Abstract
Recent evidence indicates that the gut microbiota plays a key role in the pathophysiology of obesity. Indeed, diet-induced obesity (DIO) has been associated to substantial changes in gut microbiota composition in rodent models. In the context of obesity, enhanced adiposity is accompanied by low-grade inflammation of this tissue but the exact link with gut microbial community remains unknown. In this report, we studied the consequences of high-fat diet (HFD) administration on metabolic parameters and gut microbiota composition over different periods of time. We found that Akkermansia muciniphila abundance was strongly and negatively affected by age and HFD feeding and to a lower extend Bilophila wadsworthia was the only taxa following an opposite trend. Different approaches, including multifactorial analysis, showed that these changes in Akkermansia muciniphila were robustly correlated with the expression of lipid metabolism and inflammation markers in adipose tissue, as well as several circulating parameters (i.e., glucose, insulin, triglycerides, leptin) from DIO mice. Thus, our data shows the existence of a link between gut Akkermansia muciniphila abundance and adipose tissue homeostasis on the onset of obesity, thus reinforcing the beneficial role of this bacterium on metabolism.
The digestive tract is colonized by trillions of microbes (i.e., the gut microbiota) that exert a wide range of beneficial functions for the host. In recent years, it has become apparent that alterations in gut microbiota composition is associated with the development of highly prevalent metabolic disorders such as obesity and type 2 diabetes (T2D) in both animal and human studies1,2,3,4,5,6,7. We and others have demonstrated that high-fat diet (HFD) feeding profoundly affects the gut microbial community1,8,9,10,11,12,13,14. In particular, the abundance of Bifidobacterium spp. and Akkermansia muciniphila is consistently reduced under HFD regimen8,14,15. We also observed a decrease in Lactobacillus spp. and Bacteroides/Prevotella spp. However, the reduced abundance of these genera was less consistent16,17,18 suggesting that these changes were not involved in the development of the disorders associated with diet-induced obesity (DIO). Recently, increased abundance of Bilophila wadsworthia has been associated with fat feeding and inflammation7,19.
The pathological massive expansion of adipose tissue in obesity states is associated with the development of low-grade inflammation, which is reflected by enhanced production of cytokines, chemokines and pro-inflammatory fatty acids. This causes an imbalance between pro- and anti-inflammatory factors produced by leukocytes, further promoting inflammation and adipose tissue dysfunction (e.g., β-oxidation, browning processes, adipogenesis)20,21,22,23. Despite the well-established link between DIO and gut microbiota alterations, it is currently unknown whether changes in the abundance of specific bacteria precedes or remains present during the development of obesity and related adipose tissue metabolic alterations.
In the current study we explored the potential interconnection between adipose tissue inflammation/function and changes in specific microbes during obesity and T2D development following chronic HFD feeding in mice. Our results demonstrate specific changes in gut bacteria over the progress of obesity, together with a robust negative association between Akkermansia muciniphila and inflammation and a positive association with adipose tissue browning process markers. This work also demonstrates that this bacterium declines before the onset of metabolic alterations, thereby suggesting a putative causative implication in the disease progression.
Results
Body weight and adiposity progression over 16 weeks of HFD feeding
It is well established that HFD feeding promotes weight gain and adiposity leading to obesity. Here we found that HFD administration modestly, but significantly, increased body weight and adiposity after 3 and 6 weeks of dietary treatment. However, after this time point, we found a major effect of the diet on body weight gain, which rose up to ~45% after 12–16 weeks as compared to only ~15% after 6 weeks of treatment (Fig. 1A). Adiposity followed a similar pattern, with a dramatic increase at 12 weeks of age (~3.5 fold) as compared to control diet (CT) mice. This effect was even more pronounced after 16 weeks of HFD feeding (Fig. 1B). Collectively, these results indicate that the greatest impact on body weight gain and adiposity in mice occurs after 6 consecutive weeks of HFD administration.
Figure 1. Kinetic evolution of body weight and adiposity following chronic HFD administration.
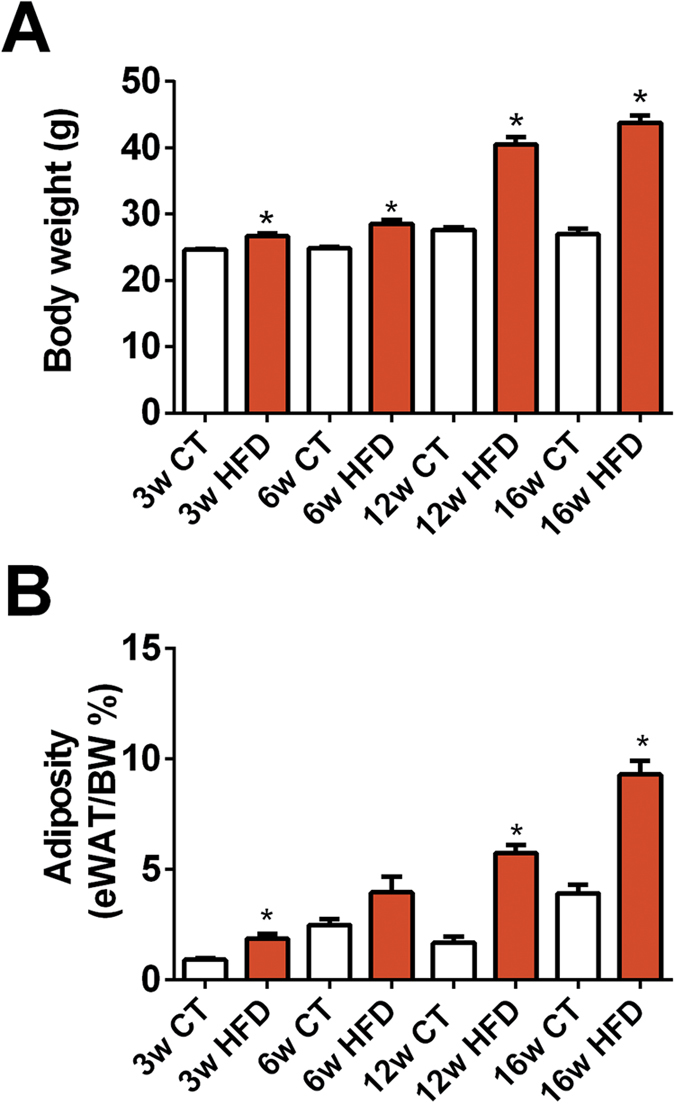
(A) Body weight gain (g) and (B) adiposity (% of eWAT on total body weight) measured after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a control diet (CT) (n = 6/group). Data are presented as the mean ± SEM. Data are significantly different (P < 0.05) according to the unpaired two-tailed Student t-test. *indicates a significant difference versus CT (P < 0.05).
Markers of adipose tissue metabolism and inflammation were altered after HFD administration
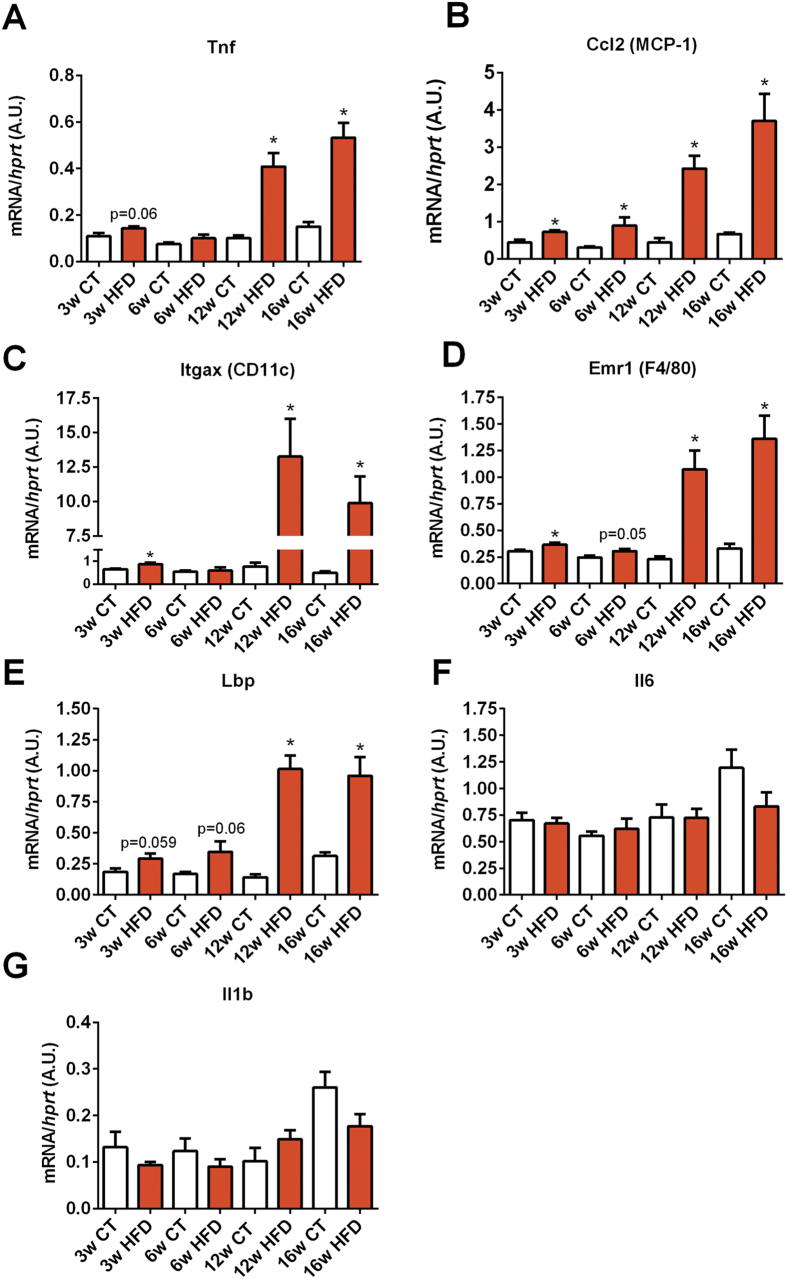
We next assessed the repercussion of chronic HFD feeding on adipose tissue metabolism by measuring several markers of metabolic inflammation (Tnf, Ccl2, Itgax, Emr1, Lbp, Il6, Il1), fatty acid oxidation (Acox1, Cpt1a, Acacb), fat browning (Cidea, Ppargc1a, Ppargc1b, Elovl3, Prdm16), lipogenesis (Dgat2, Fasn, Acaca) and adipogenesis (Pparg and Cebpa) by quantitative PCR (qPCR). Macrophage infiltration markers (Itgax and Emr1) and recruitment of immune cells (Ccl2) were significantly increased as early as 3-weeks after the beginning of the HFD treatment (Fig. 2B–D), whereas markers of inflammation such as Tnfa and Lbp exhibited a trend to be increased but did not reach significance (Fig. 2A,E). After 12 weeks of HFD, all the aforementioned markers were strongly and significantly increased. Both Il6 and Il1 mRNA expression were not significantly affected by the treatment (Fig. 2F,G).
Figure 2. Time series evolution of markers of inflammation and macrophage infiltration in white adipose tissue following HFD administration.
mRNA expression of (A) Tnf (encoding TNF-α), (B) Ccl2 (encoding MCP-1), (C) Itgax encoding CD11c), (D) Emr1 (encoding F4/80), (E) Lbp (encoding LBP), (F) Il6 (encoding Il-6) and (G) Il1b (encoding IL-1β) measured in the adipose tissue after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a control diet (CT) (n = 6/group). Data are presented as the mean ± SEM. Data are significantly different (P < 0.05) according to the unpaired two-tailed Student t-test. *indicates a significant difference versus CT (P < 0.05).
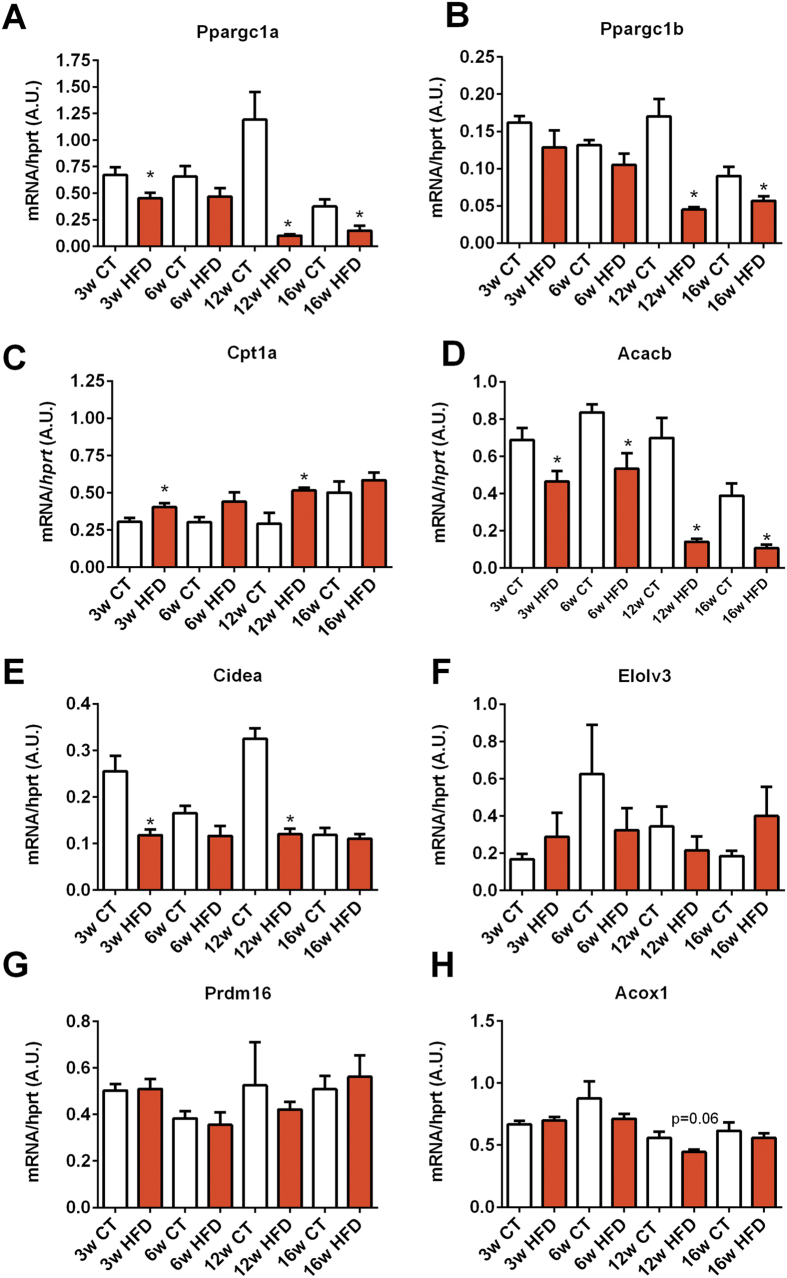
Among the different markers of lipid metabolism involved in oxidation and browning program in white adipose tissue, we found that Ppargc1a, Cidea, and Acacb mRNA expression levels were significantly decreased after only 3 weeks of HFD (Fig. 3A,E,D), whereas other key markers of β-oxidation (Acox1) were not affected (Fig. 3H). Ppargc1a, Ppargc1b and Acacb transcript expression was dramatically decreased after 12 weeks of HFD and remained lower until the end of the dietary intervention (Fig. 3A–D). Cpt1a was slightly but significantly increased after 3, 6 and 12 weeks of treatment (Fig. 3C). Elovl3 and Prdm16 mRNA expression levels were not affected by HFD throughout the experimental period.
Figure 3. Time series evolution of markers of fatty acid oxidation and browning in white adipose tissue following HFD treatment.
mRNA expression of (A) Pppargc1a (encoding PGC1-α), (B) Pppargc1b (encoding PGC1-β), (C) Cpt1a (encoding CPT-1a), (D) Acacb (encoding ACC2), (E) Cidea (encoding CIDEA), (F) Elovl3 (encoding ELOVL3), (G) Prdm16 (encoding PRDM16) and (H) Acox1 (encoding ACOX1) measured in the adipose tissue after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a control diet (CT) (n = 6/group). Data are presented as the mean ± SEM. Data are significantly different (P < 0.05) according to the unpaired two-tailed Student t-test. *indicates a significant difference versus CT (P < 0.05).
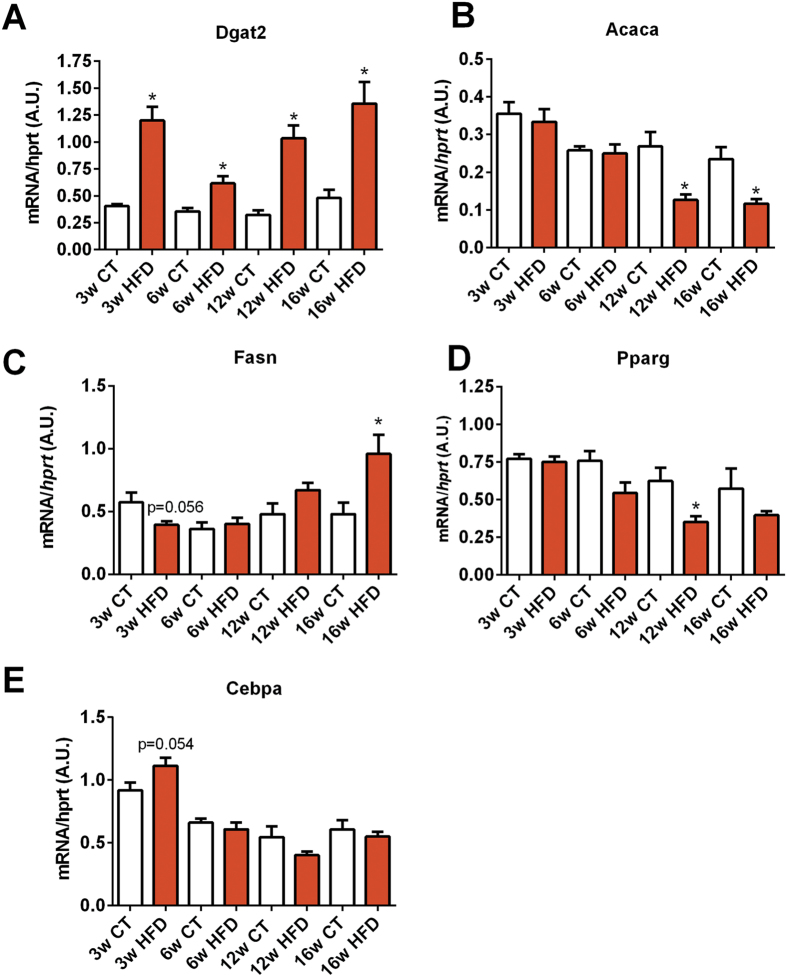
Dgat2 expression, the enzyme that catalyzes the final step in triglyceride synthesis, was increased after 3 weeks of HFD and remained high during the overall period of time (Fig. 4A). Markers of lipogenesis were inversely affected by the HFD, while Fasn mRNA expression increased, Acaca transcript expression decreased after 16 weeks of dietary intervention (Fig. 4B,C). Ppparg and Cebpa mRNA expression levels, both markers of adipocyte differentiation, showed marginal changes over the course of the diet (Fig. 4D,E).
Figure 4. Time series evolution of markers of lipogenesis and adipogenesis in white adipose tissue following HFD treatment.
mRNA expression of (A) Dgat2 (encoding DGAT2), (B) Acaca (encoding ACC1), (C) Fasn (encoding FAS), (D) Pparg (encoding PPAR-γ) and (E) Cebpa (encoding CEBP-α) measured in the adipose tissue after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a control diet (CT) (n = 6/group). Data are presented as the mean ± SEM. Data are significantly different (P < 0.05) according to the unpaired two-tailed Student t-test. *indicates a significant difference versus CT (P < 0.05).
Collectively, the gene expression changes observed in white adipose tissue indicate that HFD administration causes profound changes in key genes implicated in inflammation, fatty acid oxidation and lipogenesis. Minimal effects were observed on genes mediating adipocyte browning and differentiation. In general, and in line with the physiological results, the most obvious alterations in gene expression were recorded after prolonged HFD treatments.
Evolution of regulatory markers of food intake, diabetes and adiposity over 16 weeks of HFD
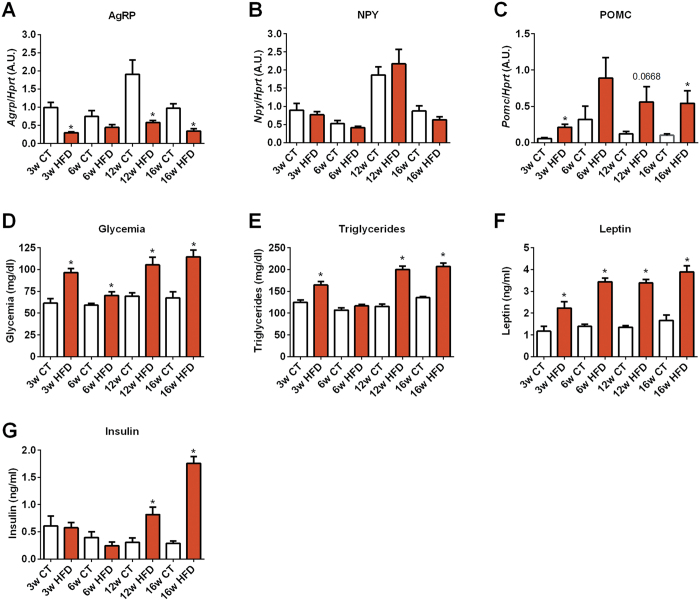
Defective energy balance is a major feature occurring during obesity. Energy balance is finely regulated by interactions between peripheral tissues, such as the intestine, and the central nervous system. In the hypothalamus, neuropeptide Y (NPY) and Agouti-related protein (AgRP) are orexigenic peptides, whereas proopiomelanocortin (POMC) precursor protein is cleaved into anorexigenic peptides. Interestingly, alterations in the expression of these neuropeptides have been extensively reported during HFD. Moreover, gut microbiota has recently been shown to be able to influence the expression of these neuropeptides in the hypothalamic arcuate nucleus of mice24. Therefore, we measured these parameters in our study. We found a decrease in the expression of orexigenic peptides such as AgRP during HFD, whereas the expression of anorexigenic peptides (POMC) were increased (at all studied time points) (Fig. 5A and C). Aging did not seem to impact on the expression of AgRP, NPY or POMC in the hypothalamus (Fig. 5A–C). We also found that markers of insulin resistance, such as fasted glycemia and insulinemia, progressively increased during prolonged HFD treatment. A similar trend was observed for both triglyceridemia and leptin levels (Fig. 5D–C).
Figure 5. Time series evolution of orexigenic and anorexigenic markers in the hypothalamus and circulating levels of metabolic parameters following HFD treatment.
mRNA expression of (A) AgRP (encoding AGRP), (B) Npy (encoding NPY), (C) Pomc (encoding POMC) measured in the hypothalamus,. Circulating levels of (D) glucose, (E) triglycerides, (F) leptin and (G) insulin measured in the serum of mice after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a control diet (CT) (n = 6/group). Data are presented as the mean ± SEM. Data are significantly different (P < 0.05) according to the unpaired two-tailed Student t-test. *indicates a significant difference versus CT (P < 0.05).
Evolution of selected gut bacteria during HFD feeding
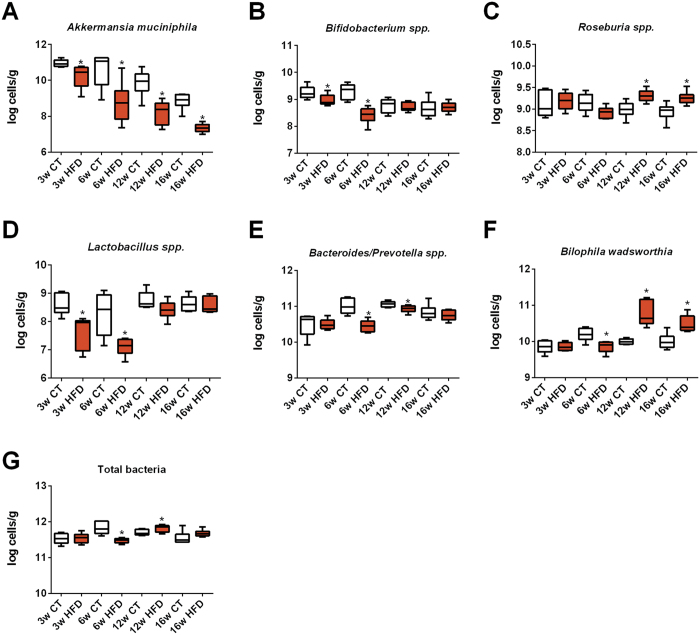
Based on previous findings8,14,15,16,17,18, we measured the abundance of selected bacteria during the progress of HFD administration. We found that Akkermansia muciniphila, Bifidobacterium spp. and Lactobacillus spp. were significantly decreased after 3 and 6 weeks on HFD, although this decrease was transient for Bifidobacterium spp. and Lactobacillus spp. (Fig. 6A,B,D). The abundance of Akkermansia muciniphila decreased gradually to finally reach a level ~10,000 times lower than the initial one. Remarkably, under control diet, the levels of Akkermansia muciniphila progressively decreased by ~100 times as compared to young CT mice. Among the different bacteria the effect of aging was observed only for this taxon (Fig. 6A). We also observed that the abundance of Roseburia spp. and the abundance of Bilophila wadsworthia increased after 12 and 16 weeks of HFD treatment (Fig. 6C,F). Interestingly, the abundance of Bilophila wadsworthia was unaffected or even decreased during earlier time points of HFD (Fig. 6F). Both Bacteroides/Prevotella spp. and total bacteria were affected by the treatment at 6 and 12 weeks of age but were similar between groups at the end of the experimental period (Fig. 5E,F). Thus these data confirm that HFD feeding affects specific gut bacteria, and highlight that the abundance of Akkermansia muciniphila progressively decline with prolonged dietary treatment in CT mice, and that this effect is exacerbated upon HFD.
Figure 6. Time series evolution of specific gut bacteria following HFD treatment.
Quantification of (A) Akkermansia muciniphila, (B) Bifidobacterium spp., (C) Roseburia spp. (D) Lactobacillus spp., (E) Bacteroides/Prevotella spp, (F) Bilophila wadsworthia and (G) total gut bacteria abundance after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a control diet (CT) and expressed as Log cells per g of caecal content (n = 6/group). Data are presented as the mean ± SEM. Data are significantly different (P < 0.05) according to the unpaired two-tailed Student t-test. *indicates a significant difference versus CT (P < 0.05).
Multifactorial analysis of adipose tissue markers and bacteria
In order to identify putative markers driving the modulation of metabolism in older animals and/or during HFD-feeding, we combined the bacterial taxa and metabolic parameters obtained at each time point in a multifactorial analysis. This multifactorial analysis takes into account all the different parameters measured in this study for each mouse, and allows us to determine if mice from the same group cluster together according to these metabolic parameters, thereby suggesting that altogether these parameters are key factors affected by age and/or HFD-feeding. The individual factor map shows the distribution of the samples according to the multifactorial analyses results (Fig. 7). All the samples from week 3 were clustered together, whereas samples from week 6 were separated along axis 2 according to the diet. We found that samples from weeks 12 and 16 were all separated along axis 1 according to the dietary treatment, thereby confirming the major effect of the diet on all the parameters. CT samples from weeks 3, 6 and 12 were all clustered together, while CT samples from week 16 were closer to the HFD samples from weeks 3 and 6. This observation suggests that older mice exhibit changes in the adipose tissue expression profile and microbiota composition similar to those observed during HFD at the early stage of obesity development (Fig. 7).
Figure 7. Individuals factor map.
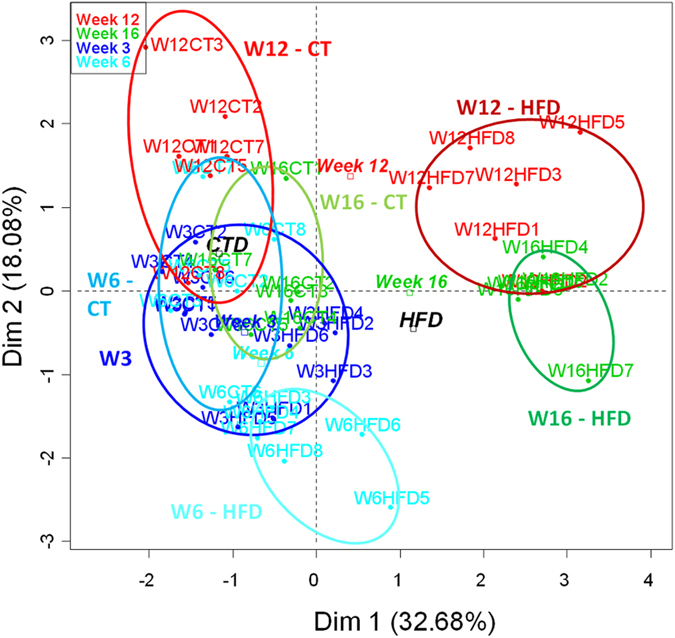
The individual factor map presents the repartition of the samples (dots) in the multifactorial analysis’ plane. Time and diet (squares) are presented as illustrative (inactive) qualitative factors. Samples are colored according to treatment’s duration. Circles regroup all samples from a specific diet for a single time point, except week 3 where all samples are grouped together.
The hierarchical clustering is another way to illustrate the similarity and the clustering between different samples. The smaller the distances (resulting from the addition of horizontal distances in dark line) linking samples are, the higher is the similarity for the parameters measured. The hierarchical clustering (Fig. 8) shows that HFD samples from weeks 12 and 16 grouped distant from the other samples. In addition, this graphical representation showed a robust clustering of CT samples from week 16 with HFD samples from week 6, emphasizing again putative similarities between HFD and aging. CT samples from weeks 6 (HFD) and 12 (CT)clustered together, reinforcing the results obtained with the individual factor map (Fig. 7).
Figure 8. Hierarchical clustering.
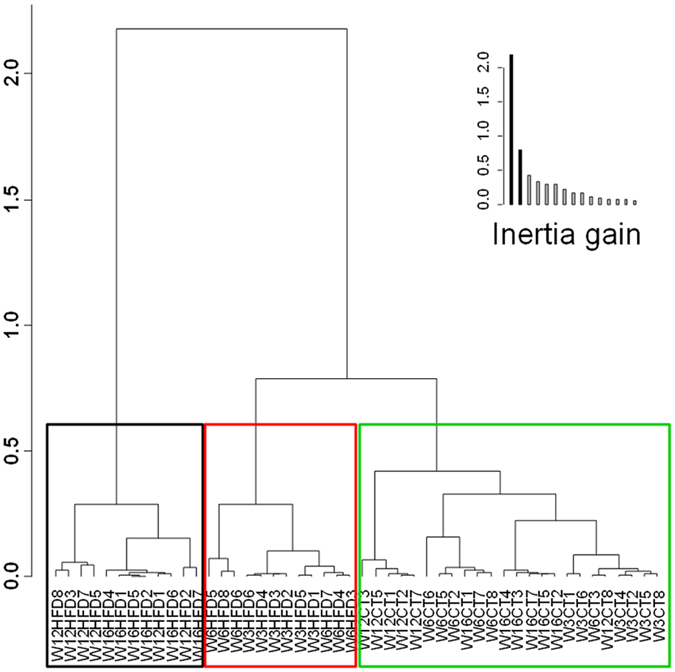
Hierarchical clustering of the samples on the principal components of the multifactorial analysis. This graph illustrates the similarities and the clustering between different samples. The smaller the distance linking samples is (resulting from the addition of horizontal distances in dark line), the more similar these samples are for the parameters measured.
Akkermansia muciniphila strongly correlates with adipose tissue metabolic parameters
To further explore whether one specific bacterial taxa was correlated with host metabolism markers, we performed a multifactorial correlation circle and correlation map. This analysis is taking into account all the parameters measured in the study and determine if specific parameters are influencing or correlating with the others. It allows to determine the strength of the relation between the different parameters measured in the study. When the arrows are pointing in the same direction, this demonstrates that the parameters are positively correlated. If the arrows are pointing in opposite directions, the parameters are inversely correlated. When they are indicating different directions, this means that the parameters are not evolving in the same way and are likely not correlated. It is also important to mention that the length of the arrows is an indicator of the strength of the correlation.
Surprisingly, we found that the levels of Akkermansia muciniphila were strongly and positively correlated with the levels of almost all the parameters involved in fatty acid oxidation and fat browning (Fig. 9A). Conversely, the levels of Akkermansia muciniphila were inversely associated with inflammatory markers, lipid synthesis, and several plasma markers of insulin resistance, cardiovascular risk and adiposity (Fig. 9A). Bifidobacterium spp. tended to exhibit a similar pattern as Akkermansia muciniphila, whereas the other taxa were not associated with adipose tissue metabolic and inflammatory parameters, except the pathobiont Bilophila wadsworthia showing a moderate but opposite trend to Akkermansia muciniphila. Interestingly, most of the inflammatory factors were strongly correlated with axis 1 on the right side (Fig. 9A), which mainly represents the direction of older mice samples (i.e., prolonged treatment time) as depicted in Fig. 7. Roseburia spp. tended to correlate moderately in this direction, while the other bacteria investigated were not associated with other parameters such as diet or aging (Figs 9A and 7). These observations were further reinforced using another mathematical model (Fig. 9B). Akkermansia muciniphila was significantly and inversely correlated with 5 out of 7 inflammatory markers, all circulating parameters (i.e., insulin, glucose, triglycerides and leptin) and positively correlated with 118 out of 13 genes involved in fatty oxidation, synthesis and browning, while Bifidobacterium spp. abundance was significantly and negatively correlated with 3 inflammatory markers, and leptin and positively correlated with only 2 gene involved in fatty oxidation. However, Bilophila wadsworthia points in an opposite direction than Akkermansia muciniphila. Although not fully correlated with the same parameters, Bilophila wadsworthia was positively associated with 4 out of 7 inflammatory markers, and negatively associated with 7 out of 13 genes involved in fatty oxidation, synthesis and browning and positively with 3 plasma parameters (i.e., insulin, triglycerides and leptin).
Figure 9. Correlation circle and heat map correlation showing associations between bacterial taxa and adipose tissue metabolic parameters.
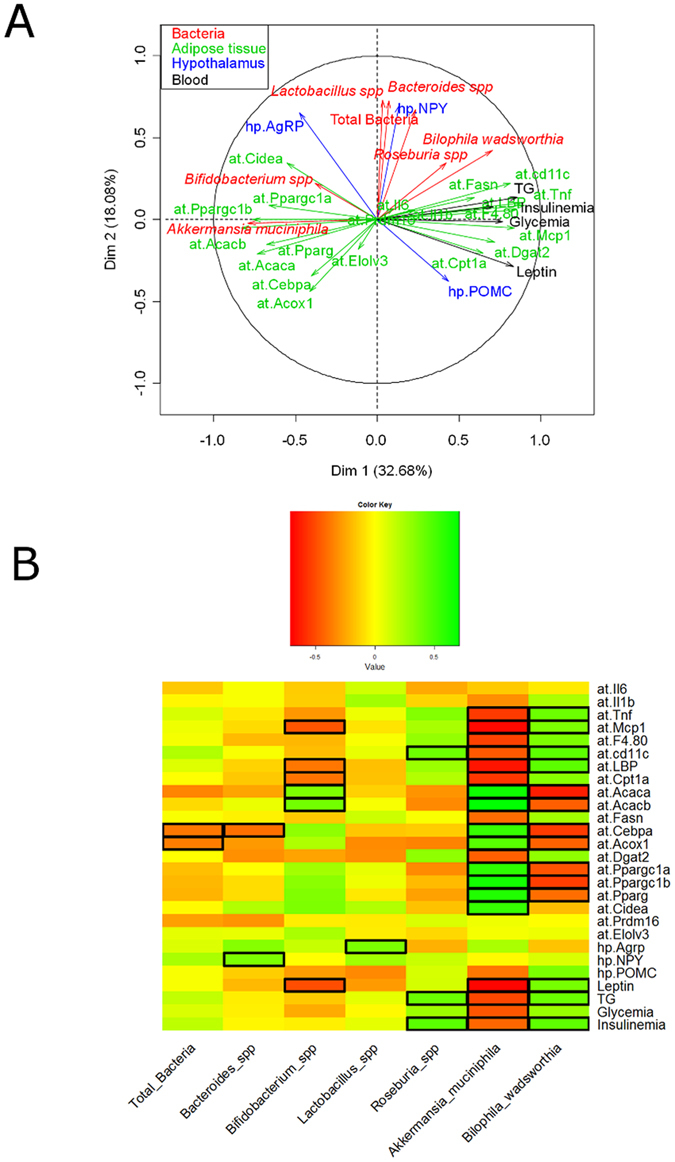
Multifactorial analysis (A) correlation map and (B) heat map of the Spearman r correlations between the bacterial genera and the metabolic parameters measured in the adipose tissue of mice after 3 weeks (3w); 6 weeks (6w); 12 weeks (12w) and 16 weeks (16w) of a high-fat diet (HFD) or a CT diet (CT) (n = 6/group). Data are presented as the mean ± SEM. Squared cells depict significance following the Spearman correlation and FDR correction for multiple comparisons *P < 0.05.
Discussion
In the present study, we used targeted approaches to investigate the impact of HFD on specific gut bacteria abundance and metabolic parameters. Importantly, we focused our adipose tissue metabolic analysis on epididymal fat since this fat depot is an appropriate surrogate of whole-body white adipose tissue. This fat pad is particularly increased after HFD administration when compared to mesenteric fat25. In addition, a detailed analysis of the general inflammatory response upon HFD administration shows an early and more dramatic activation of the inflammatory program in epididymal adipose tissue in comparison with mesenteric26. Moreover, by using this epididymal fat, we could investigate several important markers of inflammation, adipogenesis and browning, whereas this latter parameter does not occur in another visceral fat pad such as mesenteric fat for instance. The taxa we have selected are the most commonly studied, and corresponded to bacteria that have been previously associated with HFD feeding by using several culture-dependent and independent strategies1,7,8,9,13,14,15,17,19,27,28,29,30.
We found that changes in specific gut bacteria occur progressively following HFD feeding. In particular, Akkermansia muciniphila was linearly affected by the dietary treatment. By using a number of strategies, such as multifactorial analysis, we found a robust association between this bacterium and particular changes in adipose tissue inflammation and metabolic parameters. Among the 27 parameters measured, 20 of them were significantly (positively or negatively) correlated with Akkermansia muciniphila (Fig. 9A,B). This suggests that this bacterium dialogue with host to maintain adequate adipose tissue metabolism and function, but this hypothesis warrants further direct investigation. Among all other bacteria measured in this study, Bilophila wadsworthia shows opposite trend with 14 of the 27 parameters significantly correlated with this bacterium. Bifidobacterium spp. has shown similar associations, but only few parameters (6 out of 20) were significantly correlated with this bacterium. Consistent with this, it has been reported a positive impact of Bifidobacterium spp. on metabolism during fat feeding31.
We have previously demonstrated lower levels of Akkermansia muciniphila in genetic obese (ob/ob) mice and mice fed a HFD for up to 5 weeks1,14. Other recent reports also found that the abundance of this bacterium is inversely associated with obesity and diabetes development following HFD administration15,28,32, thereby supporting our past and present findings. However, it is worth noting that recent studies have shown an increased abundance of Akkermansia muciniphila upon high fat and high carbohydrate (sucrose, maltodextrin, corn starch) diet feeding33,34, thus the mechanisms promoting the bloom of Akkermansia muciniphila in these reports warrants further investigation. We recently discovered that the composition of the fatty acids per se may also strongly contribute to the modulation of the abundance of Akkermansia muciniphila. We found that mice fed with a lard-enriched diet exhibited a significant decrease in Akkermansia muciniphila, whereas fish oil-enriched diet dramatically increased Akkermansia muciniphila in the gut and this effect was associated with a better control of the gut barrier function and lower adipose tissue inflammation, a phenomenon that can be transferred to germ-free recipient mice35.
Finally, another argument suggesting that Akkermansia muciniphila protects against body weight gain and adiposity development is based on studies showing that the simultaneous administration of HFD and Akkermansia muciniphila mitigated the impact of the diet in gut barrier dysfunction and body weight and fat mass gain14,28, although the underlying mechanisms are still unknown.
We found important variations of Akkermansia muciniphila quantity with for instance a 100-fold reduction over the period studied, however, the magnitude of the difference observed between inflammatory markers was not following the same trend. Although one would have expected a linear relationship, that is less Akkermansia corresponds to more inflammation, the present study as well as previous studies does not support the idea that such simple and strict linear relationship exists14,28,36,37. Thus, although we still do not know exactly the levels required to observe a shift between a healthy versus a pathological situation, it is likely that below a certain level (i.e., a threshold) of Akkermansia muciniphila gut barrier function and other physiological parameters (e.g., altered adipogenesis/browning) become altered. In accordance with this hypothesis, we have recently demonstrated in human that below a given quantity of Akkermansia muciniphila, subjects were less prone to positively respond to caloric restriction diet in terms of the improvement of inflammatory markers, insulin resistance and glycemia. Therefore, we do not rule out that the decrease in Akkermansia muciniphila observed in older mice in this study was not enough to reach the threshold level of Akkermansia muciniphila for inducing strong inflammatory markers and diabetes.
We have also revealed a link between Akkermansia muciniphila and age, since the intestinal levels of this bacterium also declined with age upon CT diet feeding. Interestingly, HFD strongly influenced adipose tissue profile and intestinal microbiota in a way that mimicked aging, or at least older mice. For example, young HFD fed mice (6 weeks of HFD) were clustered with older CT mice (12 weeks of diet). Moreover, older CT samples (16 weeks) displayed an intermediate state between young and long-term HFD samples. In humans, it has also been suggested that Akkermansia muciniphila progressively decreases in elderly subjects38, thereby corroborating our findings. Taken together, our results indicate that HFD administration is an aggravating factor of age, although we did not further investigate this question in mice followed throughout their entire lifespan.
Although previous studies have found a putative link between caloric intake and the abundance of Akkermansia muciniphila39,40, we did not found any relationship between this taxa and markers of hypothalamic control of food intake. Since HFD diet is associated with an increase in calorie intake, the decreased expression of orexigenic peptides (AgRP) during HFD coupled with the increased expression of anorexigenic peptides (POMC) (at all studied time points) in the hypothalamus strongly suggest a resistance to negative feedback usually associated with ingestion of caloric rich diet as reported in the literature41. The available literature shows a significant variability and inconsistencies in neuropeptide expression upon HFD administration. This is likely the consequence of the rodent strain used, the percentage of fat in the diet and the length of diet exposure. Nevertheless, our results are in accordance with the accepted general idea that HFD reduces orexigenic NPY and AgRP expression while increases POMC levels41.
In conclusion, our data show that diet-induced metabolic disorders in mice progressively develop with time reaching a maximal impact after 12 weeks of treatment. Moreover, the abundance of specific taxa, such as Bifidobacterium spp. and Akkermansia muciniphila, were strongly associated with markers of lipid metabolism and negatively associated with inflammation in adipose tissue, circulating glucose, leptin, triglycerides and insulin. Although both bacteria are early affected by the HFD treatment, Akkermansia muciniphila levels robustly decreased in a sustained manner until the end of the experimental period. This effect was also present during CT diet feeding, thereby suggesting the impact of age on this bacterium, since Bilophila wadsworthia that correlates with metabolic parameters was mainly affected after a prolonged dietary intervention (i.e., 12 to 16 weeks of HFD). Thus, although future studies are required to decipher the mechanisms underlying the rapid decline of Akkermansia muciniphila during HFD and aging, our data reinforce the putative beneficial impact of this bacterium on metabolism and the need of investigating its role in humans on the development of metabolic disorders associated with fat feeding and aging.
Methods
Mice and diets
Male age-matched C57BL/6J mice were purchased from Harlan Europe. Mice were maintained on a 12:12h light–dark cycle with free access to water and standard chow (Harlan Research Laboratories) or high-fat diet (45% Kcal from fat; Research Diets) for 3, 6, 12 and 16 consecutive weeks (starting at 6 weeks of age). All experimental groups were maintained and studied in parallel to minimize environmental effects. The whole study included n = 6 mice/diet/time point. Body weights were weekly determined. Mice were overnight fasted and culled by cervical dislocation. The entire epididydmal fat was carefully dissected and tissues (blood, fat, hypothalamus and caecal content) were rapidly snapped frozen in liquid nitrogen. To avoid potential variations between groups, tissues from mice fed with standard or HFD were alternately dissected. Mice from only one time-point were processed per day. Adiposity was measured by weighting the epididymal fat depot normalized by body weight. All in vivo studies were performed with approval of the University of Barcelona Ethics Committee, complying with current Spanish and European legislation.
Quantitative PCR analysis of gene expression
Tissue mRNA isolated using Trizol (Invitrogen) following standard protocols. Total RNA was quantified using a NanoDrop spectrophotometer and cDNA generated using Taqman High-capacity Retrotranscription kit (Applied Biosystems). Quantitative PCR (qPCR) was performed as previously described42. Relative gene expression was calculated using the standard curve method. Proprietary Taqman Gene Expression assay FAM/TAMRA primers used (Applied Biosystems) were: Acetyl-CoA carboxylase 1 (Acaca; Mm01304257m1), Acetyl-CoA carboxylase 2 (Acacb; Mm01204671m1), Peroxisomal acyl-coenzyme A oxidase 1 (Acox; Mm01246834m1), chemokine (C-C motif) ligand 2 (Ccl2; Mm00441242m1), CCAAT/enhancer-binding protein alpha (Cebpa; Mm00514283s1), Cell death activator (Cidea; Mm00432554m1), carnitine palmitoyltransferase 1A (Cpt1a; Mm01231183m1), Diglyceride acyltransferase (Dgat2; Mm00499536m1), Fatty Acid Elongase 3 (Elovl3; Mm00468164m1), EGF-like module-containing mucin-like hormone receptor-like 1 (Emr1; Mm00802529m1), Fatty acid synthase (Fasn; Mm00662319m1), Interleukin-1 beta (Il1b; Mm00434228m1), Interleukin-6 (Il6; Mm00446190m1), Integrin alpha X (Itgax; Mm00498698m1), Lipopolysaccharide binding protein (Lbp; Mm00493139m1), Peroxisome proliferator-activated receptor gamma (Pparg; Mm01184322m1), Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (Ppargc1a; Mm01208835m1), Peroxisome proliferator-activated receptor gamma coactivator 1-beta (Ppargc1b; Mm00504720m1), PR domain containing 16 (Prdm16; Mm00712556m1), tumor necrosis factor alpha (Tnfa; Mm00443258m1), Agouti-related peptide (agrp; Mm00475829g1), Neuropeptide Y (npy; Mm00445771m1) and pro-opiomelanocortin (pomc; Mm00435874m1). Hypoxanthine-guanine phosphoribosyltransferase (Hprt; Mm01545399m1) was used as housekeeping gene. mRNA levels were measured using the ABI Prism 7900 HT system (Applied Biosystems).
Gut microbiota qPCR quantification
Metagenomic DNA was extracted from the caecal content using a QIAamp-DNA stool mini-kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and the adapted procedure previously described43. The primers and probes used to detect the different bacteria were based on 16S rRNA gene sequences: Total Bacteria (Bacteria Universal) F-ACTCCTACGGGAGGCAGCAG, R-ATTACCGCGGCTGCTGG; Bifidobacterium spp. F-TCGCGTCYGGTGTGAAAG, Bifidobacterium spp. R-CCACATCCAGCRTCCAC, Lactobacillus spp. F-CCTTTCTAAGGAAGCGAAGGAT, and Lactobacillus spp. R-AATTCTCTTCTCGGTCGCTCTA; Bacteroides-Prevotella spp. F-CCTTTCTAAGGAGCGAAGGAT, Bacteroides-Prevotella spp. R- AATTCTCTTCTCGGTCGCTCTA; Akkermansia muciniphila F-CAGCACGTGAAGGTGGGGAC, Akkermansia muciniphila R-CCTTGCGGTTGGCTTCAGAT; Roseburia spp. F-AAGCGACGATCAGTAGCCGA, Roseburia spp. R-.TTCTTCTTCCCTGCTGATAGAG; Bilophila wadsworthia F-, ACCCTGGTAGTCCACGCTGT Bilophila wadsworthia R- TGAGTTCAGCCTTGCGACCG; Detection was achieved with a STEP one PLUS instrument and software (Applied Biosystems, Foster City, CA, USA) using MESA FAST qPCR MasterMix Plus for SYBR Assay (Eurogentec, Verviers, Belgium). Each assay was performed in duplicate in the same run. The cycle threshold of each sample was then compared to a standard curve (performed in triplicate) made by diluting genomic DNA (five-fold serial dilution) (BCCM/LMG, Ghent, Belgium and DSMZ, Braunshweig, Germany). The data were expressed as Log bacteria/g of cecal content as described in Everard et al.14.
Statistical analysis
Data are expressed as means ± s.e.m. Differences between two groups were assessed using the unpaired two-tailed Student’s t-test. Data were analysed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). Data related to the gut microbiota were analysed using JMP 8.0.1 (SAS Institute, Inc., Cary, NC) and R 3.0.2 (The R Foundation) with the RStudio 0.97.310 packages and gplots for the heatmap. Multifactorial analysis was performed using the FactoMineR 1.28 package for R (FactoMineR: an R package for multivariate analysis). The results were considered statistically significant at P < 0.05. False Discovery Rate (FDR) corrections for multiple comparisons were performed according to the Benjamini Hochberg procedure44.
Additional Information
How to cite this article: Schneeberger, M. et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5, 16643; doi: 10.1038/srep16643 (2015).
Acknowledgments
We would like to thank A. Barrois and H. Danthinne for excellent technical assistance. PDC is a research associate at FRS-FNRS (Fonds de la Recherche Scientifique), Belgium. AE is a postdoctoral researcher at FRS-FNRS, Belgium. PDC is the recipient of grants from FNRS (convention J.0084.15, convention 3.4579.11) and PDR (Projet de Recherche, convention: T.0138.14) and ARC (Action de Recherche Concertée—Communauté française de Belgique convention: 12/17-047). This work was supported by the Fonds de la Recherche Scientifique - FNRS for the FRFS-WELBIO under Grant n° WELBIO-CR-2012S-02R. PDC is supported in part by the Funds InBev-Baillet Latour (Grant for Medical Research 2015). PDC is a recipient of ERC Starting Grant 2013 (European Research Council, Starting grant 336452-ENIGMO). This work was supported by: grants PI13/01604 (MC) and PI13/01500 (RG) from the National R + D + I (Ministerio de Economia y Competitividad, Spain) cofunded by Instituto Salud Carlos III (ISCIII) and the ERDF; 2014 SGR 659 Generalitat de Catalunya (RG); Marie Curie People Cofund Fellowship, Seventh Framework Programme of the European Commission grant 267248:DIATRAIN (AGG-V); MS is a recipient of an undergraduate grant from the UB. MC is a recipient of a Miguel Servet contract (MICINN-ISCIII, CP09/00233). This work was carried out in part at the Esther Koplowitz Centre.
Footnotes
Author Contributions M.C. and P.D.C. conceived, supervised the project and designed experiments, interpreted all the results and wrote the manuscript. M.S. and A.E. performed most of the experiments, generated figures and tables and wrote the manuscript. A.G.G.V., S.M. and S.R. performed experiments, contributed to generate figures and tables. N.M.D. and R.G. provided reagents, facilities and participated in discussions. All the authors reviewed the manuscript.
References
- Everard A. et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. The ISME journal 8, 2116–2130, doi: 10.1038/ismej.2014.45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A., Matamoros S., Geurts L., Delzenne N. M. & Cani P. D. Saccharomyces boulardii Administration Changes Gut Microbiota and Reduces Hepatic Steatosis, Low-Grade Inflammation, and Fat Mass in Obese and Type 2 Diabetic db/db Mice. mBio 5, doi: 10.1128/mBio.01011-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A. et al. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nature communications 5, 5648, doi: 10.1038/ncomms6648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A. et al. Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588, doi: 10.1038/nature12480 (2013). [DOI] [PubMed] [Google Scholar]
- Le Chatelier E. et al. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546, doi: 10.1038/nature12506 (2013). [DOI] [PubMed] [Google Scholar]
- Tremaroli V. & Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249, doi: 10.1038/nature11552 (2012). [DOI] [PubMed] [Google Scholar]
- David L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563, doi: 10.1038/nature12820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D. et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772 (2007). [DOI] [PubMed] [Google Scholar]
- Cani P. D. et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50, 2374–2383 (2007). [DOI] [PubMed] [Google Scholar]
- Cani P. D. et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481 (2008). [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Backhed F., Fulton L. & Gordon J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host.Microbe 3, 213–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M. A. et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137, 1716–1724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino M. et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 61, 543–553, doi: 10.1136/gutjnl-2011-301012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 110, 9066–9071, doi: 10.1073/pnas.1219451110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H., Bose S., Kim H. G., Han K. S. & Kim H. Fermented Rhizoma Atractylodis Macrocephalae alleviates high fat diet-induced obesity in association with regulation of intestinal permeability and microbiota in rats. Scientific reports 5, 8391, doi: 10.1038/srep08391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewulf E. M. et al. Inulin-type fructans with prebiotic properties counteract GPR43 overexpression and PPARgamma-related adipogenesis in the white adipose tissue of high-fat diet-fed mice. J. Nutr. Biochem. 22, 712–722 (2011). [DOI] [PubMed] [Google Scholar]
- Neyrinck A. M. et al. Curcuma longa Extract Associated with White Pepper Lessens High Fat Diet-Induced Inflammation in Subcutaneous Adipose Tissue. PLoS One 8, 0081252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyrinck A. M. et al. Polyphenol-rich extract of pomegranate peel alleviates tissue inflammation and hypercholesterolaemia in high-fat diet-induced obese mice: potential implication of the gut microbiota. The British journal of nutrition 109, 802–809, doi: 10.1017/S0007114512002206 (2013). [DOI] [PubMed] [Google Scholar]
- Devkota S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108, doi: 10.1038/nature11225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y. et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157, 1292–1308, doi: 10.1016/j.cell.2014.03.066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. R. et al. Meteorin-like Is a Hormone that Regulates Immune-Adipose Interactions to Increase Beige Fat Thermogenesis. Cell 157, 1279–1291, doi: 10.1016/j.cell.2014.03.065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccioli G. G. et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 6, 392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts L. et al. Adipose tissue NAPE-PLD controls fat mass development by altering the browning process and gut microbiota. Nature communications 6, 6495, doi: 10.1038/ncomms7495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schele E. et al. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology 154, 3643–3651, doi: 10.1210/en.2012-2151 (2013). [DOI] [PubMed] [Google Scholar]
- Wueest S., Yang X., Liu J., Schoenle E. J. & Konrad D. Inverse regulation of basal lipolysis in perigonadal and mesenteric fat depots in mice. American journal of physiology. Endocrinology and metabolism 302, E153–160, doi: 10.1152/ajpendo.00338.2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton M. C. et al. Inflammatory Signals shift from adipose to liver during high fat feeding and influence the development of steatohepatitis in mice. J Inflamm (Lond) 8, 8, doi: 10.1186/1476-9255-8-8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anhe F. F. et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64, 872–883, doi: 10.1136/gutjnl-2014-307142 (2015). [DOI] [PubMed] [Google Scholar]
- Shin N. R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735, doi: 10.1136/gutjnl-2012-303839 (2014). [DOI] [PubMed] [Google Scholar]
- Kim H. et al. Modulation of the intestinal microbiota is associated with lower plasma cholesterol and weight gain in hamsters fed chardonnay grape seed flour. J Agric Food Chem 63, 1460–1467, doi: 10.1021/jf5026373 (2015). [DOI] [PubMed] [Google Scholar]
- Dewulf E. M. et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut, doi: 10.1136/gutjnl-2012-303304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D. & Van Hul M. Novel opportunities for next-generation probiotics targeting metabolic syndrome. Curr Opin Biotechnol 32, 21–27, doi: 10.1016/j.copbio.2014.10.006 (2015). [DOI] [PubMed] [Google Scholar]
- Parks B. W. et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab 17, 141–152, doi: 10.1016/j.cmet.2012.12.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell host & microbe 17, 72- 84, doi: 10.1016/j.chom.2014.11.010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M. K., Boudry G., Lemay D. G. & Raybould H. E. Changes in intestinal barrier function and gut microbiota in high-fat diet-fed rats are dynamic and region dependent. American journal of physiology. Gastrointestinal and liver physiology 308, G840–851, doi: 10.1152/ajpgi.00029.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P. D. & Backhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab, doi: 10.1016/j.cmet.2015.07.026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao M. C. et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut, doi: 10.1136/gutjnl-2014-308778 (2015). [DOI] [PubMed] [Google Scholar]
- Everard A. et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60, 2775–2786 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Derrien M., Isolauri E., de Vos W. M. & Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73, 7767–7770, doi: 10.1128/AEM.01477-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoyama K. et al. Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl Environ Microbiol 75, 6451–6456, doi: 10.1128/AEM.00692-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remely M. et al. Increased gut microbiota diversity and abundance of Faecalibacterium prausnitzii and Akkermansia after fasting: a pilot study. Wiener klinische Wochenschrift, doi: 10.1007/s00508-015-0755-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs D. I., Enriori P. J., Lemus M. B., Cowley M. A. & Andrews Z. B. Diet-induced obesity causes ghrelin resistance in arcuate NPY/AgRP neurons. Endocrinology 151, 4745–4755, doi: 10.1210/en.2010-0556 (2010). [DOI] [PubMed] [Google Scholar]
- Claret M. et al. AMPK is essential for energy homeostasis regulation and glucose sensing by POMC and AgRP neurons. J.Clin.Invest 117, 2325–2336 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewulf E. M. et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 62, 1112–1121, doi: 10.1136/gutjnl-2012-303304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]