than 10-fold (p = 0.0012). MiR-182 cells were also more proficient than equivalent vector-transduced cells at closing an artificial wound created over a confluent monolayer (Fig. 4B), further reaffirming the conclusion that miR-182 enhances the migratory of LNCaP cells.
Figure 4.
MiR-182 enhances the migratory and invasive abilities of LNCaP cells in vitro. (A), Trans-well migration assay on LNCaP cells stably transduced with either empty vector (Vector) or miR-182 (miR-182) (n=3). (B) Wound-healing assay on LNCaP cells transduced with empty or miR-182 vectors. Pictures were taken upon making the wound and at day 0, day 1, and day 3 (n=2). (C), Trans-well invasion assay of LNCaP transduced with empty or miR-182 vectors (n=2). Cells were added to cell culture inserts containing a filter with pores coated with matrigel. (*p<0.05; ** p<0.01, ***p<0.001).
The invasive potential of miR-182 cells was analyzed using Matrigel-coated cell culture inserts. As seen in figure 4C, miR-182 upregulation dramatically enhanced the invasive capacity of normally poorly invasive LNCaP cells (p = 0.0006). Therefore, miR-182 upregulation enhances migratory and invasive properties of LNCaP prostate cells. Together with the observed higher miR-182 expression levels in aggressive and recurrent prostate tumors, these findings implicate miR-182 in metastatic progression.
MiR-182 negatively regulates FOXO1 in LNCaP cells and primary prostate tumors
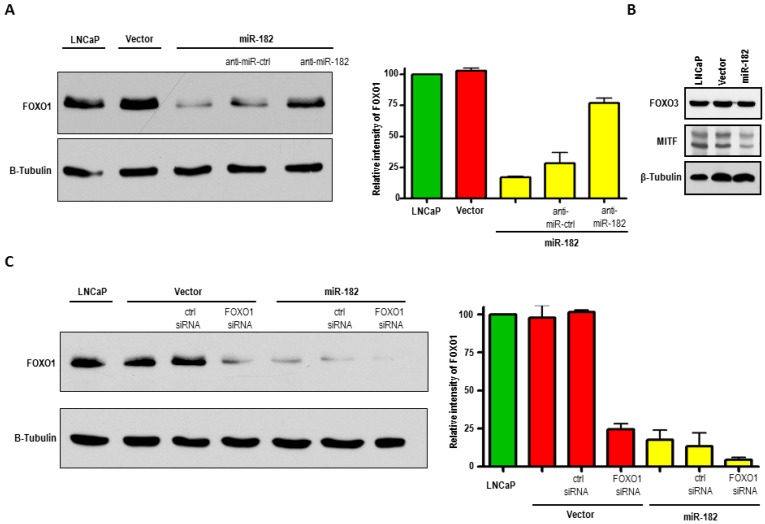
To identify targets that, when inhibited by miR-182, could result in enhanced metastatic properties that we saw in LNCaP cells, we searched the literature and prediction algorithms such as miRBase and TargetScan, and found FOXO1, FOXO3, and MITF among the putative gene targets of miR-182. FOXO1 protein is dramatically reduced in LNCaP-miR-182 cells as compared to parental and vector alone cells, as shown by Western blot analysis (Fig. 5A). We also found a slight diminishment in MITF protein levels in response to miR-182 overexpression (Fig. 5B). We did not, however, detect any changes in FOXO3 protein levels.
Figure 5.
MiR-182 negatively regulates FOXO1 in LNCaP cells. (A), Western blot analysis of FOXO1 protein expression in LNCaP cells, stably transduced LNCaP cells with empty vector (Vector), and with miR-182-expressing vector (miR-182). FOXO1 levels inversely correlate with miR-182 expression. LNCaP-miR-182 cells were also transfected with antisense miR-182 or control inhibitors. Inhibition of miR-182 increases FOXO1 levels. Densitometric quantification of FOXO1 relative to β-Tubulin from three independent experiments is graphically depicted. (B), Western blot analysis of putative targets of miR-182, FOXO3 and MITF, in LNCaP, Vector, and miR-182 cells. (C), LNCaP-miR-182 cells transfected with FOXO1 siRNA shows that miR-182-mediated downregulation of FOXO1 was comparable to that achieved by FOXO1-specific siRNA. Densitometric quantification of FOXO1 relative to β-Tubulin from three independent experiments is graphically depicted.
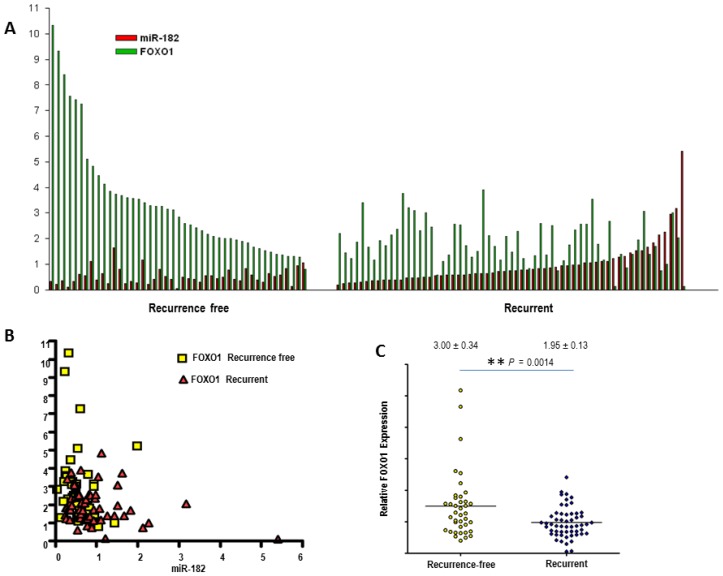
To further confirm that miR-182 blocks FOXO1expression, LNCaP-miR-182 cells were transfected with antisense miR-182 or negative control inhibitors. These are single-stranded RNA molecules which, after transfection, specifically inhibit miRNA function. As seen in Fig. 5A, inhibition of miR-182 resulted in elevation of FOXO1 protein levels, while negative control miRNA inhibitors had no effect (compare lanes 3, 4, and 5). Increased expression of FOXO1 after transfection of miR-182 inhibitor proves that miR-182 is regulating FOXO1 expression. LNCaP-miR-182 cells were also transfected with FOXO1 siRNA, and miR-182-mediated downregulation of FOXO1 was comparable to that achieved by FOXO1-specific siRNA (Fig 5C, compare lanes 4 and 5). While over-expression of miR-182 resulted in 76% knockdown of FOXO1, concomitant over-expression of miR-182 and FOXO1 siRNA increased the FOXO1 knockdown efficiency to 94%. To compare paired expression levels of FOXO1 and miR-182, we analyzed the expression of miR-182 and FOXO1 by qRT-PCR in groups of prostate cancer samples, characterized by recurrent or non-recurrent prostate cancer. As shown in Figure 6, we find that there is a significant inverse correlation between FOXO1 and miR-182 expression levels (Pearson's Test; p=0.0054). Together, these results indicate that FOXO-1 is regulated by miR-182.
Figure 6.
MiR-182 inversely correlates FOXO1 in primary prostate tumors. (A), FOXO1 levels inversely correlate with miR-182 expression in primary prostate tumor tissues, organized in aggressive and non-aggressive subgroups. Relative expression levels of miR-182 and FOXO1 were measured by qRT-PCR. (B) Nonlinear regression (curve fit) and Pearson product-moment correlation coefficient were used for correlation analysis. (C) mRNA expression levels of FOXO1 were assessed using qRT-PCR in groups of recurrent (n=61) and non-aggressive (n=45) prostate tumors. All samples were normalized to β-actin expression.
We then used qRT-PCR analysis on primary PCa specimens to assess whether FOXO1 expression levels are associated with clinicopathological features. We found that the expression of FOXO1 mRNA was significantly lower (p=0.0014) in tumors from patients who developed biochemical recurrence (1.95 ± 0.13) compared to tumors from patients who were recurrence-free five years after their radical prostatectomy (3.00 ± 0.34) (Fig. 6C).
Discussion
MiRNAs have been implicated in the post-transcriptional regulation of gene expression in diverse cellular processes. Recent studies have evaluated miRNAs as potential biomarkers of disease formation and progression in numerous cancer subtypes, in part based on the recognition that some miRNAs are abnormally expressed in tumor versus normal tissue12, and in many cases, in metastatic and aggressive disease11. One of the miRNAs that shows potential as a regulator of cancer metastasis is miR-182. In addition to studies of the immune system, melanoma, and breast cancer which suggest that miR-182 is an important oncogenic miRNA closely linked to tumor progression and metastasis, recent work has shown that miR-182 may be involved in prostate cancer progression10. While we were unable to validate the role of miR-182 as a prognostic biomarker, we demonstrated several novel findings regarding its role in prostate cancer progression.
First, we demonstrate the miR-182 expression was higher in cell lines known to exhibit more aggressive behaviour. This finding is in keeping with the hypothesis that miR-182 expression levels may be prognostically important10. We then showed that ectopic expression of miR-182 induced dramatic morphological changes and anchorage-independent growth of LNCaP prostate cancer cells. These cells also show increased migratory and invasive abilities. Finally, we identified FOXO1 as regulated by miR-182 in prostate cancer. This was done by first identifying specific gene targets of miR-182 through which it can promote invasive behaviour. We searched publically-available target prediction algorithms for putative target genes of miR-182 whose down-regulation could mediate the observed effects of miR-182 over-expression. These computational target prediction programs point to 3' UTR of FOXO1 mRNA as a direct target of miR-182, and this has already been confirmed by several groups using luciferase assay8,9,13. We then confirmed this relationship in a series of in vitro experiments as well as by analyzing 147 prostate cancer specimens. In particular, we showed that upregulation of miR-182 levels resulted in the reduction of FOXO1 protein levels in prostate cancer cells to the same extent as FOXO1-specific siRNA. Conversely, miR-182 inhibition rescued FOXO1 protein levels. The inverse correlation between the expression levels of mir-182 and FOXO1 was also observed in primary prostate tumors. These findings are in keeping with previous work which has demonstrated FOXO1 mRNA levels to be significantly decreased in cancerous as compared with normal prostate tissue14-17.
The Forkhead Box O family of transcription factors comprises three main members (FOXO1, FOXO3, and FOXO4) which are involved in many important cellular processes such as cell cycle regulation, cell differentiation, glucose metabolism, and other cellular functions18. FOXO proteins are mainly located in the nucleus and regulate expression of an extensive array of tumor suppression genes. FOXO1 is itself a tumor suppressor that negatively regulates the highly oncogenic phosphatidylinositol 3-kinase (P13K)/AKT signalling pathway19. It is located on the q14 band of chromosome 13 (13q14), where chromosomal deletions in cell lines, xenografts, and clinical specimens of PCa are common20. Here, we report that FOXO1 is also post-transcriptionally down-regulated via miRNA regulation. Our qRT-PCR analysis of FOXO1 and miR-182 mRNA levels in primary samples of prostate carcinoma show a direct correlation between over-expression of miR-182 and reduction of FOXO1 mRNA. This inverse correlation is especially intriguing when considering the established role of FOXO1 as a tumor suppressor in PCa. Recent data suggests that FOXO1 binds to and suppresses another transcription factor, Runx2, which plays a critical role in osteoblast maturation, bone formation, and prostate cancer cell metastasis21. While our studies validate FOXO1 as a target of miR-182, future studies are needed to identify and functionally evaluate downstream targets of FOXO1 that are related to prostate cancer progression and metastasis, and are affected by miR-182 expression levels.
There are indeed promising breakthroughs in the field of miRNA research, such that we can now envision miRNA-based therapeutics in the near future. In prostate cancer, for example, systemic delivery of a synthetic mimic of miR-34a has been used to inhibit prostate cancer metastasis and extend survival of tumor-bearing mice22. However, more research is needed in order to elucidate the interplay of various miRNAs involved in prostate cancer development and progression, and the exact mechanism of their action through their target genes. Our study presents a model of prostate cancer progression, in which increased miR-182 expression enhances cancer aggressiveness by promoting oncogenic and invasive characteristics. According to this model, up-regulation of miR-182 promotes metastasis by affecting tumor suppressor FOXO1. These findings demonstrate that miR-182 and its downstream effectors could prove to be valuable in future exploitation of miR-182 as a prostate cancer biomarker and therapeutic agent.
Acknowledgments
We thank Anthony Chow and Dr. Yutaka Amemiya for their technical assistance. Additionally, we thank Carlos S. Moreno for providing the Lentiviral pLemiR construct used in this research. This work was funded by Cancer Research Society and the Canadian Cancer Society Research Institute, grant no. 019038.
References
- 1.Siadat F, Sykes J, Zlotta AR, Not all gleason pattern 4 prostate cancers are created equal: A study of latent prostatic carcinomas in a cystoprostatectomy and autopsy series. The Prostate; 2015. [DOI] [PubMed] [Google Scholar]
- 2.Jung JW, Lee JK, Hong SK, Byun SS, Lee SE. Stratification of patients with intermediate-risk prostate cancer. BJU international. 2015;115(6):907–912. doi: 10.1111/bju.12703. [DOI] [PubMed] [Google Scholar]
- 3.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer research. 2009;69(19):7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 5.Eulalio A, Rehwinkel J, Stricker M. et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21(20):2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagga S, Bracht J, Hunter S. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. The Journal of biological chemistry. 2007;282(34):25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 8.Segura MF, Hanniford D, Menendez S. et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. The Journal of biological chemistry. 2009;284(35):23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanova-Salas I, Rubio-Briones J, Calatrava A. et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. The Journal of urology. 2014;192(1):252–259. doi: 10.1016/j.juro.2014.01.107. [DOI] [PubMed] [Google Scholar]
- 11.Gordanpour A, Stanimirovic A, Nam RK. et al. miR-221 Is down-regulated in TMPRSS2:ERG fusion-positive prostate cancer. Anticancer Res. 2011;31(2):403–410. [PMC free article] [PubMed] [Google Scholar]
- 12.Ambs S, Prueitt RL, Yi M. et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer research. 2008;68(15):6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stittrich AB, Haftmann C, Sgouroudis E. et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11(11):1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes DR, Yu J, Shanker K. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapointe J, Li C, Higgins JP. et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaTulippe E, Satagopan J, Smith A. et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer research. 2002;62(15):4499–4506. [PubMed] [Google Scholar]
- 17.Tomlins SA, Mehra R, Rhodes DR. et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39(1):41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120(Pt 15):2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 19.Kau TR, Schroeder F, Ramaswamy S. et al. A chemical genetic screen identifies inhibitors of regulated nuclear export of a Forkhead transcription factor in PTEN-deficient tumor cells. Cancer cell. 2003;4(6):463–476. doi: 10.1016/s1535-6108(03)00303-9. [DOI] [PubMed] [Google Scholar]
- 20.Dong XY, Chen C, Sun X. et al. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer research. 2006;66(14):6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Pan Y, Zheng L. et al. FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer research. 2011;71(9):3257–3267. doi: 10.1158/0008-5472.CAN-10-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Kelnar K, Liu B. et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature medicine. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]