Abstract
Single-molecule-localization-based superresolution microscopy requires accurate sample drift correction to achieve good results. Common approaches for drift compensation include using fiducial markers and direct drift estimation by image correlation. The former increases the experimental complexity and the latter estimates drift at a reduced temporal resolution. Here, we present, to our knowledge, a new approach for drift correction based on the Bayesian statistical framework. The technique has the advantage of being able to calculate the drifts for every image frame of the data set directly from the single-molecule coordinates. We present the theoretical foundation of the algorithm and an implementation that achieves significantly higher accuracy than image-correlation-based estimations.
Introduction
Single-molecule-localization-based superresolution microscopy has revolutionized the optical microscopy field by pushing spatial resolution to the scale of nanometers (1, 2, 3). The remarkable improvement in spatial resolution comes at the cost of a more complicated imaging procedure: instead of taking simple snapshots of the sample, tens of thousands of images are taken from the same sample, in which random subsets of the target molecules are turned on to be imaged and localized. The final image from the process is in the form of a histogram describing the frequency of the molecules being localized to certain spatial pixels. Sample drift during the data collection process can be minimized, but is generally unavoidable. The popularity of this imaging method has resulted in extensive research on localization algorithms to process single-molecule imaging data, and the efficiency and accuracy of various algorithms have been discussed in significant detail (4, 5). However, without accurate sample drift correction, the spatial resolution in the final reconstructed image will be poor even with the best localization accuracy.
Current sample drift-correction techniques can be categorized into two groups. The first group attempt to directly measure the drift with hardware implementations. A popular technique is to add bright fiducial markers into the sample, which are coimaged with the target molecules (1, 6). Other related techniques include the use of a secondary image of the sample (7, 8). These techniques introduce extra complexities into the experimental procedure and are not always straightforward to implement. For example, fiducial markers often themselves photobleach gradually, which could result in shifting of their centroid positions and thus in errors in the drift measurements. The second group of techniques are based on the idea of estimating drift directly from the single-molecule data using image correlation (3, 9, 10, 11). In general, drift compensation of this type involves computing coarse superresolution images based on substacks of the total data set and computing sample drift of those substacks using image correlation. Although simple to implement, the technique has the disadvantage that the drift is estimated at a coarse time resolution. Furthermore, although the technique works well for drifts that are smooth, it could be problematic if mechanical creeps, which are sudden and large jumps in sample positions due to build-up of mechanical strain, existed in the drift.
To offer a better approach for drift compensation, we treat it as a statistical inference problem. According to the Bayesian statistics framework, the estimation of the drift, d, from the single-molecule data set, , is the problem of obtaining a maximum a posteriori (MAP) estimation:
| (1) |
Here, the data set is a three-dimensional matrix representing all N frames of individual superresolution images. The size of each image is W × H pixels. For raw experimental data, the intensity values of each pixel can really only be either 0 or 1, depending on whether a molecule is detected at that pixel or not. However, here we will deal with a slightly more general case, in which the intensity can be any natural number, i.e., 0, 1, 2 …. This allows us to deal with special cases where the raw frames were binned every few frames before drift inference, which is useful for reducing computational time for extremely large data sets. The drift, , is a matrix representing time-dependent sample positions of all image frames. Although this article focuses on two-dimensional imaging, extension to the three-dimensional case should be straightforward. Furthermore, since the final constructed images are in pixelized form, it is unnecessary to compute drifts at unlimited resolution. Thus, we model drift in the integer domain, assuming they are in units of pixels.
Importantly, the term P(d) reflects our prior knowledge about the drift. Incorporating prior knowledge is a powerful way to achieve accurate inference from noisy data. In fact, the success of the image-correlation method is at least in part due to the fact that it implicitly incorporates a prior knowledge—the drift should be smooth in time. We explicitly model the prior distribution (see Materials and Methods) as a Markovian process; the displacements between adjacent frames were assumed to be approximately normally distributed, which favors smooth drifting traces.
Direct optimization based on Eq 1 is, unfortunately, difficult, because the computation of the probability term involves high dimensional integration. However, we noted that if we had guessed a final superresolution image, , the probability distribution of the drift could be estimated relatively easily according to the statistical modeling of . Based on this insight, we can iteratively make a better estimation of d and using the so-called expectation maximization algorithm (12) and finally reach cooptimization of both the most likely drift trace and the compensated superresolution image, . Details of the statistical model and derivation of the convergence formula are outlined in the next section. We call our method Bayesian sample drift inference (BaSDI).
Materials and Methods
BaSDI overview
Equation 1 is difficult to solve directly. However, we note that the difficulty is because we do not know anything about how the molecules are spatially distributed in the sample. In other words, we don’t have a good estimation of the true final superresolution image, , where is proportional to the molecular density at the spatial coordinate (i, j). For convenience, we can make sure that θ is normalized:
| (2) |
If we know the value of θ, the conditional probability can be expressed in relatively simple analytical forms. To see that, we note first that since θ is normalized, the probability of observing a single localization event at pixel location (i, j) is simply . In a single image frame, , there are multiple molecules detected at various coordinates. The joint probability of observing all these localization events is therefore
| (3) |
where denotes the total number of localization events in f and the factorial term is to account for the permutations of all the sequences of the molecules. Nevertheless, the expression ignores the effect of drift. If the current image frame is captured with a drifted sample position (x, y), then we need to shift the indices of the observed image. Furthermore, it is often convenient to compute probability distribution in log terms:
| (4) |
Here, we changed the equal sign to an approximate sign because the index shifting may have moved some of the observed localization events outside the reference frame, altering the total number of localization events considered. In practice, this approximation should not introduce large errors if we simply add an empty border of a certain amount of pixels, s, to the images. Then, as long as the maximum drift does not exceed s, we can still perform the computation to obtain reasonable values of the a posteriori distribution. In the real imaging experiment, data with too much drift (i.e., >s) are going to be problematic anyway. Therefore, the approximate approach should not introduce extra limitations.
Finally, the likelihood of observing the whole data set is simply the joint probability of all individual image frames:
| (5) |
where
| (6) |
Here, we dropped the explicit expression of the permutation terms because they are independent of the parameters we are trying to optimize.
We propose that the optimization can be realized with an iterative computational algorithm based on the framework of expectation maximization (EM) (12).
E-Step: Given a guessed matrix, compute the conditional distribution of , that is, what is the likelihood of certain drift traces if we already guessed the final superresolution image? A reasonable initial guess for the is simply the summed image of all frames without drift correction.
M-Step: Based on the computed distribution of , perform a new optimization of :
| (7) |
where means the expectation value, and D is the set of all possible configurations of d. It can be proven (12) that the new estimation, , is guaranteed to be a better one than the older estimation, .
To perform the optimization shown in Eq. 6, we first expand it by plugging in the result from Eq. 5.
| (8) |
where the definitions of i′ and j′ are the same as in Eq. 6. Next, we shift the summation over image frames out of the expectation function and express the expectation function as explicit summations:
| (9) |
Note that the optimization is constrained by the condition specified in Eq. 2. To obtain the maximum value of this function, we use the standard Lagrangian optimization technique, by solving for the root of the derivative of the target function in addition to a Lagrangian term:
| (10) |
and it’s easy to see from there that
| (11) |
Equation 11 now allows us to compute for the M-step. The E- and M-steps iterate until the algorithm converges. The most updated source code for the implementation is available to download from https://github.com/jiyuuchc/BaSDI/releases/.
Prior distribution for the drift, d
To apply the EM algorithm, we need to compute , which is a marginal probability that can be calculated by integrating over all frames that are not k. Note that this integration depends on the choice of the prior probability, P(d). However, brute-force integration would be typically too time-consuming to be practical, so the prior probability distribution needs to be designed carefully to satisfy two requirements: a) it needs to enforce some level of smoothness in the drift traces so as to agree with our physical intuitions about the drift, and b) it should allow easier computation of the marginal probability. In our case, we use a simple function based on the random-walk model, where the stepping probability distribution is a truncated Gaussian distribution:
| (12) |
| (13) |
where denotes normal distribution and is a hyperparameter corresponding to the speed of the drift. The value is used to account for the small probability of a creep—a rare, sudden jump with larger amplitude—in the system. We kept ε at 1/N for all our calculations in this article.
Because of the Markovian characteristics of p(d), the marginal probability can be computed using the well-known forward-backward algorithm,
| (14) |
where both the and the functions are calculated iteratively, which is much more efficient than the brute-force integration. The values are computed from the first frame to the last frame:
| (15) |
and the values are calculated in reverse, starting from the last image frame:
| (16) |
In both Eqs. 15 and 16, denotes the observed data in the kth image frame. Its probability calculation follows Eq. 4.
Efficiency of convergence
Although the EM algorithm guarantees the increase of the likelihood with each iteration, it does not guarantee convergence to the global maximum (i.e., it can converge to a local maximum). We found that although the algorithm performs very efficiently when the estimated image is smooth across most pixels, it tends to be trapped at a local maximum when contains many pixels of zero value. This suggests that for a data set of low sampling rate—i.e., when the total number of detected molecules is too low to construct a smooth final photoactivated localization microscopy (PALM) image—it is more efficient to compute at lower resolution (or use a larger pixel size), which has the effect of smoothing out the global-search-space function, allowing more efficient convergence to the global optimum. To accommodate all data-set scenarios, we designed the EM iterations to follow a multiround optimization schedule: The initial rounds reduce the effective resolution of by applying a smoothing filter. The size of the filter is gradually reduced in the later rounds, which iteratively refine the drift estimation until at the final round, no smoothing filter is applied.
The default optimization schedule we implemented in the accompanying software performs very well for all varieties of data set and has not failed once in all the simulated data sets we have generated for testing so far. Alternatively, to save computational time, one could skip earlier rounds of iteration if the data-set sampling rate is high, and can skip later rounds if the sampling rate is low. In all the testing cases, we found that the convergence of the BaSDI algorithm was typically reached in 5–10 iterations. For estimating 1000 drift frames from a data set of 1000 × 1000 pixels, each iteration takes less than 30 s to finish on a laptop personal computer. The computational time scales linearly with respect to the number of image frames and quadratically with respect to image size (Fig S1 in the Supporting Material), indicating good scalability to different sizes of data sets.
Simulation and algorithm validation
PALM data sets for algorithm validation were generated with Monte Carlo simulation. To do that, we first produce a ground-truth image with certain characteristic features. Molecules of interest are assumed to be concentrated within these features and much less abundant outside (the background). The values of the ground-truth image were assumed to represent the concentration of molecules within the area of each image pixel. To generate individual frames of localization images, we simply produced Bernoulli samples for each pixel position, for which the Bernoulli probabilities are assumed to be proportional to the intensity of the ground-truth image. Bernoulli sampling ensures that each pixel can have no more than one molecule in the localization image, similar to real experiments where molecules are localized individually. After a stack of multiple images was generated, each image was shifted according to a drift trace, also produced from Monte Carlo simulation (see below). The Bernoulli probabilities are assumed to be unchanged for all image frames. It should be noted that based on this generation process, all images were produced independent of each other. In other words, in our simulation, the detected molecules will generally not persist over multiple image frames. In real experiments, the switching off of a single molecule is not instantaneous; thus, signal from a single molecule sometimes persists over consecutive frames. This type of temporally correlated data provides extra information about the drift represented in the lateral translocations of those individual molecules. However, experimentally, it is also desirable to switch off any detected molecules as quickly as possible to increase data acquisition speed. Therefore, we opted to not utilize this extra temporal information, and the simulation is designed to test the validity of the algorithm in the most general case possible.
Drifts were generated according to a random-walk model, i.e., . Values of were chosen randomly from 0.05 to 0.5, unless otherwise specified, to simulate various rates of drift, although this value does not change within the same drift trace. To generate persistent drift traces, we model the velocity as a random walk, . Regardless of the ways the drifts were generated, for all simulations, the inferences by BaSDI were performed with the fixed prior hyperparameter of .
For image correlation analysis, the data were grouped into various numbers (4–20) of substacks and the best groupings were empirically chosen by comparing the results to the known input drift traces. This is of course impossible for real experimental data, where the true drift is unknown. Thus, for real experimental data, we used the grouping that performed best based on our simulations.
BaSDI application to experimental PALM microscopy data
The human Crk cDNA sequence, a gift from Dr. Bruce Mayer (University of Connecticut Health Center, Farmington, CT), was subcloned into a mammalian expression vector carrying mEos3, producing the mEos3-Crk fusion construct. MCF-7 or MEF cells were transfected with 500 ng of DNA using 1.5 μL of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 6 h, before being replated on plasma-cleaned glass-bottom dishes (MatTek, El Segundo, CA) overnight. Cells were then fixed with 4% paraformaldehyde, washed twice with phosphate-buffered saline, and imaged with PALM with 561-nm total-internal-reflection laser illumination and 405-nm photoactivation. Data acquisition was essentially the same as previously described (13). Image processing and single-molecule localization were performed using the octane software (13). The localization data were then used as input to BaSDI, which computed the optimized drift traces, as well as the final constructed superresolution image, in an unsupervised manner.
Results and Discussion
Algorithm validation with simulated image data
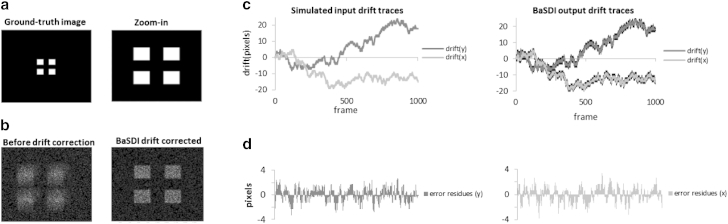
To validate the BaSDI algorithm, we used Monte Carlo simulation to generate various testing data sets and tested whether BaSDI correctly estimated drift from those data sets (Fig. 1). The ground-truth image is designed so that the features (central squares) represent ∼10% of the total probability mass; thus, ∼90% of the detected molecules will be in the background. To mimic the localization microscopy data, frame-by-frame molecular coordinates were simulated by random sampling of single-molecule coordinates from a ground-truth image (Fig. 1 a), assuming that 50 molecules were detected on average for each image and 1000 total images were acquired. Each image was shifted according to a two-dimensional drift trace according to the random-walk model to mimic experimental sample drifts. We then reconstructed the superresolution image (Fig. 1 b) either without any drift correction, or performing drift correction by feeding the simulated imaging data to BaSDI. To estimate statistical error, the test was repeated 30 times by resampling the same ground-truth image to produce new randomized data sets. It can be seen that the reconstructed PALM image without drift compensation is distorted and differs significantly from the ground truth image (Fig. 1 b). However, BaSDI was able to correctly estimate the drift traces (Fig. 1, c and d) and, after drift correction, produce a high-resolution image matching the ground-truth image (Fig. 1 b).
Figure 1.
Assessing BaSDI’s performance with simulated single-molecule localization data. (a) The ground-truth image of 500 × 500 pixels (left) and a zoom-in view of its key feature (four squares at the center) (right). (b) Simulated PALM images before (left) and after (right) drift correction by BaSDI. The input drift traces were simulated according to a random-walk model. Single-molecule coordinates are generated by randomly sampling the ground-true image in (a), assuming detection of 50 molecules per frame on average and 1000 frames per data set. (c) The input drift traces (left) and the corresponding BaSDI estimated drift traces (right). Sampling noise in the BaSDI calculation, represented by the line width (right), were estimated by rerunning the simulation and the corresponding drift recalculation 30 times. (d) Error residues obtained from the differences between the traces in (c) in the y and x directions.
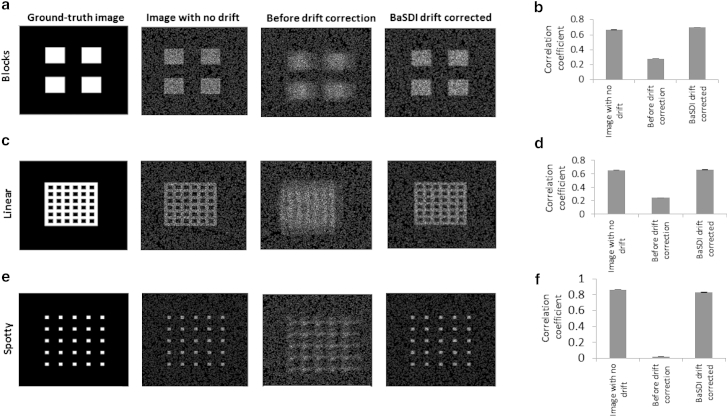
Real biological samples can exhibit many different types of spatial features. For example, cytoskeletal structures are made of mostly linear elements, and cell surface receptor clusters are spotty structures. Thus, we next tested how the underlying image features affect BaSDI performance (Fig. 2). Besides the blocky image shown in Fig. 1, we also generated two new ground-truth images, one with predominately linear features (Fig. 2 c) and the other with spotty features (Fig. 2 e). All ground truth images have the same integrated signal/background ratio, i.e., 10% of the total molecules residing within the features. We found, again with the simulated localization image data, that BaSDI was able to significantly improve the quality of the reconstructed image by correcting the drift in the data set. To quantitatively compare the performances in all three cases, we used the Pearson correlation coefficient to evaluate the similarity between the reconstructed images and the ground-truth image (Fig. 2, b, d, and f). Unsurprisingly, the uncorrected images had very low correlation with the ground-truth images, and BaSDI correction significantly improved the results. Interestingly, since we knew the exact drift used to produce the data, we could also construct images with perfect drift correction (Fig. 2, a, c, and e) and compute their correlation with the ground-truth images. Even with no residue drift, these images do not exhibit perfect correlation due to noise rising from sampling. More importantly, we observed no significant differences between their correlation values and the values of the BaSDI-corrected images, indicating that in these simulations, the BaSDI-corrected images are not statistically distinguishable from images with no drift. By comparing results from different ground-truth images, we found that the best results were obtained from the spotty images, whereas results from the blocky and linear images were similar. This is also not surprising, as intuitively one would expect that higher spatial frequency would allow higher confidence in reconstructing drift-corrected images. The blocky image likely represented the hardest problem; all the subsequent simulation tests will be performed on this particular ground-truth image.
Figure 2.
Assessing BaSDI’s performance for drift correction given different types of image features. Simulations were performed on (a and b) blocky, (c and d) linear, and (e and f) spotty ground-truth images. Images in (a), (c), and (e) are, from left to right, the ground-truth images and the reconstructed images with perfect drift correction, no drift correction, and BaSDI drift correction. Images in (b), (d), and (f) show the correlation coefficients for each of the reconstructed images with their respective ground-truth images. Error bars represent the mean ± SE, n = 30.
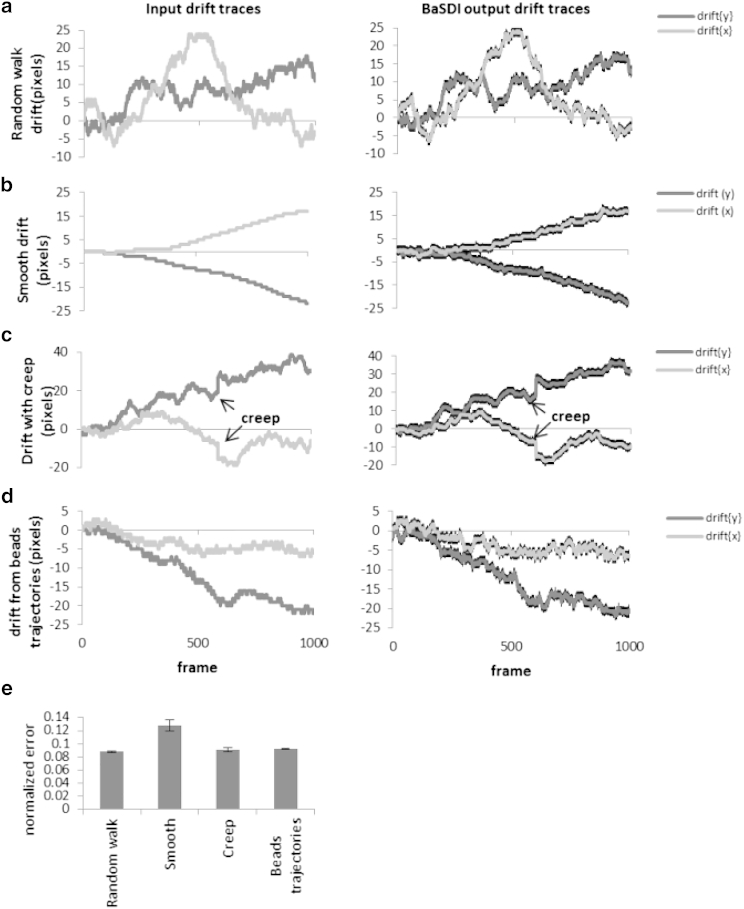
In our statistical model, we assumed a Markovian-type prior-distribution function for the drift. However, different microscopes may have very different drift characteristics due to differences in their mechanical properties. Therefore, we further tested whether the BaSDI algorithm can handle drift traces generated with other drift models (Fig. 3). In addition to the random-walk model (Fig. 3 a), similar to data shown earlier, we further examined two additional drift-generation models, a model in which the drift velocity (instead of drift position) undergoes random walk (Fig. 3 b), which results in a more persistent drift trace, and a model in which random mechanical creeps (sudden jumps of large amplitude due to the release of built-up stress) were introduced in a random-walk drift (Fig. 3 c). Finally, realistic drift characteristics can only be obtained by examining the drift from a real microscope. Therefore, we also obtained real experimental drift traces by tracking fluorescent beads from our own PALM microscope (Fig. 3 d), and we produced a simulated PALM data set using these experimentally obtained drift traces. We found that in all of these scenarios, BaSDI was able to estimate the drifts incorporated into the data set (Fig. 3). To quantify BaSDI’s performance in estimating drift traces from all different drift input scenarios, we calculated the root mean-square (RMS) error of the estimated drifts normalized to the standard deviation of the input drifts (Fig. 3 e). As expected, the largest error was observed in the case of persistent drift, probably because it deviated the most from our model assumptions. We found no significant differences in the RMS errors of the remaining three drift inputs.
Figure 3.
Assessing BaSDI’s drift estimation of different drift types. BaSDI was tested against (a) random-walk drift, (b) smooth drift, (c) drift with mechanical creep, and (d) drift from fluorescent bead trajectories. The input drift traces in both the y (vertical) and x (horizontal) directions are shown on the left (a–d), and the BaSDI estimated output drift traces for each type are shown on the right. (e) Normalized root mean-square error for BaSDI estimates drift for each type of input drift. Error bars are represented as the mean ± SE, n = 30.
Algorithm validation with experimental image data
A true test of the algorithm has to be based on its performance on real PALM experimental data. We performed PALM imaging experiments on MCF-7 cells expressing mEos3-Crk. Crk is an adaptor protein molecule that can induce cell transformation (14, 15). It contains an Src homology 2 (SH2) domain that allows it to bind to various tyrosine-phosphorylated proteins in cells. In mammalian cells, Crk is known to localize to focal adhesions (FAs) (15), which typically contain concentrated phosphotyrosines, but also to cell-surface receptor tyrosine kinases (16), which can be phosphorylated as well. The competition between the two and the difference in phosphorylation dynamics between them result in complicated Crk-localization variations. The Crk localization is further complicated by the fact that the majority of Crk localizes to cytosol, which complicates PALM imaging. Even with total internal reflection imaging, the high amount of cytosolic background reduces the effective sampling rate of membrane-localized Crk, increases the data collection time, and makes it more challenging to estimate drift directly from the single-molecule data.
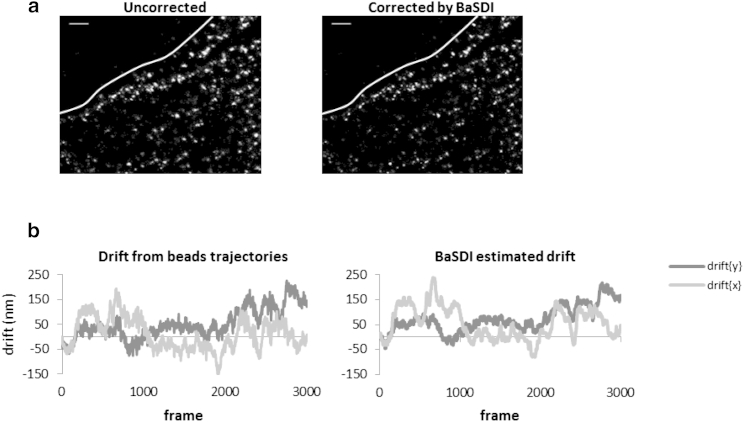
To validate the BaSDI algorithm with an experimental Crk PALM data set, we added fluorescent-bead-based fiducial markers into the cell samples during the PALM data collection and experimentally measured the drift of the sample stage during data collection using the positions of the fluorescent beads. The drift is then also estimated with BaSDI using the single-molecule coordinates, excluding any information from the fluorescent beads. The comparison between the experimentally measured drift and the BaSDI-estimated drift (Fig. 4) showed that they are highly consistent with each other. The effective difference between the two, measured in the RMS sense, is 25 nm, which is roughly equal to the expected single-molecule localization accuracy for these experiments. Thus, we conclude that BaSDI is able to correctly estimate drift from an experimental PALM data set.
Figure 4.
Comparison of BaSDI estimated drifts with experimentally measured drifts. (a) PALM images of mEos3-Crk in an MCF-7 cell before (left) and after (right) drift correction by BaSDI. Scale bars are 2 μm. (b) Drift traces measured by imbedded fluorescent beads (left) or estimated by BaSDI (right).
Performance of BaSDI algorithm
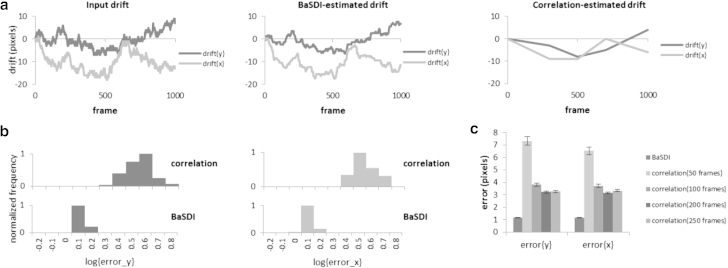
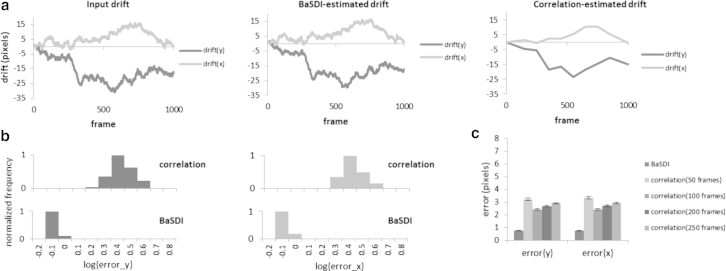
Next we tested whether BaSDI achieves better accuracy over the existing image-correlation methods. For comparisons, we generated 100 sets of simulated drift traces and the accompanying PALM data sets, each including 1000 individual image frames, based on the ground-truth image in Fig. 1. We then estimated the drift using either BaSDI or the image correlation method (Figs. 5 and 6). The accuracy of the methods was evaluated by computing the error of the estimated drift (by comparing to the input drift) in the mean-square error sense. Furthermore, to account for different image conditions, we generated simulated data in two different scenarios, a low-sampling-rate scenario, in which an average of 50 molecules were detected in each frame, and a high-sampling-rate scenario, where 200 molecules per frame were detected. Because image correlation only directly computes drift for a subset of key frames, we computationally generated the rest of the drift traces using linear interpolation (Figs. 5 a and 6 a). Another factor that we needed to consider when carrying out the image correlation was deciding how many substacks the whole data set should be divided into. Using too many substacks will cause high error in drift estimation between substacks, whereas too few substacks will lead to fewer data points and a worse temporal resolution (9). For real experimental data, the optimal number can be difficult to determine objectively. For our simulation, we computed the error for four different image-correlation groupings (50, 100, 200, and 250 frames). The groupings with the lowest error were selected to be compared with BaSDI, for both the low-sampling-rate (Fig. 5) and the high-sampling-rate scenarios (Fig. 6). As expected, the high sampling rate is correlated with lower error for both methods (Figs. 5 c and 6 c). Nevertheless, in both cases, we found that BaSDI outperforms image correlation by about three- to fourfold (Figs. 5 b and 6 b). Interestingly, BaSDI also seems to have a more stable performance, as indicated by the narrow distribution of errors in the histograms (Figs. 5 b and 6 b) in comparison to the image-correlation method.
Figure 5.
Comparing drift estimations by BaSDI and image correlation at a low sampling rate (50 molecules/frame). (a) Simulated input drift (left) and drift traces estimated by either BaSDI (middle) or image correlation using the optimal frame grouping with the least error (right). (b) Histograms comparing the root mean-square error distributions of BaSDI and image correlation. The distributions of error were computed from 100 independent runs with randomly generated drifts. (c) Comparing the averaged errors of BaSDI and image correlation given the different frame groupings. Error bars are represented as the mean ± SE, n = 100.
Figure 6.
Comparing drift estimations by BaSDI and image correlation at a high sampling rate (200 molecules/frame). (a) Simulated input drift (left) and drift traces estimated by either BaSDI (middle) or image correlation using the optimal frame grouping with the least error (right). (b) Histograms comparing the root mean-square error distributions of BaSDI and image correlation. The distributions of error were computed from 100 independent runs with randomly generated drifts. (c) Comparing the averaged errors of BaSDI and image correlation given the different frame groupings. Error bars are represented as the mean ± SE, n = 100.
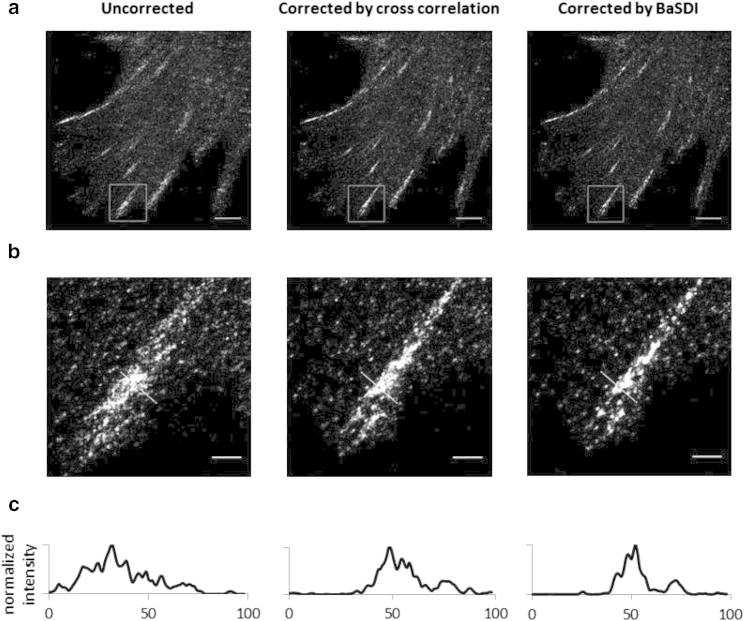
Finally, we evaluated the relative performance of BaSDI and image correlation by comparing the image quality from real PALM experimental data (Fig. 7). To do so, we imaged mEos3-Crk in propagating mouse embryonic fibroblast cells. In this condition, Crk localizes to the classical strip pattern typical of FA structure. We then applied either BaSDI or image correlation to carry out the drift compensation (Fig. 7) based on estimating drift traces of 18,000 points. Both methods improved the PALM image sharpness in comparison to the uncompensated result (Fig. 7). To choose a hyperparameter, , we utilized the existing fluorescent-bead tracking data measured separately on the same microscope and calculated the variance of position drifts between frames. We note that for small values, the accuracy of the drift calculation is insensitive to its exact value (Fig S2). Therefore, the variance measurement does not need to be very accurate, and the day-to-day variations in the drift characteristics of the microscope are expected to have little impact on the inference results. Thus, we did not perform recalibration for PALM data acquired on different dates. Comparison between the estimated drift from BaSDI and from the correlation method showed that the two methods generally agree with each other, except that BaSDI generated not only estimation of key frames but also detailed traces between key frames, resulting in better-resolved image features in the final constructed PALM images (Fig. 7 c).
Figure 7.
Comparing the performances of the BaSDI and image-correlation methods on experimental PALM data. (a) Uncorrected (left), image-correlation-corrected (middle), and BaSDI-corrected (right) PALM images of mEos3-Crk in a mouse embryonic fibroblast cell. Data showed aggregated localization patterns consistent with focal adhesions. Scale bars are 5 μm. (b) Zoomed-in view of the corresponding PALM images at the top, giving a detailed view of the focal adhesion structure outlined by the white box in (a). Scale bars are 1 μm. (c) Intensity line scans from (b) showing the improved image sharpness of the PALM images after drift corrections.
In conclusion, we have devised an algorithm to estimate sample drift from single-molecule-based superresolution imaging methods. The algorithm is validated by both computer simulation and experimental data. The algorithm outperforms previous analysis methods and, more importantly, is based on a statistical principle, not on heuristics. We believe that it will become a useful addition to the tool set for researchers working on superresolution imaging.
Author Contributions
A.E. performed experiments and analyzed data. J.Y. designed the project, performed experiments, and wrote the article.
Acknowledgments
This study is supported in part by NIH grant U01CA146843.
Editor: Paul Wiseman.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00954-6.
Supporting Material
References
- 1.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Hess S.T., Girirajan T.P.K., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang B., Wang W., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sage D., Kirshner H., Unser M. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods. 2015;12:717–724. doi: 10.1038/nmeth.3442. [DOI] [PubMed] [Google Scholar]
- 5.Small A.R., Parthasarathy R. Superresolution localization methods. Annu. Rev. Phys. Chem. 2014;65:107–125. doi: 10.1146/annurev-physchem-040513-103735. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.H., Baday M., Selvin P.R. Using fixed fiduciary markers for stage drift correction. Opt. Express. 2012;20:12177–12183. doi: 10.1364/OE.20.012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates M., Huang B., Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y., Wang X., Dai L. Sub-nanometer drift correction for super-resolution imaging. Opt. Lett. 2014;39:5685–5688. doi: 10.1364/OL.39.005685. [DOI] [PubMed] [Google Scholar]
- 9.Geisler C., Hotz T., Egner A. Drift estimation for single marker switching based imaging schemes. Opt. Express. 2012;20:7274–7289. doi: 10.1364/OE.20.007274. [DOI] [PubMed] [Google Scholar]
- 10.Mlodzianoski M.J., Schreiner J.M., Bewersdorf J. Sample drift correction in 3D fluorescence photoactivation localization microscopy. Opt. Express. 2011;19:15009–15019. doi: 10.1364/OE.19.015009. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Schnitzbauer J., Huang B. Localization events-based sample drift correction for localization microscopy with redundant cross-correlation algorithm. Opt. Express. 2014;22:15982–15991. doi: 10.1364/OE.22.015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dempster A.P., Laird N.M., Rubin D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Series B Stat. Methodol. 1977;39:1–38. [Google Scholar]
- 13.Tatavarty V., Kim E.J., Yu J. Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One. 2009;4:e7724. doi: 10.1371/journal.pone.0007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuda M., Tanaka S., Shibuya M. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol. Cell. Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J., Machida K., Mayer B.J. Proteins that bind the Src homology 3 domain of CrkI have distinct roles in Crk transformation. Oncogene. 2010;29:6378–6389. doi: 10.1038/onc.2010.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abassi Y.A., Vuori K. Tyrosine 221 in Crk regulates adhesion-dependent membrane localization of Crk and Rac and activation of Rac signaling. EMBO J. 2002;21:4571–4582. doi: 10.1093/emboj/cdf446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.