Abstract
Leucine-rich repeat (LRR) is a versatile motif widely present in adhesive proteins and signal-transducing receptors. The concave structure formed by a group of LRRs is thought to facilitate binding to globular protein domains with increased affinities. However, little is known about the conformational dynamics of LRRs in such a structure, e.g., whether and how force induces conformational changes in LRRs to regulate protein binding and signal transduction. Here we investigated the platelet glycoprotein Ibα (GPIbα), a demonstrated mechanoreceptor with known crystal structures for the N-terminal domain (GPIbαN), as a model for LRR-containing proteins using a combined method of steered molecular dynamics simulations and single-molecule force spectroscopy with a biomembrane force probe. We found that force-induced unfolding of GPIbαN starts with LRR2–4 and propagates to other LRRs. Importantly, force-dependent lifetimes of individual VWF-A1 bonds with GPIbα are prolonged after LRR unfolding. Enhancement of protein-protein interactions by force-induced LRR unfolding may be a phenomenon of interest in biology.
Main Text
Binding of von Willebrand factor (VWF) to platelet membrane receptor glycoprotein Ibα (GPIbα) initiates hemostasis and thrombosis under high shear conditions, mostly seen in arteries and restricted vessels (1). VWF is a multimeric protein composed of identical 250-kDa subunits. Each subunit has a single 24-kDa A1 domain that binds to the 45 kDa, ∼280 residues, N-terminal domain of GPIbα (GPIbαN). The crystal structures of the A1-GPIbαN complex (2, 3) reveal that GPIbαN consists of eight leucine-rich repeats (LRRs, K19–L208) that form an elongated shape (cf. Fig. 1 A). The concave LRR framework grabs the A1 in a pincerlike grip with an N-terminal contact (involving the β-finger motif and LRR1) and a C-terminal contact (involving LRR5–8 and the β-switch loop). Intriguingly, the middle portion of the GPIbαN, from LRR2–4, makes no contacts to the A1 in the crystal structures; yet, these LRRs are required for ristocetin-dependent and shear dependent VWF binding (4). Because crystal structures were obtained in the absence of force, they provide little insight into any possible conformational change induced by shear.
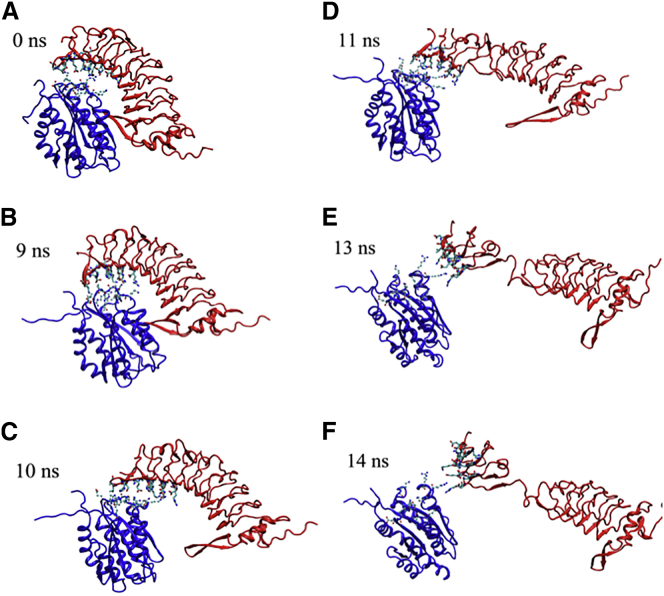
Figure 1.
SMD simulation of GPIbαN dissociation from the A1 domain. Sequential snapshots of representative SMD-simulated structures revealing that force induces GPIbαN unfolding in the middle region of GPIbαN (LRR2–4). (Blue, A1; red, GPIbαN.) The equilibrated structure (A) was used as the starting structure for SMD simulations, which generated the structures in (B)–(F) at the indicated times. To see this figure in color, go online.
Single-molecule force spectroscopic studies using atomic force microscopy, an optical trap, and a biomembrane force probe (BFP) suggested that at least two conformational states may exist upon formation of the receptor-ligand complex (5). A recent crystal structure of a gain-of-function mutant A1 complexed with GPIbαN revealed A1 interaction with LRR3-5 that further enhances the A1-GPIbαN affinity (6). These findings suggest that force may induce a conformational change that alters the central A1-GPIbα LRR contact to enhance affinity.
Force induces LRR conformational change on A1-GPIbα bond in simulations
To investigate whether the force applied on the A1-GPIbα complex can cause significant protein conformational change, we carried out 11 independent steered molecular dynamics (SMD) simulations and observed in four trajectories unfolding of GPIbαN before dissociation from A1. At the starting point (t = 0), GPIbαN contacts A1 via the β-switch and β-finger (Fig. 1 A). At t > 0, the Cα atom of GPIbαN C-terminal residue L267 was pulled to move horizontally at a constant speed to the right with the Cα atom of the A1 N-terminal residue H1268 harmonically constrained. Upon loosening of the β-switch from the A1 central β-sheet at 9 ns (Fig. 1 B), the GPIbαN started to tilt about the β-finger–A1 hinge point, a counterclockwise rotation that continued throughout the remainder of the simulations. At 10 ns, the GPIbα β-switch dissociated from the A1 central β-sheet, but the β-finger remained bound to A1, although interacting with different A1 residues from those seen at 0 ns (Fig. 1 C). At 11 ns, several hydrophobic interactions near the GPIbαN β-finger were ruptured by further pulling (Fig. 1 D). Surprisingly, a few residues spanning the LRR3, T68–L89, uncoupled from each other and became loose. At 12 ns, the LRR3 was completely unfolded and extended by force (Fig. 1 E). At 14 ns, the region below LRR3, e.g., LRR4, started getting loose; however, the GPIbαN dissociated from A1 before complete unfolding of LRR4 was observed (Fig. 1 F). Additional simulations with different force loading rates and anchor points showed similar LRR unfolding behavior (Fig. S1 in the Supporting Material). Regardless of the anchor point, GPIbαN unfolding started at LRR2-3 in a fast pulling process, suggesting that rapid loading facilitates LRR extension (Fig. S1, B and C).
Experimental characterization of force-induced conformational change in individual A1-GPIbα complexes
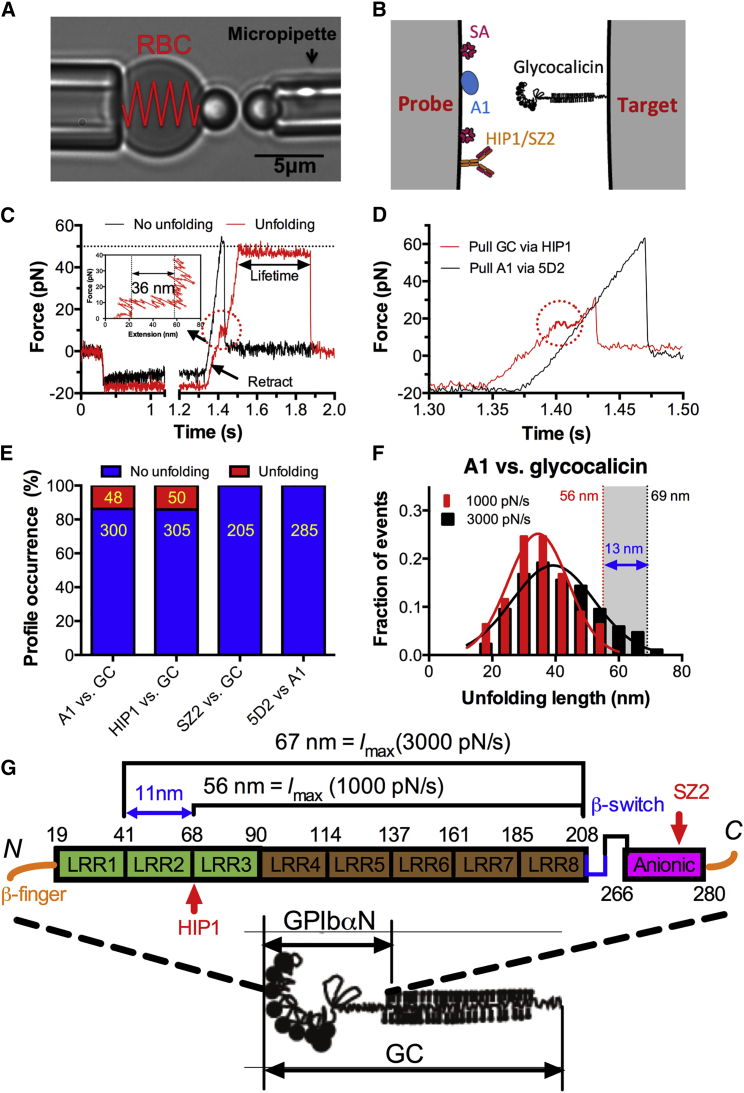
We next used the previously described BFP force spectroscopy (7, 13) to observe conformational change in single A1-GPIbα complexes and its impact on force-dependent bond lifetimes (Fig. 2, A and B). In the BFP force traces, a sudden force drop occurring at 5–20 pN, caused by an abrupt length increase between the probe and the target, was consistently (∼14% occurrence) observed in the retraction phase when a ramp force (1000 pN/s) was applied to the extracellular portion of GPIba, glycocalicin (GC), via A1 (Fig. 2, C and E). Pulling GC via a monoclonal antibody (mAb) HIP1 yielded a similar unfolding occurrence as pulling via A1 (Fig. 2 E). In contrast, unfolding event was eliminated when HIP1 was replaced by another mAb SZ2 on the probe or when GC was replaced by an anti-A1 mAb 5D2 on the target (Fig. 2, D and E). This indicates that it was GC, not A1, that was being unfolded, and that unfolding occurred within the LRR region, because the binding epitope of SZ2 is mapped to the anionic region below all LRR domains (8) (Fig. 2 G).
Figure 2.
GPIbαN unfolding observed by BFP force-clamp assay. (A) BFP photomicrograph. A micropipette-aspirated RBC with a bead (left, probe) attached to the apex formed a picoforce sensor. It was aligned with another bead (right, target) aspirated by an apposing micropipette. (B) BFP functionalization with indicated molecules. (C and D) Force versus time traces from representative force-clamp cycles for A1-GC bond lifetime measurement (C) and antibody-protein stretch assay (D). (Inset) The putative unfolding event is circled and zoomed-in to show the details. (E) Occurrence frequencies of unfolding-versus-no-unfolding events (numbers are indicated in yellow) from all binding events mediated by the indicated interactions. (F) Normalized histograms (bar) and their Gaussian fits of unfolding length. (G) Glycocalicin schematic (left) and GPIbαN domain organization (right). Starting and ending residues for each domain as well as the binding epitopes of two anti-GPIbα mAbs are all indicated. To see this figure in color, go online.
The force-extension analysis showed that the extension lengths range from the minimum of 18 nm (lmin = length of one LRR, assuming a contour length of 4 Å per residue) to the maximum of 56 nm (lmax = length of six LRRs) (Fig. 2, F and G). Surprisingly, the A1–GC unfolding length histogram right-shifted when force loading rate was increased from 1000 to 3000 pN/s (Fig. 2 F). The unfolding length difference between the lmax(3000 pN/s) (= 69 nm, i.e., LRR2-8) and lmax(1000 pN/s) (= 56 nm, i.e., LRR3–8) is ∼13 nm (Fig. 2 G), matching the length difference (∼11 nm) between the unfolding starting points (LRR2 to LRR3) by different SMD ramping processes (Fig. S1). The combined SMD simulations (Figs. 1 and S1) and the BFP results indicated that force can unfold multiple repeats of the LRR region starting from the noncontact LRR2-4.
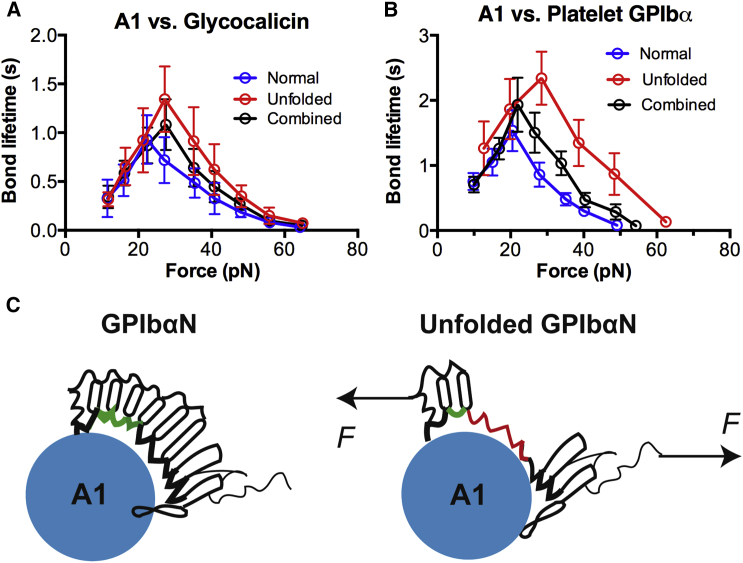
To investigate the functional consequence of the force-induced GPIbα unfolding, we measured lifetimes of A1 bonds with GC on a bead (Fig. 3 A) or GPIbα on a platelet (Fig. 3 B). Consistent with our previous reports (7, 9, 13), force counterintuitively prolongs bond lifetime (catch bond) from 10 to 25 pN. Further increase in force reduces bond lifetime (slip bond). To delineate the role of force-induced unfolding, we segregated the lifetimes into two subgroups based on whether an unfolding event was observed in the force-ramp phase (Fig. 2 C). In >20 pN forces, however, bond lifetimes with unfolding events became longer than those without unfolding events (Fig. 3, A and B). This indicates that GPIbα unfolding can immediately prolong the lifetime of its bond with VWF-A1.
Figure 3.
Unfolding of GPIbαN prolongs the bond lifetime. (A and B) Plots of lifetime (mean ± SE of >20 measurements per point) versus force of VWF-A1 bonds with GC (A) or GPIbα on platelets (B). (C) Schematic of the A1-GPIbαN complex in normal state (left) and force, induced a longer-lived state (right). (Horizontal arrows) Tensile forces. To see this figure in color, go online.
Most studies on the effect of force on the structure of the VWF-GPIbα axis focus on shear-induced conformational changes in the VWF. As a long molecule (>40 nm above the membrane) (10), however, GPIbα may also appear susceptible to the conformational change upon force application. A recent study identified a mechanosensitive domain in the juxtamembrane region below the GC portion that can be unfolded by pulling force (11). The authors suggest that no other GPIbα region can be unfolded, as mutations in this mechanosensitive domain abolished the force-induced unfolding they observed. Here we show GPIbαN conformational changes that enhance binding to VWF-A1, indicating the functional relevance of the LRR unfolding. Our results suggest that force extends the GPIbαN concave structure by unfolding a few LRRs, allowing the globular A1 domain to fit better into the enlarged binding pocket and reach a longer-lived state (Fig. 3 C). This may explain platelets agglutination via VWF-GPIbα bonds without activation or binding of integrin GPIIb/IIIa at sites of atherothrombosis where severe vessel narrowing produces extremely high shears (>10,000 s−1) (12). Whether GPIbα mechanosensitivity endows this molecule with the capacity of mechanosensing requires future studies.
Author Contributions
L.J. and C.Z. designed experiments; L.J., J.L., Y.C., and Z.L. performed experiments and analyzed the data; and all authors contributed to writing.
Acknowledgments
We thank Renhao Li and the Emory blood bank for providing outdated platelets, and Jing-fei Dong and Vince F. Fiore for purifying glycocalicin.
This work was supported by National Institutes of Health grants No. HL091020 and No. HLR01HL093723 (to C.Z.).
Editor: Nathan Baker.
Footnotes
Supporting Materials and Methods and one figure are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00946-7.
Contributor Information
Lining Ju, Email: arnold.ju@sydney.edu.au.
Cheng Zhu, Email: cheng.zhu@bme.gatech.edu.
Supporting Material
References
- 1.Gardiner E.E., Andrews R.K. Structure and function of platelet receptors initiating blood clotting. Adv. Exp. Med. Biol. 2014;844:263–275. doi: 10.1007/978-1-4939-2095-2_13. [DOI] [PubMed] [Google Scholar]
- 2.Huizinga E.G., Tsuji S., Gros P. Structures of glycoprotein Ibα and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 3.Dumas J.J., Kumar R., Mosyak L. Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibα complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J. Biol. Chem. 2004;279:23327–23334. doi: 10.1074/jbc.M401659200. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y., Cranmer S.L., Andrews R.K. Leucine-rich repeats 2–4 (Leu60-Glu128) of platelet glycoprotein Ibα regulate shear-dependent cell adhesion to von Willebrand factor. J. Biol. Chem. 2006;281:26419–26423. doi: 10.1074/jbc.M604296200. [DOI] [PubMed] [Google Scholar]
- 5.Liu B., Chen W., Zhu C. Molecular force spectroscopy on cells. Annu. Rev. Phys. Chem. 2015;66:427–451. doi: 10.1146/annurev-physchem-040214-121742. [DOI] [PubMed] [Google Scholar]
- 6.Blenner M.A., Dong X., Springer T.A. Towards the structural basis of regulation of von Willebrand factor binding to glycoprotein Ib. J. Biol. Chem. 2014;289:5565–5579. doi: 10.1074/jbc.M113.511220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ju L., Dong J.-F., Zhu C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibα. J. Biol. Chem. 2013;288:32289–32301. doi: 10.1074/jbc.M113.504001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y., Romo G.M., Andrews R.K. Requirement of leucine-rich repeats of glycoprotein (GP) Ibα for shear-dependent and static binding of von Willebrand factor to the platelet membrane GP Ib-IX-V complex. Blood. 2000;95:903–910. [PubMed] [Google Scholar]
- 9.Yago T., Lou J., Zhu C. Platelet glycoprotein Ibα forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J. Clin. Invest. 2008;118:3195–3207. doi: 10.1172/JCI35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox J.E., Aggerbeck L.P., Berndt M.C. Structure of the glycoprotein Ib⋅IX complex from platelet membranes. J. Biol. Chem. 1988;263:4882–4890. [PubMed] [Google Scholar]
- 11.Zhang W., Deng W., Li R. Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood. 2015;125:562–569. doi: 10.1182/blood-2014-07-589507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggeri Z.M., Orje J.N., Reininger A.J. Activation-independent platelet adhesion and aggregation under elevated shear stress. Blood. 2006;108:1903–1910. doi: 10.1182/blood-2006-04-011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju L., Chen Y., Cruz M.A. Von Willebrand factor-A1 domain binds platelet glycoprotein Ibα in multiple states with distinctive force-dependent dissociation kinetics. Thromb. Res. 2015;136:606–612. doi: 10.1016/j.thromres.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.