Abstract
Detecting intracellular calcium signaling with fluorescent calcium indicator dyes is often coupled with microscopy techniques to follow the activation state of non-excitable cells, including lymphocytes. However, the analysis of global intracellular calcium responses both at the single-cell level and in large ensembles simultaneously has yet to be automated. Here, we present a new software package, CalQuo (Calcium Quantification), which allows the automated analysis and simultaneous monitoring of global fluorescent calcium reporter-based signaling responses in up to 1000 single cells per experiment, at temporal resolutions of sub-seconds to seconds. CalQuo quantifies the number and fraction of responding cells, the temporal dependence of calcium signaling and provides global and individual calcium-reporter fluorescence intensity profiles. We demonstrate the utility of the new method by comparing the calcium-based signaling responses of genetically manipulated human lymphocytic cell lines.
Cellular calcium signaling is involved in most aspects of the cell’s life cycle1,2,3. Signaling occurs when the cell is stimulated to release calcium ions (Ca2+) from intracellular compartments and/or when Ca2+ enters the cell through calcium permeable channels4,5. Examples range from strongly localized Ca2+ sparks or spikes accompanying contraction in cardiac muscle cells or synaptic signaling in neuronal cells, to more global responses in non-excitable cells such as lymphocytes. In view of the large extracellular Ca2+ levels (>1 mM), cells must invest significant resources to maintain and to drive changes in the relatively low cytoplasmic Ca2+ levels (10–100 nM). This is especially true when calcium channels in the endoplasmic reticulum (ER) or the plasma membrane open during signaling, and the cytoplasmic Ca2+ concentration increases 10–100 fold5. In the case of non-excitable cells, triggering of cell surface protein receptors often leads to a sudden increase in cytoplasmic calcium levels5. Triggering of G-protein-coupled receptors, tyrosine kinase-coupled receptors, such as growth factor receptors and, in the case of leukocytes, non-catalytic tyrosine-phosphorylated receptors, such as B- and T-cell receptors activate phospholipase C (PLC) which hydrolyzes the membrane phospholipid PIP2 to form inositol 1,4,5-triphospate (IP3). IP3 then diffuses to the ER where it binds to the IP3 calcium channel triggering the release of Ca2+ 2,4,6.
Despite the identification and extensive characterization of the signaling pathways initiated by receptor triggering, and the central importance calcium plays in these events, the automated real-time quantification of calcium dynamics is generally not straightforward. Calcium responses are rapid and usually transient, follow many different pathways, and can vary significantly from cell to cell. Such dynamic and variable responses place considerable demands on the quantitative analysis of stimulus-elicited changes in cytoplasmic Ca2+ levels. While various automated analysis methods have been proposed to quantify Ca2+ levels or to resolve strongly localized Ca2+ sparks or spikes in e.g. the cytoplasm of cardiac muscle cells or neurons7,8,9,10,11,12, similar approaches have not yet been applied to study IP3-mediated Ca2+ responses triggered by cell surface receptors expressed by non-excitable cells, such as leukocytes. Here, Ca2+ “responses” are defined as global intracellular Ca2+ level increases that are transient on the sub-second to second time scale. Preferably, it needs to be possible to allow different routes of stimulation (e.g. via functionalized surfaces, cell-cell contact or controlled addition of small molecules) and to detect and distinguish calcium responses by individual cells within cell populations, at several micrometer spatial resolution and sub-second temporal resolution non-invasively. Common live-cell approaches rely on the use of fluorescence reporters that change emission properties upon Ca2+ binding13,14. Typically, quantitation of Ca2+ responses involves observing a small number of cells (<100) under a microscope, or in a flow cytometer or micro-plate reader. These approaches are limited in scope and struggle especially to quantitate individual cell behavior within large ensembles of cells. Additional issues that often arise include the detection sensitivity and/or sampling noise4,15. Recently, quantification of global Ca2+ responses of individual cells within an ensemble has been achieved by using fluorescence microscopy with single-cell resolution in combination with different software packages16,17. However, automation has yet to be fully incorporated into these analyses, and instead time-consuming, user-dependent, manual procedures had to be used, supported occasionally by commercial software packages of limited capability such as Microsoft Excel13. To overcome these limitations and to facilitate the automated quantification of global intracellular Ca2+ responses we developed a bespoke MATLAB-based software for Calcium Quantification (CalQuo). CalQuo allows automated, real-time determination of global calcium responses in hundreds of individual cells simultaneously, using conventional fluorescence microscopes, in this case a spinning-disk confocal scanning microscope.
We exemplified our approach by examining the calcium responses of T-cells settling on functionalized microscope cover glass. The release of intracellular Ca2+ is one of the earliest steps in T-cell activation and a well-established maker for early T-cell receptor (TCR) triggering13,14. Because early T-cell activation events modulate the course of the adaptive immune response, the quantification of cytoplasmic Ca2+ has been an important tool in the study of these processes13. We measured changes in cytoplasmic Ca2+ concentration using the fluorescein-based tetracarboxylate chelator Fluo-4, a Ca2+ fluorescent reporter, which upon binding to divalent cations increases its fluorescence emission, in the case of calcium, up to 100 fold18,19. We graphically visualize the Fluo-4 calcium transients corresponding to the calcium responses by monitoring the fluorescence intensity profiles of individual T-cells. Equipped with a low 10 x magnification and high numerical-aperture (NA = 0.45) objective and a 50 μm sized pinhole, the spinning disc microscope allows high-speed imaging of a large field of view (900 × 900 μm2) with hundreds of cells at single-cell spatial resolution simultaneously. In the present experiments the focal plane was placed at the glass surface and we recorded 870 images at a frame rate of 2 Hz (Supplementary Materials). In addition to image acquisition speed, the spinning disk microscope has the advantage of minimizing detection of out-of-focus fluorescence signals, allowing cells at and above the glass surface to be distinguished.
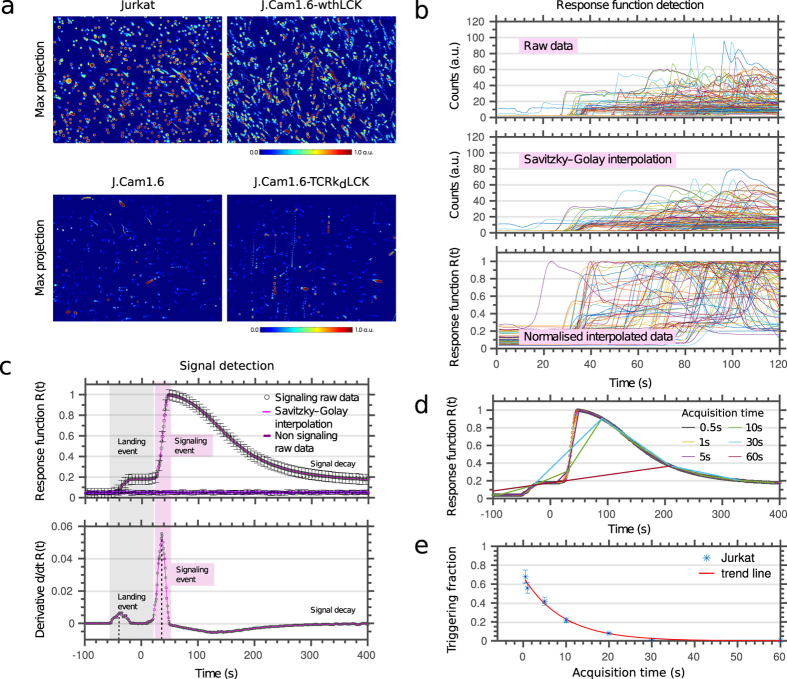
CalQuo uses feature recognition and distance regularized level set evolution (DRLS) algorithms20,21,22 to segment the raw-data image stacks and uses the calcium-dependent fluorescence signal to detect both calcium levels and cell features (see Supplementary Materials). Specifically, CalQuo detects cell motion, including the moment cells interact with, or “land” on the surface. To visualize the calcium responses following these landing events, we generated maximum projections of the time-lapse images with the detected fluorescence intensity color-coded from blue (low) to red (high calcium), as shown in Fig. 1a. The images reveal the motion of individual cells over time in suspension, allowing landing cells to be identified by an immediate cessation of movement, with the subsequent calcium response represented by a sudden increase in the fluorescence signal.
Figure 1. Observing global calcium release in T-cells using CalQuo.
(a) Maximum projections of fluorescence intensity over time from Fluo-4 loaded Jurkat (upper left), J.Cam1.6 (lower left), J.Cam1.6-wthLCK with restored signaling ability (upper right), and J.Cam1.6-TCRbkd-Lck with reduced TCRβ chain (lower right) when landing on the activating antibody-coated microscope cover glass, as measured by a spinning disc confocal fluorescence microscope, whose sampling plane was put on the cover glass. Low to warm colors represent low to high fluorescence intensities. Moving and triggering T-cells can be identified by a red dot with a blue tail. (b) Raw response data R(t) from different individual cells demonstrating the different analysis step: raw data (upper panel), data following Savitzky–Golay interpolation (middle panel) and normalised interpolated data (lower panel). (c) Representative intensity profiles R(t) (upper panel) and their derivatives dR(t)/dt (lower panel); average over 200-650 individual T-cells (black circles: raw data, mangenta line: Savitzky–Golay interpolation for signaling T-cells, purple line: data of non-signaling cells). CalQuo determines the characteristic times for the landing (grey shaded area) and signaling event (magenta shaded area) from the characteristic peaks in dR(t)/dt. Error bars representing s.d.m. (d) Representative response functions R(t) of a calcium releasing cell analyzed for different time resolutions 0.5 s–60 s, as indicated. The ability to identify calcium response decreases with decreasing time resolution, as revealed by (e) the fraction of signaling, i.e. calcium-releasing cells determined from the same data set for different time resolutions as shown in (e).
Cells that reached the surface in the first 300 s and were within the field of view (and therefore not closer than one cell diameter from the outer edges) were identified and selected by CalQuo for further analysis. We specifically excluded events where cell boundaries overlap at the same position. Such events were <2% due to choosing a suitable cell density. In this way, CalQuo was able to record the calcium responses for virtually every cell in the field, i.e. the fluorescence intensities I(t) over time t (Fig. 1b and Fig. S1). For our analysis, we interpolated and normalized the individual raw data curves using the Savitzky-Golay and normalised-data interpolation (see Supplementary Materials), defining the relative fluorescent intensity R(t) = I(t)/Imax, where Imax is the maximum intensity value of the response curve (Fig. 1b). To follow the calcium responses of individual cells, R(t) was averaged across each cell. In the next step, CalQuo identified the landing time and the beginning of the calcium response (described in more detail in Supplementary Materials). Both events led to an increase in fluorescence signal and therefore became prominent as two separate peaks in the derivative dR(t)/dt of the response function (Fig. 1c). The increase in fluorescence signal due to landing was relatively small and was the result of the weakly fluorescent cells reaching the focal plane of the objective located at the glass surface. In contrast, cell signaling resulted in the strong calcium-dependent increase of the fluorescence emission of Fluo-4. Signaling cells were thus identified by a characteristic sharp increase in fluorescence (after landing) followed by a slow decrease, while non-signaling cells exhibited little or no change in fluorescence signal. To allow direct comparisons of the responses of individual cells, the time axes of all the response curves R(t) were aligned, with time t = 0 set to 2 s after the landing event (Fig. 1c). CalQuo therefore allowed us to (1) identify the number and fraction of triggered cells, i.e. those showing a >500% increase above background (arbitrarily set by the user) and subsequent decrease in R(t); and (2) determine a response profile R(t) averaged over all activated cells, with error bars reflecting the cell-to-cell variation in responses (Fig. 1c).
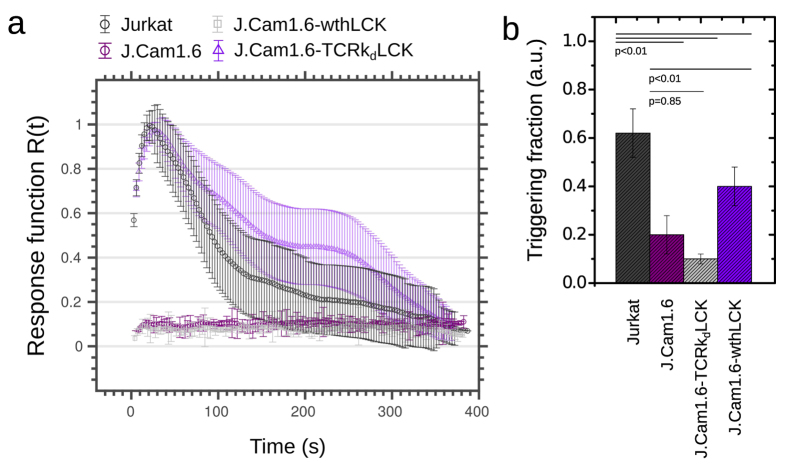
To demonstrate the capabilities of CalQuo, we characterized the calcium responses of Jurkat T-cells and a calcium signaling-deficient derivative, J.Cam1.6, which is a T-cell line deficient in the Lck kinase due to partial deletion of the Lck gene23,24,25. J.Cam1.6 cells are unresponsive to T-cell receptor (TCR) activation and therefore exhibit impaired calcium responses upon TCR-ligand binding23,25. Both Jurkats and J.Cam1.6 cells were labeled with the Fluo-4 dye and allowed to interact with a glass surface coated with stimulating anti-CD3ε (OKT3) and anti-CD28 (CD28.2) antibodies at 37 °C for ~400 s25,26. CalQuo detected clear differences in the fractions of responding Jurkats and J.Cam1.6 cells (Fig. 1a, left panels): whereas 60% of Jurkat T-cells exhibited an increase in Fluo-4 intensity upon surface contact and thus antibody binding (Fig. 2 and Movie S1), only 20% of the J.Cam1.6 cells showed a similar response (Fig. 2 and Movie S2). Moreover, the average response curve R(t) obtained for the Jurkat T-cells exhibited the characteristic sharp increase in fluorescence followed by a slow decrease, while that of J.Cam1.6 cells was essentially flat (Fig. 2a). Upon transfection of wild-type Lck, 40% of signaling competent J.Cam1.6 cells showed the characteristic calcium response following TCR triggering (J.Cam1.6 wthLCK, Fig. 1a right top panel, Fig. 2 and Movie S3)23. Conversely, Lck-expressing J.Cam1.6 cells where the expression of the TCRβ chain, and therefore of the TCR complex, was reduced using shRNA exhibited severely compromised calcium responses upon stimulation (J.Cam1.6 TCRkdLCK, Fig1a right lower panel, 10% responses, Fig. 2 and Movie S4).
Figure 2. CalQuo output for the different types of T-cells.
Average response functions R(t) (a) and fraction of signaling, i.e. calcium-releasing cells (b). Error bars as s.d.m. over 200–650 cells (see Table 1).
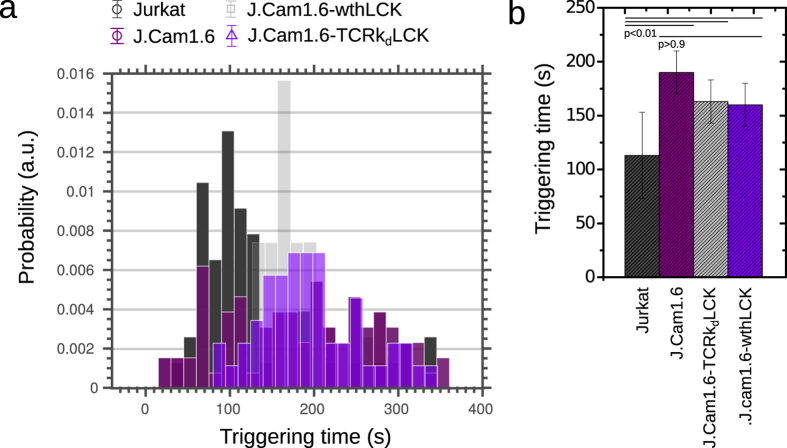
By determining the exact times of stimulation (i.e. landing) and signaling (i.e. calcium release) using R(t), CalQuo allowed us to measure the time interval between these two events, T = ttriggering – tlanding, which is another parameter characterizing the calcium signaling response, and is particularly useful for characterizing the agonist potential of putative receptor ligands. In Fig. 3a a histogram showing T for the four sets of T-cells responding to stimulating conditions is shown, and Fig. 3b depicts boxplots of the triggering intervals. Overall, Jurkat T-cells showed the highest triggering fraction and the fastest TCR triggering responses (Table 1), followed by signaling-restored J.Cam1.6 cells (J.Cam1.6 wthLCK). As expected both J.Cam1.6 and J.Cam1.6 cells lacking TCRβ chains (J.Cam1.6 TCRkdLCK) exhibited significant delays in TCR triggering (Table 1).
Figure 3. CalQuo output for the different types of T-cells:
Triggering time T (i.e. time between landing and signaling) for the fraction of signaling cells; histogram (a) and corresponding boxplots (b). Error bars as s.d.m. over 200–650 cells (see Table 1).
Table 1. Average and s.d.m. of the fraction of calcium responding cells and the triggering times T for the different types of T-cells (N = number of cells investigated).
| Experiment | Triggeringfraction | Triggeringtime | N |
|---|---|---|---|
| Jurkat | 0.6 ± 0.1 | 113 ± 40 | 366 |
| J.Caml.6 | 0.2 ± 0.1 | 191 ± 30 | 629 |
| J.Caml.6-wthLCK | 0.4 ± 0.1 | 158 ± 20 | 302 |
| J.Caml.6-TCRkdLCK | 0.1 ± 0.01 | 163 ± 20 | 213 |
We also tested the impact of time resolution on the analysis. We analyzed our data mimicking different time resolutions from 0.5 s to 60 s (Fig. 1d). The fraction of positive Ca2+ responses was significantly lower at the slower acquisition times, with >1 s fast recordings as used elsewhere13,17 partially or even fully failing to detect responses (Fig. 1e).
While CalQuo allowed us to quantify with high statistical accuracy fractions of Ca2+ responding cells within a cell ensemble, the investigated responses were averaged over the whole cell. CalQuo is therefore ideally suited to characterize receptor triggering in non-excitable immune cells. In its current formulation, CalQuo is unable to resolve Ca2+ “sparks”, usually occurring locally within a cell (sub-micrometer) within milliseconds, or quantify absolute Ca2+ concentrations. However, CalQuo allowed us to determine the contribution of differences in Fluo-4 dye loading and/or background levels to the global responses under analysis. To this end, we determined absolute (i.e. non-normalized) fluorescence intensity levels of the basal and maximum signal, I0 and dI = Imax – I0, respectively, at single-cell level. Values of dI/I0 did not change in the course of our study, i.e. Jurkat, J.Cam 1.6, J.Cam1.6 wthLCK and J.Cam1.6 TCRkdLCK (Fig. S2), indicating that our readout was robust against dye loading or background levels. Furthermore, the determined response curves were largely unaffected by collecting 2500 to 20 camera pixels per cell (Fig. S3), suggesting that, in principle, CalQuo could be used with a 5 x magnification objective lens. Indeed, the fraction of triggered cells was unaffected by moving from the 10 x objective lens to a 40 x or even 60 x objective lens on our spinning-disc microscope (Fig. S4).
In conclusion, we have developed a robust method for the automated quantification of global calcium responses of individual cells within relatively large populations. Combining feature detection and the novel DRLS algorithms with our response curve filtering algorithms offers unprecedented accuracy in segmenting the raw images for cell feature detection and reliable quantification of the calcium responses. The use of CalQuo in combination with fast spinning-disc microscopy and fluorescent calcium reporters allowed the quantification of the number (or fraction) of calcium-releasing cells, including the time-dependent global calcium responses, and the interval between stimulation and calcium release. At present the software requires a minimal level of MATLAB-based operating knowledge.
CalQuo is well suited to studying other cell signaling processes, such as the release of proteins or other molecules (e.g. toxins), if these molecules can be detected using fluorescent labels. We expect CalQuo to contribute to a more facile and thus better understanding of the molecular basis of global signaling in eukaryotic cells.
Additional Information
How to cite this article: Fritzsche, M. et al. CalQuo: automated, simultaneous single-cell and population-level quantification of global intracellular Ca2+ responses. Sci. Rep. 5, 16487; doi: 10.1038/srep16487 (2015).
Supplementary Material
Acknowledgments
We thank Alain Townsend for kindly providing the lentiviral plasmid used for shRNA delivery. Funding: This research was supported by funding from the Wellcome Trust, the Medical Research Council (grant number MC_UU_12010/unit programmes G0902418 and MC_UU_12025), BBSRC and ESPRC (grant number MR/K01577X/1), the Wolfson Foundation, and institutional funding from the University of Oxford.
Footnotes
Author Contributions M.F. designed, performed the research, and developed the custom-written analysis software in matlab. R.F. and H.C. contributed to code writing. M.F., R.F. and A.S. performed experiments. C.B.L. and S.L.L. helped setting up the experiments. M.F. and R.F. wrote the manuscript. S.D. and C.E. supervised the project, and wrote the manuscript.
References
- Berridge M. J., Bootman M. D. & Lipp P. Mechanisms for T cell receptor triggering. Nature. 395, 645–648 (1998). [DOI] [PubMed] [Google Scholar]
- Clapham D. E. Calcium Signaling. Cell. 131, 10471–59 (2007). [DOI] [PubMed] [Google Scholar]
- Bootman M. D. Calcium Signaling. Cold Spring Harbor Perspectives in Biology. 4, a011171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G. S., DeHaven W. I., Smyth J. T. & Putney J. W. Jr. Methods for studying store-operated calcium entry. Methods 46, 1046–2023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J., Lipp P. & Bootman M. D. The versatility and universality of calcium signaling. Nature Reviews 1, 11–21 (2000). [DOI] [PubMed] [Google Scholar]
- Khan A. A., Steiner J. P., Klein M. G., Schneider M. F. & Snyder S. H. IP3 receptor: localization to plasma membrane of T cells and cocapping with the T cell receptor. Science 7, 815–8 (1992). [DOI] [PubMed] [Google Scholar]
- Picht E., Zima A. V., Blatter L. A. & Bers D. M. SparkMaster: automated calcium spark analysis with ImageJ. Am J Physiol Cell Physiol. 293, C1073–C1081 (2007). [DOI] [PubMed] [Google Scholar]
- Kong C. H. T., Soeller C. & Cannell M. B. Increasing sensitivity of Ca2+ spark detection in noisy images by application of a matched-filter object detection algorithm. BiophyJ, 95, 6016–6024 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele E. M. & Steele D. S. Automated Detection and Analysis of Ca2+ Sparks in x–y Image Stacks Using a Thresholding Algorithm Implemented within the Open-Source Image Analysis Platform ImageJ. BiophyJ, 106, 566–576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis et al. Automated region of interest analysis of dynamic Ca2+ signals in image sequences. Am J Physiol Cell Physiol, 303, C236–C243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock J. T., Ellefsen K. L., Settle B. & Parker I. Smith, Imaging local Ca2+ signals in cultured mammalian cells. IFJ. Vis. Exp. 97, e52516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C. & Konnerth A. Imaging calcium in neurons. Neuron, 73, 862–885 (2012). [DOI] [PubMed] [Google Scholar]
- Wülfing C. et al. Kinetics and Extent of T Cell Activation as Measured with the Calcium Signal. J Exp Med. 19, 185(10): 1815–1825 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D. J. et al. Direct observation of ligand recognition by T cells. Nature 419, 845–849 (2002). [DOI] [PubMed] [Google Scholar]
- Albrecht T., Zhao X., Nguyen T. H., Campbell R. E. & Johnson J. D. Fluorescent biosensors illuminate calcium levels within defined beta-cell endosome subpopulations. Cell Calcium. 57, 263–274 (2015). [DOI] [PubMed] [Google Scholar]
- Christo S. N. et al. Scrutinizing calcium flux oscillations in T lymphocytes to deduce the strength of stimulus. Sci Rep. 5, 7760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp A. M. et al. Lck Mediates Signal Transmission from CD59 to the TCR/CD3 Pathway in Jurkat T Cells. PLoS ONE. 9, e85934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minta A., Kao J. P. Y. & Tsien R. Y. Journal of Biological Chemistry 264 (14), pp. 8171–8178 720 (1989). [PubMed] [Google Scholar]
- Gee K. R., Brown K. A., Chen W.-N. U., Bishop-Stewart J., Gray D. & Johnson I. Cell Calcium 27, 97–106 (2000). [DOI] [PubMed] [Google Scholar]
- Li C., Xu C., Gui C. & Fox M. D. Distance Regularized Level Set Evolution and its Application to Image Segmentation. IEEE Trans. Image Processing, 19, 12 (2010). [DOI] [PubMed] [Google Scholar]
- Gao Y. & Kilfoil M. L. Accurate detection and complete tracking of large populations of features in three dimensions. Opt Express. 16, 17(6), 4685–704 (2009). [DOI] [PubMed] [Google Scholar]
- O’Haver T. C. Version 5.1, MATLAB routine, http://terpconnect.umd.edu/~toh/spectrum/ PeakFindingandMeasurement.htm (1995).
- Straus D. & Weiss A. Genetic evidence for the involvement of the lck tyrosine kinase in signal transduction through the T cell antigen receptor. Cell 70, 585–593 (1992). [DOI] [PubMed] [Google Scholar]
- Karnitz L. et al. Effects of p56lck deficiency on the growth and cytolytic effector function of an interleukin-2-dependent cytotoxic T-cell line. Mol. Cell. Biol. 12, 4521–4530 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettgen H. C., Terhorst C., Cantley L. C. & Rosoff P. M. Stimulation of the T3-T cell receptor complex induces a membrane-potential-sensitive calcium influx. Cell 40, 583–590 (1985). [DOI] [PubMed] [Google Scholar]
- Davis M. M. et al. Ligand recognition by alpha beta T cell receptors. Annual Rev. Immunol. 16, 523–544 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.