Abstract
Monodisperse lipid nanodiscs are particularly suitable for characterizing membrane protein in near-native environment. To study the lipid-composition dependence of photocycle kinetics of bacteriorhodopsin (bR), transient absorption spectroscopy was utilized to monitor the evolution of the photocycle intermediates of bR reconstituted in nanodiscs composed of different ratios of the zwitterionic lipid (DMPC, dimyristoyl phosphatidylcholine; DOPC, dioleoyl phosphatidylcholine) to the negatively charged lipid (DOPG, dioleoyl phosphatidylglycerol; DMPG, dimyristoyl phosphatidylglycerol). The characterization of ion-exchange chromatography showed that the negative surface charge of nanodiscs increased as the content of DOPG or DMPG was increased. The steady-state absorption contours of the light-adapted monomeric bR in nanodiscs composed of different lipid ratios exhibited highly similar absorption features of the retinal moiety at 560 nm, referring to the conservation of the tertiary structure of bR in nanodiscs of different lipid compositions. In addition, transient absorption contours showed that the photocycle kinetics of bR was significantly retarded and the transient populations of intermediates N and O were decreased as the content of DMPG or DOPG was reduced. This observation could be attributed to the negatively charged lipid heads of DMPG and DOPG, exhibiting similar proton relay capability as the native phosphatidylglycerol (PG) analog lipids in the purple membrane. In this work, we not only demonstrated the usefulness of nanodiscs as a membrane-mimicking system, but also showed that the surrounding lipids play a crucial role in altering the biological functions, e.g., the ion translocation kinetics of the transmembrane proteins.
Introduction
Among the widely used membrane-mimicking systems, monodisperse lipid nanodiscs provide a native-like membrane environment with great stability and flexibility in lipid composition. Therefore, it has been successfully employed to solubilize functional membrane proteins (1, 2, 3) and the major light-harvesting complex II (4). Subsequently, biophysical functions of many membrane proteins have been characterized in this near-native membrane-mimicking system, including phosphorylation study of rhodopsin (5), signaling pathway of the G protein-coupled receptors (6, 7), and high-resolution protein structure determination (8, 9). Lipid nanodiscs are small disk-shaped lipid bilayers, wrapped with two copies of membrane scaffold proteins (MSP) (8, 10, 11, 12). By adjusting the length of MSP constructs, the size of nanodiscs can range from 8.5 to 13 nm in diameter (8, 11, 12). It has been reported that nanodiscs can be assembled using various lipids, including the zwitterionic, anionic, and synthetic cationic lipids, and even native lipids from membrane extracts (3, 10, 12, 13, 14, 15).
Bacteriorhodopsin (bR), a transmembrane protein in the halophile Halobacterium salinarum (16), has been successfully reconstituted into nanodiscs composed of zwitterionic lipids (9, 17, 18). The functional inner core of bR was found to be almost unperturbed in nanodiscs (9) with its photocycle activity reserved (18). Initiated by an exposure to a yellow-green illumination, the photocycle of bR was characterized in terms of the status of the retinal Schiff base (19),
| (1) |
The first process is the photochemical trans-to-cis isomerization at C13=C14 of the retinal, in association with the generation of intermediate K (20). A consequent proton migration from the protonated Schiff base to Asp85 in the bR interior leads to the L-to-M transition (21, 22, 23), coinciding with the proton release at the extracellular side (24) through a hydrogen-bonding network involving the residues Glu194, Glu204, and Arg82 (21, 25). The following reprotonation of the deprotonated 13-cis Schiff base from the proton donor Asp96 leads to the decay of intermediate M, i.e., the generation of intermediate N (23, 26). Afterwards, the transition from intermediate N to intermediate O is attributed to the retinal reisomerization from 13-cis to all-trans (27) and the reprotonation of Asp96 through the proton collecting funnel, comprising Asp36, Asp102, Asp104, and Glu161 (28). The decay of intermediate O is determined by the reprotonation of Glu204 from Asp85 and the relaxation of the twisted retinal (29). The structures of the intermediates were also characterized using x-ray diffraction (30).
A primitive unit of purple membrane (PM) in hexagonal crystalline lattice is composed of 3 bRs and 10 lipids per bR (31, 32), including two-to-three PGP-Me (phosphatidylglycerophosphate methyl ester lipids), three glycolipid sulfates, two SQs (squalenes), one PG (phosphatidylglycerol), one archaeal GlyC (glycocardiolipin), small amounts of PGS (phosphatidylglycerosulfate), BPG (bisphosphatidylglycerol), and a negligible amount of vitamin MK8 (33). To study the effect of chemical environments on structural stability and photocycle kinetics of bR, bR has been reconstituted into lipid bilayer liposomes (34), delipidated PM (35), and detergent micelles (36, 37). The results from these studies showed that the individual lipids, especially squalene and PGP, played essential roles in the conventional photocycle of bR and light-driven proton pump activity (31, 37, 38).
In this work, we used the advantages of nanodiscs to provide bR with a homogeneous lipid environment with exact control of ratios of zwitterionic lipid PC (phosphatidylcholine) and anionic lipid PG composed of saturated (DM, dimyristoyl) and unsaturated (DO, dioleoyl) hydrocarbon tails (Fig. 1). Suggested by the study of proteorhodopsin in nanodiscs (39), MSP1E3D1 was chosen to be the membrane scaffold protein because it yields larger nanodiscs, which impart less structural confinement than the smaller nanodiscs assembled using MSP1D1. The photocycle kinetics of bR reconstituted in nanodiscs composed of different lipid ratios served as an indicator to understand the effects of the surrounding lipids of bR. Through the systematic study of monomeric bR reconstituted in nanodiscs composed of different lipid ratios, our results showed that lowered content of the negatively-charged lipid PG resulted in decelerated kinetics of intermediate M, correlating to the retarded proton translocation and the decrease in the transient populations of intermediates N and O. Herein, we demonstrated that the bR photocycle kinetics is controllable by systematically tuning the composition of the lipids in the nanodiscs.
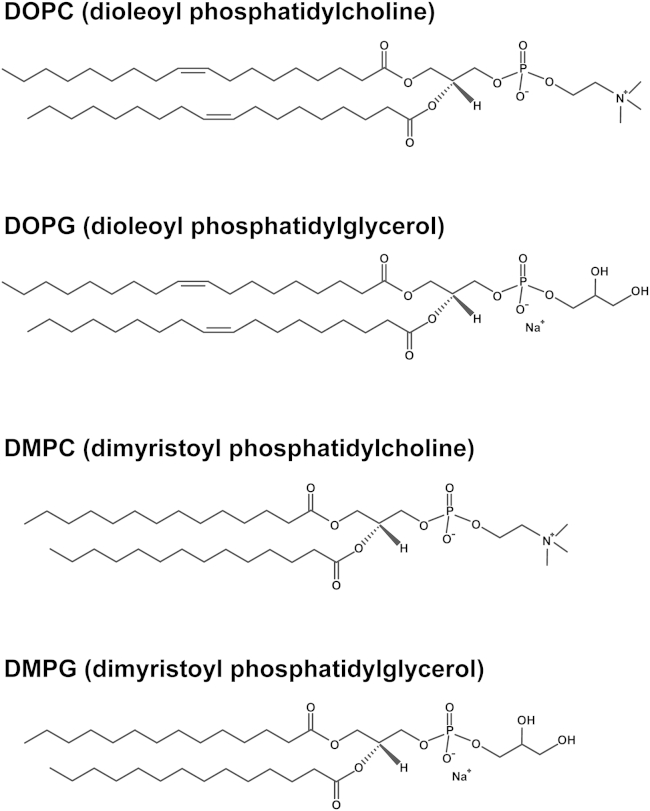
Figure 1.
The molecular structures of lipids used for nanodiscs assembly, including DOPC (dioleoyl phosphatidylcholine), DOPG (dioleoyl phosphatidylglycerol), DMPC (dimyristoyl phosphatidylcholine), and DMPG (dimyristoyl phosphatidylglycerol).
Materials and Methods
Materials preparation
Preparation of monomeric bR in the micelle of Triton X-100
The PM from H. salinarum S9 strain was prepared and purified following a standard method (40). The detergent-solubilized monomeric bR (mbR) was prepared by mixing purified PM with detergent Triton X-100 at a ratio of 1:7 (w/w) (41). The solubilizing process was carried out in a dark environment for 72 h at 25°C. The completely solubilized bR was collected from the supernatants after three centrifugations at 18,400 × g for 30 min and was light-adapted for 30 min before further experiments. The absorption maximum of the retinal of light-adapted mbR in micelle shifted from 568 to 553 nm (41). The concentration of bR was controlled to be 10 μM in a buffer containing 0.5× phosphate buffer (Sigma-Aldrich, St. Louis, MO) and additional 100 mM NaCl at pH 5.8.
Preparation of monomeric bR in lipid nanodiscs
The plasmid used to express membrane scaffold protein MSP1E3D1, was obtained from Professor Wagner of Harvard Medical School. MSP1E3D1 was expressed using Escherichia coli Rosetta 2(DE3) (Novagen, Billerica, MA) and grown in 37°C in 2 L of LB medium with a Winpact Bench-Top Fermenter (Major Science, Taipei, Taiwan). The dissolved-oxygen level was maintained at 60 ± 7.5% with maximum and minimum agitation of 400 and 50 rpm, respectively. The protein expression was induced with 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) when OD600 reached >0.6 and was harvested after 5 h of induction. The cells were collected by centrifugation and stored at –80°C until purification. The purification procedure was outlined in the literature (10, 12).
DMPC (dimyristoyl phosphatidylcholine), DOPC (dioleoyl phosphatidylcholine), DMPG (dimyristoyl phosphatidylglycerol), and DOPG (dioleoyl phosphatidylglycerol) were purchased from Avanti Polar Lipids (Alabaster, AL) and were solubilized to stock solution using a buffer solution containing Triton X-100. The concentrations of lipids and Triton X-100 in the stock solution were 50 and 200 mM, respectively. They were then made into different molar ratios of DOPC/DOPG, DMPC/DMPG, or DMPC/DOPG mixtures. The appropriate lipid/triton mixture, solubilized monomeric bacteriorhodopsin and MSP1E3D1 solution, were combined and incubated for 2 h at 4°C, with the desired final molar ratios for each mixture lipids shown in Table 1. With the sample preparation condition we provided, <4% of lipids in nanodiscs could potentially come from the remaining native lipids from PM, whose effect is therefore not significant. Detergent was removed by overnight treatment at 4°C with 800 mg of BioBeads SM-2 (Bio-Rad Laboratories, Hercules, CA) per 1 mL of assembly mixture, after which the BioBeads were separated from the assembled nanodisc solutions using centrifugation. Additional precipitate in the solutions of assembled nanodiscs was removed by fast centrifugation using a model No. 5424R centrifuge (Eppendorf, Hamburg, Germany) at a top speed of 15,000 rpm (21,130 × g). The sample was applied to a Resource Q anion exchange column (GE Healthcare, Buckinghamshire, UK) at 4°C for further purification. Using a gradient of linearly increasing salt concentration and pH, from 0 to 1 M NaCl and from pH 6.5 to 8, respectively, in elution buffer, containing 25 mM Tris-HCl and 0.5 mM EDTA, the sample components were eluted from the column, with the peak elution monitored at an absorption of 280 nm.
Table 1.
The molar ratios of lipid/bR in different bR-embedded lipid nanodiscs
| 100:0 | 75:25 | 50:50 | 25:75 | 0:100 | |
|---|---|---|---|---|---|
| DMPC/DMPG | |||||
| Lipid/MSP | 90:1 | 67:1 | 60:1 | 50:1 | 50:1 |
| MSP/mbR | 8:1 | 6:1 | 6:1 | 6:1 | 6:1 |
| DOPC/DOPG | |||||
| Lipid/MSP | 70:1 | 65:1 | 47:1 | 40:1 | 40:1 |
| MSP/mbR | 6:1 | 6:1 | 6:1 | 6:1 | 6:1 |
| DMPC/DOPG | |||||
| Lipid/MSP | 90:1 | 70:1 | 55:1 | 50:1 | 40:1 |
| MSP/mbR | 8:1 | 4:1 | 4:1 | 6:1 | 6:1 |
After anion exchange purification, the samples of assembled nanodiscs were collected and concentrated to an appropriate volume for further purification by size-exclusion chromatography (SEC). Solutions of assembled nanodiscs were injected onto Superdex 200 prep grade gel filtration column (GE Healthcare) at 4°C to separate unwanted aggregation from the nanodiscs. The size-exclusion buffer consisted of 25 mM Tris-HCl, 100 mM NaCl, and 0.5 mM EDTA at pH 7.5. Fractions expected to contain assembled nanodiscs were taken to run SDS (sodium dodecyl sulfate)-polyacrylamide gel electrophoresis 5–13% Tris-glycine gel to confirm the existence of both bacteriorhodopsin and MSP1E3D1, thus verifying the incorporation of bacteriorhodopsin into nanodiscs, shown in Fig. S1 of the Supporting Material. The concentration of nbR was controlled to be 10 M in a buffer containing 0.5× phosphate buffer (Sigma-Aldrich) and additional 100 mM NaCl at pH 5.8.
Steady-state spectroscopic measurements
The steady-state ultraviolet-visible absorption spectra were monitored with a model No. USB4000-UV-VIS spectrometer (Ocean Optics, Ostfildern, Germany). The circular dichroism (CD) spectra were recorded with a model No. J-815 spectrometer (JASCO, Fukuoka, Japan) and averaged for 1.2 s at 1-nm intervals in 700–400 nm at 28°C. A cuvette of optical path length of 1 cm was employed in these measurements.
Transient absorption in 380–710 nm
A basiScan M/120/HE optical parametric oscillator (Spectra-Physics, Santa Clara, CA) pumped by a model No. INDI-40-10 frequency-tripled Nd:YAG laser (Spectra-Physics) provided the 6-ns pulses at 532 nm with the incident excitation fluence of 0.4 mJ cm–2. A model No. 10Q20BB1 broadband beamsplitter (Newport, Irvine, CA) was mounted in the midst of the 532-nm excitation laser toward the sample compartment for the concurrent energy monitor. In addition, a model No. VS25S2T1 mechanical shutter (Uniblitz, Rochester, NY) was used to decouple the 10-Hz laser irradiation to 5, 2.5, 2, 1, and 0.5 Hz to optimize the repetition rates to prevent overshooting. Depending on the recovery kinetics of different monomeric bR in lipid nanodiscs (nbRs), an appropriate repetition rate was chosen for each sample. A dim deuterium/tungsten halogen lamp module, model No. DH-2000 (Ocean Optics), combined with different color filters, served as the stationary probing light. After passing through the cell compartment, the probe beam was further introduced to a monochromator to define the detection wavelengths in the range of 380–710 nm at 10-nm intervals. The excitation laser beam was aligned perpendicularly with respect to the probe beam and overlapped in the center of the sample cell. A model No. XNF-532.0-25.0M 532-nm notch filter (CVI Laser Optics, Albuquerque, NM) was positioned in front of the photomultiplier to reduce the light scattering from the excitation laser. The optical modulation was monitored by a model No. R928 photomultiplier (Hamamatsu, Hamamatsu City, Japan) and further recorded by a model No. 24MXs-B 200 MHz digital oscilloscope (Teledyne LeCroy, Chestnut Ridge, NY). Temporal profiles upon 550–1200 laser excitations were averaged to increase the signal-to-noise ratio. The evolution of the absorbance difference, ΔAt, was derived using
| (2) |
where St and S0 represent the dc-coupled voltages in the presence and absence of the excitation laser, respectively.
Results and Discussion
The bR-embedded lipid nanodiscs were characterized using SEC and anion-exchange chromatography. Steady-state absorption and CD spectra were collected to examine the conformations of the retinal Schiff base in the retinal pocket and the monomeric configurations of nbR, respectively. The time-resolved difference spectra provided the kinetics of the photocycle intermediates in different nanodiscs as the composition of the lipids was changed.
Determination of size and surface charges of bR-embedded nanodiscs
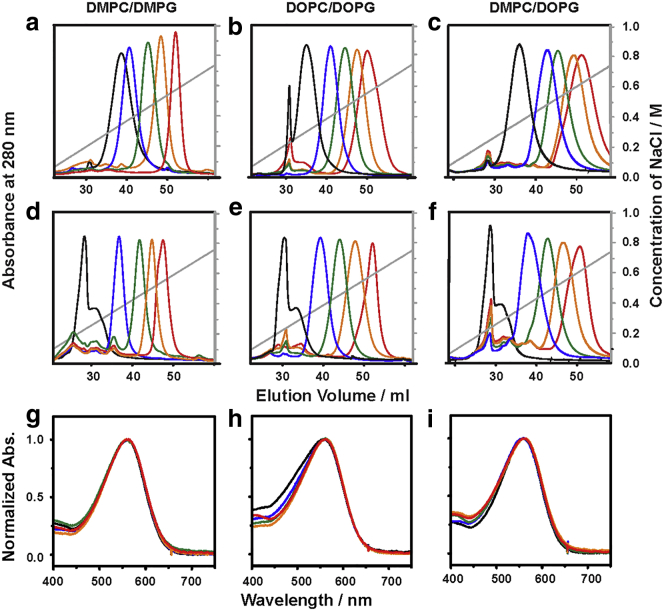
The sizes of bR-embedded lipid nanodiscs were estimated to be 140 ± 25 kDa using SEC. Besides using SEC to estimate the sizes of the bR-embedded nanodiscs, anionic-exchange chromatography was performed to evaluate the surface charges of each sample, shown in Fig. 2, a–f. With a gradient of linearly increasing salt concentration applied to elute the samples from Resource Q (GE HealthCare), greater elution volume indicates nanodiscs with greater surface charges. The nanodiscs having higher content of PG (negatively charged heads) results in greater elution volumes, as shown in red traces in Fig. 2, a–c, for the bR-embedded nanodiscs and Fig. 2, d–f, for the empty nanodiscs. The analyses of surface charges of nanodiscs using anion-exchange chromatography indicated that the overall charge in the peripherals of bR was negative. In addition, the overall surface charge increased with the increasing amount of negatively charged lipids incorporated, which was consistent with the ζ-potential measurement, as shown in Fig. S4. We demonstrated that the tuning of the lipid composition using PC and PG alters the extent of the surface charges, and the alteration in the surface charge might lead to the change in the photocycle kinetics in the later steps, referring to intermediate M, N, and O as proton translocations are assisted by the surrounding lipids (42).
Figure 2.
Anionic exchange chromatography profiles of (a–c) bR embedded nanodiscs, (d–f) empty nanodiscs, and (g–i) corresponding normalized steady-state absorption spectra of bR-embedded nanodiscs, composed of (a, d, and g) DMPC/DMPG, (b, e, and h) DOPC/DOPG, and (d, f, and i) DMPC/DOPG. The molar ratios of the PC/PG lipids are 100:0 (black), 75:25 (blue), 50:50 (green), 25:75 (orange), and 0:100 (red).
Steady-state absorption spectra
The mbR possesses a characteristic band at 553 nm (36, 43), whereas the observed contours of nbRs exhibited a slight red shift to 560 ± 1 nm and were almost independent of the content of the PG and PC lipids after 30 min light adaptation, as shown in Fig. 2, g–i. Although many reports demonstrated that no significant change in the secondary structure of bR when PM was solubilized with Trion X-100 (36), we observed only a minute spectral shift of nbR relative to the trimeric bR in PM, which suggests that the tertiary structure of bR in nanodiscs is more similar to bR in PM than mbR in Triton X-100. The observed minor shift of the absorption contours of bR in nanodiscs, with respect to bR in PM, suggests a slight change of the structure of bR, in association with the electronic energy levels of the retinal Schiff base. Hoffmann et al. (44) have studied the spectral properties of the retinal bands in highly structural similar of the microbial rhodopsins, bacteriorhodopsin, and sensory rhodopsin. Their results showed that the spectral difference could be attributed to the interaction of the retinal and the amino acids in the retinal binding pocket at the extracellular side. Brouillette et al. (45) demonstrated that adjusting the packing between the nonnative amphiphiles (SDS, DMPC, and CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate)) and bR by changing the composition of the detergents in the micelles may loosen the helical packing because the crystalline lattice is absent in monomeric bR.
Alternatively, one could also attribute the spectral shifts of nbR to the different population of the 13-cis and all-trans retinal in the light-adapted form. An early study by Casadio et al. (46) found different ratios of 13-cis/all-trans retinal in light-adapted form in PM and Triton X-100 micelles. The percentage of the all-trans retinal in PM (82%) is higher than that in Triton X-100 solubilized bR (62%) (46). Comparing with the absorption spectra of the light-adapted PM and mbR (43), we have concluded that the richer the all-trans retinal, the more red-shifted the absorption maximum. As a result, the red-shifted band of nbR suggests that there is a larger all-trans retinal population in the light-adapted nbR compared to that in mbR. Moreover, no spectral shifts were observed as the lipid composition is changed, indicating that the nanodiscs provided a milder and more native environment than Triton X-100 micelles regardless of the types of lipids in the nanodiscs.
Besides the absorption contours of the retinal moiety, the monophasic CD spectra, shown in Fig. S2, indicate that nbRs were in the monomeric form in these chemical microenvironments (17, 47). The CD spectroscopic results were consistent with a previous study that showed bR was in the monomeric form as the DMPC/bR ratio is higher than 260 in a 12.1-nm nanodisc (17). Johnson et al. (18) prepared the bR trimer in the 12.9-nm nanodiscs using a lipid/bR ratio of 33. When we replaced PC with PG, the nbR could still possess the monomeric CD feature although the lipid/bR ratios were slightly lowered compared to DMPC, shown in Table 1. Therefore, the distinct difference in the photocycle kinetics in the following measurements was mainly ascribed to the content of negatively charged lipids in single bR-embedded nanodiscs, instead of the bR trimers in the nanodiscs.
Analyses of the time-resolved difference absorption contours
Before collecting the difference spectra over the detection wavelengths in 420–650 nm, the repetition rates of the 532-nm pulses were confirmed to prevent the overshooting by monitoring the recovery of the parent state at 560 nm, shown in Fig. S3. Comparing the temporal profiles, the repetition rates operated for the acquisitions of the transient absorption spectra of each nbR sample were listed in Table S1 in the Supporting Material.
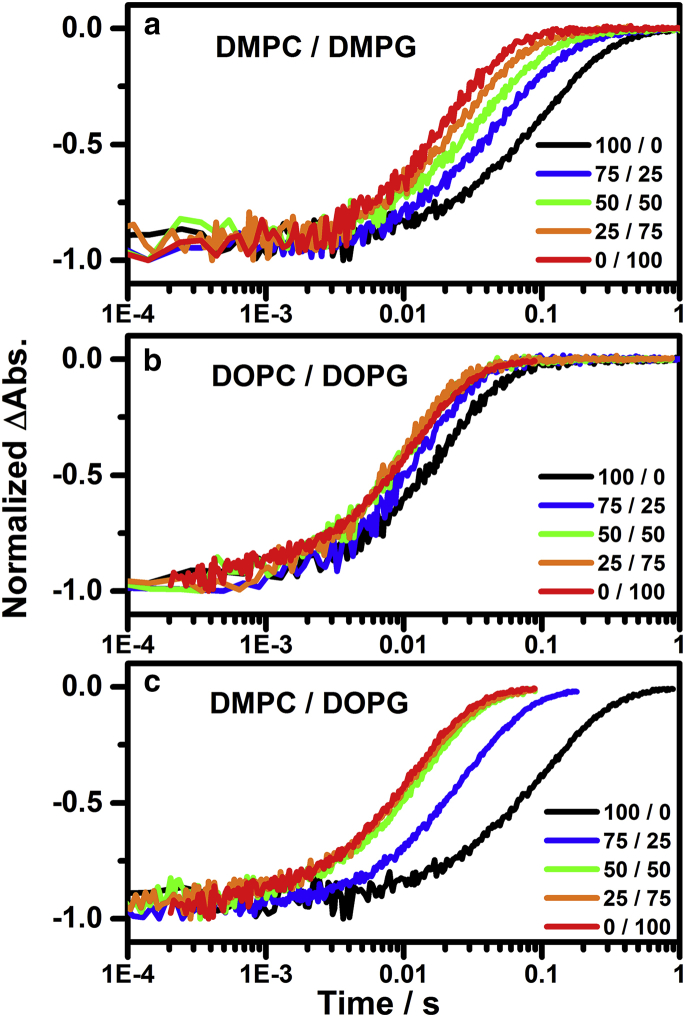
Surveying the temporal profiles of the recovery of parent state at 560 nm in Fig. 3, we found an increase in the rate of the recovery when the content of PG was increased whether the hydrophobic tails were composed of saturated (DM) or unsaturated (DO) hydrocarbons. Surveying the native lipid composition in PM, the percentage of PGP-Me, PG, and PGS is ∼40% (33). In our measurements, the increase in the recovery rate approached a maximum as the percentile of DOPG is >50% in DOPC/DOPG experiments (Fig. 3 b). This is consistent with the content of the major lipid composition in the PM configuration. The increasing percentile of DMPG in DMPC/DMPG experiments (Fig. 3 a) also leads to an accelerated recovery of parent state. The intrinsic differences in the kinetics of the recoveries of bR parent state suggest altered kinetics pathways in different nanodisc environments. However, the entire photocycle activity, which includes the intermediates M, N, and O, should be taken into consideration. We collected the time-resolved difference spectra in 380–710 nm upon 532 nm excitation to unravel the correlation of the kinetics of individual intermediates and the lipids.
Figure 3.
The normalized temporal profiles of the recovery of bR depletion at 560 nm upon 532-nm excitation of monomeric bR in nanodiscs consisted of PC/PG molar ratios of 100:0 (black), 75:25 (blue), 50:50 (green), 25:75 (orange), and 0:100 (red) for (a) DMPC/DMPG, (b) DOPC/DOPG, and (c) DMPC/DOPG.
With the temporal resolution of ∼1 μs, we were able to record the thermal events after the photoexcitation, referring to the proton translocation and retinal reisomerization in the transition from intermediate M and its cascading processes. The observed contours of the time-resolved difference spectra of monomeric bR in different nanodiscs of PC/PG compositions are shown in Fig. 4. The upward and downward features in the difference spectrum were shown in red and blue, respectively, indicating the production of new species and depletion of the parent state. Clearly, the contours of nbR with high PG content contained features of intermediate M at 410 nm (upward), intermediate N characterized by an asymmetric recovery at 450–500 nm (downward) overlapped with the depletion of parent state, and the intermediate O at 650 nm (upward). The characteristics of the intermediate N and O are strongly preserved in the contours of the difference spectra when negatively charged PG is included in nanodiscs, referring to the conventional photocycle. However, the contours of nbR with low PG content nanodiscs exhibited two symmetric features at 410 (upward) and 560 nm (downward) without the contribution of intermediates N and O, which can be characterized using the pathway bR568→M412→bR568 where bR568 denotes the parent state. In addition to the hydrophilic charged heads, the hydrophobic tails also play a role in perturbing the kinetics. The amount of the transient population of intermediate O at 650 nm for bR embedded in nanodiscs of 100% DMPG is less than that in nanodiscs of 100% DOPG, implying DM is less effective than DO in preserving the photocycle activity. As shown in Figs. 4 and S5, the parent state recovery of bR embedded in 100% DMPC is slower than that in 100% DOPC. Accounting for the apparent difference in DM and DO, the melting points of lipids could also play a role because the melting temperatures of DMPC (24°C) and DMPG (23°C) are much higher than that of DOPC (–17°C) and DOPG (–18°C). This suggests that in the absence of negatively charged lipids, the melting temperature of lipids, in addition to the lipid headgroup and the lipid tail length, could affect the photocycle kinetics in a moderate fashion.
Figure 4.
Time-resolved difference absorption spectra in two-dimensional contours of mbR in nanodiscs consisted of different PC/PG molar ratios for (a) DMPC/DMPG, (b) DOPC/DOPG, and (c) DMPC/DOPG. The flux of the 532-nm pulsed laser was controlled at 0.4 mJ cm–2. Untruncated spectra were supplemented in Fig. S5.
The monophasic features of CD spectra of nbR shown in Fig. S2 refer to the monomeric bR, therefore the observed kinetics difference will not be ascribed to the cooperative effect of the trimeric bRs in the crystalline PM (48). Scherrer et al. (49) demonstrated that the proton diffusion along the micellar surface proceeded faster than their equilibration from the surface to the bulk. They also found that an increase in the CHAPS and/or DMPC concentration resulted in a slower M decay, but the rise of intermediate M is not dependent on the bR/lipid/detergent ratio (49). The rise of intermediate M is associated with the deprotonation of the Schiff base in the protein interior, suggesting that the surrounding lipids might not play a significant role in perturbing the rise kinetics. However, the decay of intermediate M correlated to the reprotonation of the Schiff base, accompanied by the proton uptake from the cytoplasmic side of the membrane (49). The native lipids in PM assisted in the lateral proton relay between the cytoplasmon and extracellular side (42). As a result, the replacement of the lipids surrounding bR would alter the decay and it is not surprising that the hydrophilic heads of CHAPS and DMPC, which are zwitterions, could play similar roles in decelerating the decay of intermediate M. Moreover, Joshi et al. (38) proposed that high levels of PGP alone can restore full photocycle activity. Regarding the hydrocarbon tails, the double bond on the monounsaturated hydrocarbon tail of DO might provide more flexibility and space for the permeability of small molecules such as protons (hydronium ion) in the lipid bilayers than DM, which is composed of saturated hydrocarbons. Accordingly, we demonstrated that the tuning of the content of the PG analogs, DOPG, in the nanodiscs is capable of controlling the proton pump kinetics and preserving most of the bR photocycle activity.
Conclusions
We employed various lipid nanodiscs, composed of different ratios of the zwitterionic lipids (DMPC and DOPC) to negatively-charged lipids (DOPG and DMPG) and MSP1E3D1 membrane scaffold proteins, to mimic the lipid bilayer environment for monomeric bacteriorhodopsin. The steady-state spectroscopy indicated that the retinal pocket of bR did not significantly alter when changing the composition of the lipids. The ζ-potential and ion-exchange chromatography measurements showed that the surface charge of the bR-embedded nanodiscs increased the negativity as the content of the anionic lipid DOPG was increased. The main observed difference in the kinetics of the proton translocation and retinal thermal reisomerization can be attributed to the change in the capability of the proton relay via the anionic lipids, in terms of the content of DOPG in the nanodiscs. We can therefore demonstrate that the kinetics of the bR photocycle can be manually controlled when using lipid nanodiscs to mimic the lipid bilayer environments. It is advantageous for further application in bioelectric devices using bR as the building blocks. In addition, our observations also indicated that care should be taken when utilizing nanodiscs to mimic the lipid bilayer in the studies of the ion pump transmembrane proteins because the charge translocation processes could be perturbed in different lipid environments.
Acknowledgments
This work is supported by the Ministry of Science and Technology of Taiwan (grants No. NSC 101-2113-M-007-016-MY2, No. MOST 103-2113-M-007-010-MY2, and No. MOST 103-2113-M-001-001-MY2). In addition, T.-Y.Y. and V.Y. acknowledge the support from Academia Sinica Nano Program.
Editor: Andreas Engel.
Footnotes
Tsung-Yen Lee and Vivien Yeh contributed equally to this work.
Five figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00947-9.
Contributor Information
Li-Kang Chu, Email: lkchu@mx.nthu.edu.tw.
Tsyr-Yan Yu, Email: dharmanmr@gate.sinica.edu.tw.
Supporting Material
References
- 1.Bayburt T.H., Sligar S.G. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee S., Huber T., Sakmar T.P. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 3.Nath A., Atkins W.M., Sligar S.G. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 4.Pandit A., Shirzad-Wasei N., de Grip W.J. Assembly of the major light-harvesting complex II in lipid nanodiscs. Biophys. J. 2011;101:2507–2515. doi: 10.1016/j.bpj.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayburt T.H., Vishnivetskiy S.A., Gurevich V.V. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whorton M.R., Bokoch M.P., Sunahara R.K. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra N., Liu Y., Yan E.C.Y. Calcium-dependent ligand binding and G-protein signaling of family B GPCR parathyroid hormone 1 receptor purified in nanodiscs. ACS Chem. Biol. 2013;8:617–625. doi: 10.1021/cb300466n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagn F., Etzkorn M., Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etzkorn M., Raschle T., Wagner G. Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility. Structure. 2013;21:394–401. doi: 10.1016/j.str.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt T.H., Grinkova Y.V., Sligar S.G. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 11.Bayburt T.H., Sligar S.G. Membrane protein assembly into nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denisov I.G., Grinkova Y.V., Sligar S.G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 13.Wadsäter M., Maric S., Cardenas M. The effect of using binary mixtures of zwitterionic and charged lipids on nanodisc formation and stability. Soft Matter. 2013;9:2329–2337. [Google Scholar]
- 14.Denisov I.G., McLean M.A., Sligar S.G. Thermotropic phase transition in soluble nanoscale lipid bilayers. J. Phys. Chem. B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernández-Rocamora V.M., Reija B., Vicente M. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J. Biol. Chem. 2012;287:30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oesterhelt D., Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 17.Bayburt T.H., Grinkova Y.V., Sligar S.G. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Johnson P.J.M., Halpin A., Dwayne Miller R.J. The photocycle and ultrafast vibrational dynamics of bacteriorhodopsin in lipid nanodiscs. Phys. Chem. Chem. Phys. 2014;16:21310–21320. doi: 10.1039/c4cp01826e. [DOI] [PubMed] [Google Scholar]
- 19.Lanyi J.K. Molecular mechanism of ion transport in bacteriorhodopsin: insights from crystallographic, spectroscopic, kinetic, and mutational studies. J. Phys. Chem. B. 2000;104:11441–11448. [Google Scholar]
- 20.Sharkov A.V., Pakulev A.V., Matveetz Y.A. Primary events in bacteriorhodopsin probed by subpicosecond spectroscopy. Biochim. Biophys. Acta. 1985;808:94–102. [Google Scholar]
- 21.Zimányi L., Váró G., Lanyi J.K. Pathways of proton release in the bacteriorhodopsin photocycle. Biochemistry. 1992;31:8535–8543. doi: 10.1021/bi00151a022. [DOI] [PubMed] [Google Scholar]
- 22.Rothschild K.J. FTIR difference spectroscopy of bacteriorhodopsin: toward a molecular model. J. Bioenerg. Biomembr. 1992;24:147–167. doi: 10.1007/BF00762674. [DOI] [PubMed] [Google Scholar]
- 23.Váró G., Lanyi J.K. Kinetic and spectroscopic evidence for an irreversible step between deprotonation and reprotonation of the Schiff base in the bacteriorhodopsin photocycle. Biochemistry. 1991;30:5008–5015. doi: 10.1021/bi00234a024. [DOI] [PubMed] [Google Scholar]
- 24.Morgan J.E., Vakkasoglu A.S., Maeda A. Coordinating the structural rearrangements associated with unidirectional proton transfer in the bacteriorhodopsin photocycle induced by deprotonation of the proton-release group: a time-resolved difference FTIR spectroscopic study. Biochemistry. 2010;49:3273–3281. doi: 10.1021/bi901757y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phatak P., Ghosh N., Elstner M. Amino acids with an intermolecular proton bond as proton storage site in bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 2008;105:19672–19677. doi: 10.1073/pnas.0810712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerwert K., Souvignier G., Hess B. Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc. Natl. Acad. Sci. USA. 1990;87:9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith S.O., Pardoen J.A., Mathies R. Chromophore structure in bacteriorhodopsin’s O640 photointermediate. Biochemistry. 1983;22:6141–6148. [Google Scholar]
- 28.Riesle J., Oesterhelt D., Heberle J. D38 is an essential part of the proton translocation pathway in bacteriorhodopsin. Biochemistry. 1996;35:6635–6643. doi: 10.1021/bi9600456. [DOI] [PubMed] [Google Scholar]
- 29.Richter H.-T., Needleman R., Lanyi J.K. Relationship of retinal configuration and internal proton transfer at the end of the bacteriorhodopsin photocycle. Biochemistry. 1996;35:15461–15466. doi: 10.1021/bi9612430. [DOI] [PubMed] [Google Scholar]
- 30.Lanyi J.K. X-ray diffraction of bacteriorhodopsin photocycle intermediates. Mol. Membr. Biol. 2004;21:143–150. doi: 10.1080/09687680410001666345. (Review) [DOI] [PubMed] [Google Scholar]
- 31.Dracheva S., Bose S., Hendler R.W. Chemical and functional studies on the importance of purple membrane lipids in bacteriorhodopsin photocycle behavior. FEBS Lett. 1996;382:209–212. doi: 10.1016/0014-5793(96)00181-0. [DOI] [PubMed] [Google Scholar]
- 32.Cartailler J.-P., Luecke H. X-ray crystallographic analysis of lipid-protein interactions in the bacteriorhodopsin purple membrane. Annu. Rev. Biophys. Biomol. Struct. 2003;32:285–310. doi: 10.1146/annurev.biophys.32.110601.142516. [DOI] [PubMed] [Google Scholar]
- 33.Corcelli A., Lattanzio V.M.T., Fanizzi F. Lipid-protein stoichiometries in a crystalline biological membrane: NMR quantitative analysis of the lipid extract of the purple membrane. J. Lipid Res. 2002;43:132–140. [PubMed] [Google Scholar]
- 34.Yokoyama Y., Negishi L., Mitaku S. Effect of lipid phase transition on molecular assembly and structural stability of bacteriorhodopsin reconstituted into phosphatidylcholine liposomes with different acyl-chain lengths. J. Phys. Chem. B. 2010;114:15706–15711. doi: 10.1021/jp108034n. [DOI] [PubMed] [Google Scholar]
- 35.Heyes C.D., El-Sayed M.A. The role of the native lipids and lattice structure in bacteriorhodopsin protein conformation and stability as studied by temperature-dependent Fourier transform-infrared spectroscopy. J. Biol. Chem. 2002;277:29437–29443. doi: 10.1074/jbc.M203435200. [DOI] [PubMed] [Google Scholar]
- 36.Dencher N.A., Heyn M.P. Formation and properties of bacteriorhodopsin monomers in the non-ionic detergents octyl-β-D-glucoside and Triton X-100. FEBS Lett. 1978;96:322–326. doi: 10.1016/0014-5793(78)80427-x. [DOI] [PubMed] [Google Scholar]
- 37.Barnett S.M., Dracheva S., Levin I.W. Lipid-induced conformational changes of an integral membrane protein: an infrared spectroscopic study of the effects of Triton X-100 treatment on the purple membrane of Halobacterium halobium ET1001. Biochemistry. 1996;35:4558–4567. doi: 10.1021/bi952258l. [DOI] [PubMed] [Google Scholar]
- 38.Joshi M.K., Dracheva S., Hendler R.W. Importance of specific native lipids in controlling the photocycle of bacteriorhodopsin. Biochemistry. 1998;37:14463–14470. doi: 10.1021/bi980965j. [DOI] [PubMed] [Google Scholar]
- 39.Roos C., Zocher M., Bernhard F. Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E. coli MraY translocase. Biochim. Biophys. Acta. 2012;1818:3098–3106. doi: 10.1016/j.bbamem.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Oesterhelt D., Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 41.Wang J., Link S., El-Sayed M.A. Comparison of the dynamics of the primary events of bacteriorhodopsin in its trimeric and monomeric states. Biophys. J. 2002;83:1557–1566. doi: 10.1016/S0006-3495(02)73925-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heberle J., Riesle J., Dencher N.A. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 1994;370:379–382. doi: 10.1038/370379a0. [DOI] [PubMed] [Google Scholar]
- 43.Heyes C.D., El-Sayed M.A. Proton transfer reactions in native and deionized bacteriorhodopsin upon delipidation and monomerization. Biophys. J. 2003;85:426–434. doi: 10.1016/S0006-3495(03)74487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann M., Wanko M., Elstner M. Color tuning in rhodopsins: the mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. J. Am. Chem. Soc. 2006;128:10808–10818. doi: 10.1021/ja062082i. [DOI] [PubMed] [Google Scholar]
- 45.Brouillette C.G., McMichens R.B., Khorana H.G. Structure and thermal stability of monomeric bacteriorhodopsin in mixed phospholipid/detergent micelles. Proteins. 1989;5:38–46. doi: 10.1002/prot.340050106. [DOI] [PubMed] [Google Scholar]
- 46.Casadio R., Gutowitz H., Stoeckenius W. Light-dark adaptation of bacteriorhodopsin in triton-treated purple membrane. Biochim. Biophys. Acta. 1980;590:13–23. doi: 10.1016/0005-2728(80)90142-5. [DOI] [PubMed] [Google Scholar]
- 47.Cassim J.Y. Unique biphasic band shape of the visible circular dichroism of bacteriorhodopsin in purple membrane: excitons, multiple transitions or protein heterogeneity? Biophys. J. 1992;63:1432–1442. doi: 10.1016/S0006-3495(92)81701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Váró G., Needleman R., Lanyi J.K. Protein structural change at the cytoplasmic surface as the cause of cooperativity in the bacteriorhodopsin photocycle. Biophys. J. 1996;70:461–467. doi: 10.1016/S0006-3495(96)79589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherrer P., Alexiev U., Heyn M.P. Covalently bound pH-indicator dyes at selected extracellular or cytoplasmic sites in bacteriorhodopsin. 1. Proton migration along the surface of bacteriorhodopsin micelles and its delayed transfer from surface to bulk. Biochemistry. 1994;33:13684–13692. doi: 10.1021/bi00250a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.