Abstract
The cellular process of intrinsic apoptosis relies on the peroxidation of mitochondrial lipids as a critical molecular signal. Lipid peroxidation is connected to increases in mitochondrial reactive oxygen species, but there is also a required role for mitochondrial cytochrome c (cyt-c). In apoptotic mitochondria, cyt-c gains a new function as a lipid peroxidase that catalyzes the reactive oxygen species-mediated chemical modification of the mitochondrial lipid cardiolipin (CL). This peroxidase activity is caused by a conformational change in the protein, resulting from interactions between cyt-c and CL. The nature of the conformational change and how it causes this gain-of-function remain uncertain. Via a combination of functional, structural, and biophysical experiments we investigate the structure and peroxidase activity of cyt-c in its membrane-bound state. We reconstituted cyt-c with CL-containing lipid vesicles, and determined the increase in peroxidase activity resulting from membrane binding. We combined these assays of CL-induced proapoptotic activity with structural and dynamic studies of the membrane-bound protein via solid-state NMR and optical spectroscopy. Multidimensional magic angle spinning (MAS) solid-state NMR of uniformly 13C,15N-labeled protein was used to detect site-specific conformational changes in oxidized and reduced horse heart cyt-c bound to CL-containing lipid bilayers. MAS NMR and Fourier transform infrared measurements show that the peripherally membrane-bound cyt-c experiences significant dynamics, but also retains most or all of its secondary structure. Moreover, in two-dimensional and three-dimensional MAS NMR spectra the CL-bound cyt-c displays a spectral resolution, and thus structural homogeneity, that is inconsistent with extensive membrane-induced unfolding. Cyt-c is found to interact primarily with the membrane interface, without significantly disrupting the lipid bilayer. Thus, membrane binding results in cyt-c gaining the increased peroxidase activity that represents its pivotal proapoptotic function, but we do not observe evidence for large-scale unfolding or penetration into the membrane core.
Introduction
Intrinsic apoptosis, one of the pathways for programmed cell death, is initiated by signaling events in mitochondria (1, 2). The mitochondrial apoptotic pathway is implicated in various diseases and represents an important target for drug design. In neurodegenerative diseases, including Huntington’s Disease, mitochondrial apoptosis leads to neuronal degradation and mitochondrially targeted antioxidant drugs have been shown to be protective (3, 4). Conversely, the triggering of apoptosis is considered a potential anticancer treatment (1). As a result, there is a need to understand the underlying mitochondrial signaling events. Early stages of the cell death process require functionally important changes in mitochondrial proteins, lipids, and membranes, with a particularly central role for the mitochondrial lipid cardiolipin (CL). This unique lipid is normally found in the matrix-facing inner leaflet of the inner mitochondrial membrane (IMM) (5, 6). As a precursor to apoptosis, this tightly controlled distribution is disrupted and CL becomes increasingly exposed to the intermembrane space (IMS) and the outer mitochondrial membrane (OMM) (7, 8, 9, 10, 11). CL is then recognized, in both locations, by different proteins that trigger lipid peroxidation, permeabilization of the OMM, and release of proapoptotic factors (12, 13). Crucially, the arrival of CL in the OMM and the peroxidation of CL polyunsaturated fatty acid (PUFA) chains are pivotal mitochondrial signals that trigger apoptosis or mitophagy (7, 14, 15). The proapoptotic peroxidation reaction is mediated by mitochondrial reactive oxygen species (ROS), but also requires the involvement of mitochondrial cytochrome c (cyt-c) (7, 16). As the normal asymmetric distribution of CL in the IMM is disrupted, there is an increase in the normally very low IMS-facing CL concentration. As this occurs, cyt-c becomes able to interact in new ways with the IMM, most likely through an interaction with multiple CL lipids. A CL-cyt-c complex is then formed, which turns cyt-c into a potent lipid peroxidase with high selectivity for CL over other lipids (17, 18, 19). Despite substantial interest and effort, it has proved challenging to gain insight into the CL-induced structural changes in cyt-c and how they trigger the lipid peroxidase activity. Of particular biological importance are the initial interactions of cyt-c with CL-containing membrane domains under conditions where the accessibility of CL and ROS (H2O2) represents a rate-limiting bottleneck that triggers the entire process.
Mechanistically, oxidation of the lipid acyl chains requires access of the PUFA to the catalytic reactive intermediates of cyt-c. These intermediates involve the protein’s covalently attached heme as well as protein-associated radicals, recently localized to Tyr-67 (14). These sites are buried in cyt-c’s native fold, which is visualized in Fig. 1 a. Some studies have suggested that accessibility of the catalytic site upon CL binding results from cyt-c acquiring a largely unfolded or molten globule-like state within the membrane (20, 21, 22, 23, 24, 25). In contrast, the extended lipid anchorage model (26, 27, 28, 29) proposes that cyt-c stays on the bilayer surface, but extracts one or two CL acyl chains from the membrane. Hydrophobic interactions between the extracted acyl chains and a hydrophobic cleft on the protein surface then facilitate the necessary proximity to cyt-c’s catalytic site. Solution NMR and x-ray crystallographic studies have yielded high-resolution structures of cyt-c mutants with enhanced peroxidase activity due to a local structural change, without a loss of native tertiary fold (30, 31). However, these structures were obtained in the absence of lipid bilayers, and the relevance of the data to the peroxidase activity gains induced by CL-binding remains unclear.
Figure 1.
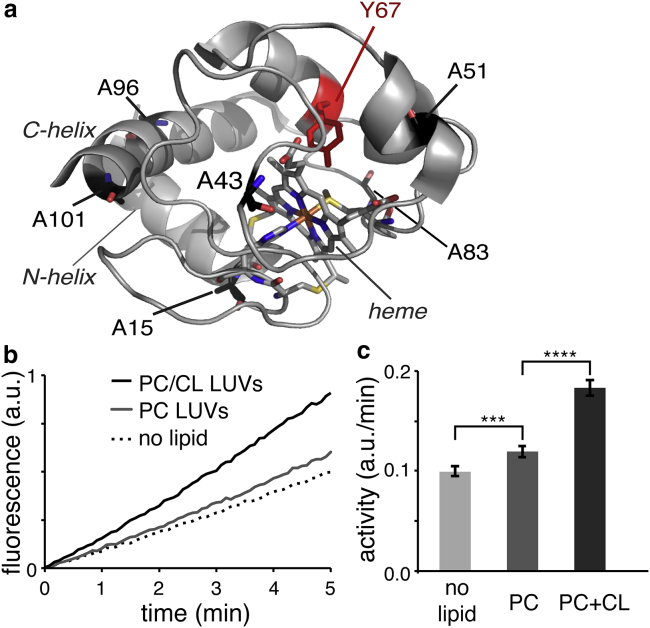
Cytochrome c structure and peroxidase activity. (a) X-ray structure of oxidized cyt-c (44). Labels mark the catalytic sites of peroxidase-active protein (heme and Tyr-67). Cyt-c’s six alanines are also indicated. (b) Generation of the fluorescent oxidation product resorufin by 0.5 μM cyt-c. Measurements were performed at room temperature in the absence (dashed line) and presence of DOPC LUVs (gray solid line) or DOPC/TOCL (4:1) LUVs (black solid line). (c) Relative peroxidation rates obtained from linear fits to the data in (b). To see this figure in color, go online.
Experimental characterization of membrane-bound cyt-c has proved challenging. Applicable techniques tend to yield relatively low-resolution structural insights and often also require the introduction of potentially disruptive fluorescent or spin labels. Solid-state NMR (ssNMR) spectroscopy permits the site-specific determination of structure and dynamics in proteins associated with lipid bilayers (32, 33, 34, 35, 36). Labeling for ssNMR involves nondisruptive stable isotopes, which can be uniformly applied because site-specific information is accessible via multidimensional spectra (35, 37). Here, we apply multidimensional MAS ssNMR to uniformly U-13C,15N-labeled cyt-c in its membrane-bound and peroxidase-active form. Prior ssNMR studies have provided important insights into the effect of bound, unlabeled cyt-c on the membrane, revealing for instance a propensity to disrupt the lipid bilayer structure (20, 21, 32, 38, 39, 40, 41, 42, 43). Previously, ssNMR on cyt-c itself was limited due to challenges with the isotopic labeling, with prior reports relying on methionine 13Cε labeling or 2H back exchange (20, 40). These studies revealed dynamics and disorder in the membrane-bound protein. Using peroxidase assays, Fourier-transform infrared (FTIR) spectroscopy, and advanced MAS NMR we probe the CL-induced peroxidase activation, and the structural features of both the membrane and the membrane-bound protein. Cyt-c is found to bind tightly to the CL-containing unilamellar vesicles, which causes the expected increase in peroxidase function. Static and MAS NMR spectra show no evidence of lipid bilayer disruption and indicate that the protein is present at the membrane interface. The membrane-bound cyt-c experiences substantial dynamics, but in room temperature FTIR studies as well as low-temperature two-dimensional (2D) and three-dimensional (3D) MAS ssNMR experiments we find that the secondary structure and much of the tertiary fold are retained.
Materials and Methods
Expression and purification of 13C,15N-cytochrome c
Uniform 13C,15N labeling of horse heart cyt-c was performed by overexpression in Escherichia coli. In line with previous reports, an expression-optimized mutant (H26N; H33N) was employed that eliminates kinetic folding traps, but is structurally similar to the wild-type protein (14, 45, 46). A pETDuet plasmid (EMD Millipore, Billerica, MA) was used to coexpress cyt-c and the heme lyase required for covalent attachment of the heme (45, 47). The respective cDNAs were cloned in the NcoI/EcoRI and NdeI/XhoI sites of the vector. For U-13C,15N isotopic labeling, the protein was expressed in minimal media with 0.1% (w/v) 15NH4Cl and 0.4% (w/v) 13C-labeled d-glucose, as well as 0.5 mM FeCl3 and 580 μM of unlabeled 5-aminolevulinic acid hydrochloride. At an optical density of ∼1.1, overexpression was induced by addition of IPTG, along with supplementary 15NH4Cl and 13C-d-glucose (0.1 and 0.2 g/L, respectively). After ∼16 h at 18°C, cells were recovered by centrifugation, resuspended in 25 mM sodium phosphate buffer (pH 6.5) with 0.02% sodium azide, and lysed. Cell debris was removed centrifugally, yielding a pink-colored supernatant. Purification of the labeled protein used a cation exchange HiTrap SP HP column, using a 25 mM sodium phosphate running buffer (pH 6.5) with 10% (v/v) glycerol and 0.02% sodium azide, and a 0–1 M NaCl salt gradient. The eluate was further purified on a gel filtration Superdex 75 26/60 column, using 25 mM sodium phosphate buffer at pH 6.5, with 150 mM NaCl, 5% glycerol and 0.02% sodium azide. Fully oxidized protein was obtained by treatment with a fivefold molar excess of potassium hexacyanoferrate (III), which was removed by extensive dialysis. Buffer exchanges to the experimental buffer (freshly degassed 20 mM HEPES buffer at pH 7.4) were performed via dialysis or use of 10 kDa Amicon Ultra-15 centrifugal filters. Typically, the yield of U-13C,15N-labeled cyt-c was 2.4 mg/L of cell culture.
Preparation of samples for ssNMR
Di-oleoylphosphatidylcholine (DOPC) and tetra-oleoyl-cardiolipin (TOCL) were obtained from Avanti Polar Lipids (Alabaster, AL). The lipids (in chloroform) were mixed in a 4:1 molar ratio, after which the solvent was removed under N2 flow and by drying under vacuum overnight. Next, 1 ml of freshly degassed 20 mM HEPES buffer (pH 7.4) was added, followed by gentle vortexing for 20–30 min. After freeze-thawing of the lipid mixture 10 times, 11 extrusion steps were performed using a mini-extruder (Avanti Polar Lipids) outfitted with 100 or 200 nm polycarbonate membranes, to yield large unilamellar vesicles (LUVs). Vesicle sizes were evaluated using dynamic light scattering (DLS), performed at room temperature on a DynaPro Plate Reader (Wyatt Technology, Santa Barbara, CA). The DLS data were analyzed using Wyatt’s DYNAMICS software (version 7) and plotted with Prism 6 (GraphPad, La Jolla, CA) and Microsoft Excel. When applicable, stock solution of cyt-c (in 20 mM HEPES pH 7.4) was added to a 1:40 protein/lipid molar ratio (P/L ratio), followed by gentle vortexing and incubation at 4°C. For MAS ssNMR, an ultracentrifugal sample packing device was used to pellet the LUVs or protein-lipid complexes into 3.2- and 4-mm MAS rotors. Centrifugation was either 3 h at ∼143,000 × g and 4.5 h at ∼175,000 × g, or ∼143,000 × g for 5 h, in a Beckman Coulter Optima L-100 XP ultracentrifuge with a SW-32 Ti rotor. The packing devices were designed and built in-house, and resemble sedimentation devices (48). The supernatant was removed and ultraviolet-visible light (UV-VIS) measurements on the supernatant were used to determine the amount of cyt-c bound to the pelleted, red-colored LUVs. MAS rotors were from Bruker Biospin (Billerica, MA) and CortecNet (Mill Valley, CA).
ssNMR spectroscopy
All ssNMR spectra were acquired on a wide-bore 600 MHz (14.1 T) spectrometer (Bruker Biospin, Billerica, MA), using 3.2 mm MAS probes with a HCN EFree coil or HFCN-tuned solenoid coil, unless otherwise noted. One-dimensional (1D) 13C spectra were obtained at 8.333 kHz MAS, using either ramped 1H-13C cross-polarization (CP) (1 ms contact time) (49), the rotor-synchronized refocused insensitive nuclei enhanced by polarization transfer (INEPT) scheme with τ1 = 1.44 ms and τ2 = 0.72 ms (50), or a 50 kHz 90° 13C excitation pulse. During acquisition, two-pulse phase modulation (TPPM) 1H decoupling was applied (51). 2D 13C-13C spectra were obtained using dipolar assisted rotational resonance (DARR) 13C-13C mixing (52), and a 3D 15N-13C NCACX spectrum was recorded using a standard NCA(CX) pulse sequence. Additional experimental details for the various ssNMR experiments are listed in Table S1 in the Supporting Material. Static 31P NMR was performed with a Bruker 31P/1H ssNMR probe outfitted with a horizontal EFree reduced electric field coil, using high-power 1H decoupling. The sample temperature was regulated at the indicated temperatures with cold cooling gas. The temperatures indicated throughout this article reflect internal sample temperatures that were calibrated using both external reference samples and internal referencing, following previously described protocols (53, 54, 55). Spectra were processed using Bruker Topspin and NMRPipe software (56). Except where specified otherwise (see Figs. 2 and 4), processing involved apodization with a cosine-bell function, and zero-filling of the direct and indirect dimensions with once or twice the original data size, phase correction, and Fourier transformation. Spectral analysis, plotting, and assignments were done using the CcpNmr Analysis program developed by the Collaborative Computation Project for the NMR community (CCPN) (57, 58). 31P NMR spectra were analyzed using the line shape fitting routines of Topspin. The resulting chemical shift anisotropy (CSA) parameters are reported as CSA span values (Ω, as defined in (59)), which are here equivalent to the shielding anisotropy Δδ. Lipid peaks were assigned based on reference samples and previous reports (33, 42, 60). 31P isotropic chemical shifts were referenced to phosphoric acid based on MAS NMR on hydroxylapatite (from Acros Chemicals, Geel, Belgium) (61). 1H, 13C, and 15N chemical shifts were referenced to dilute aqueous 2,2-dimethylsilapentane-5-sulfonic acid and liquid ammonia, via external indirect referencing of adamantane 13C chemical shifts (59, 62). Synthetic spectra were generated using utilities from the NMRPipe toolkit, and chemical shifts were predicted from a published crystal structure (44) using the SPARTA+ program (63).
Figure 2.
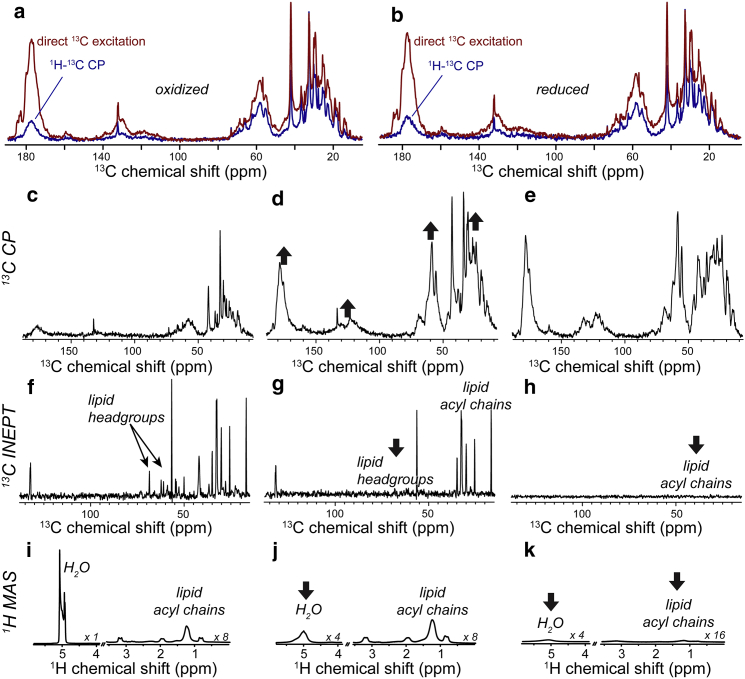
1D MAS NMR spectra of oxidized and reduced U-13C,15N cyt-c bound to TOCL/DOPC LUVs. (a and b) 1H-13C CP and direct excitation 13C spectra of oxidized and reduced membrane-bound cyt-c, at 271 K. The spectra in each panel are plotted with absolute intensities, and were processed with 25 Hz exponential line broadening and fourfold zero-filling. (c) 1H-13C CP spectrum at 257 K, showing mostly lipid signals. (d) CP spectrum at 250 K, and (e) 236 K, with much increased protein signals (up arrows). (f) 1D 1H-13C refocused INEPT spectrum at 257 K, with mostly lipid peaks (headgroups marked). (g and h) INEPT data at 250 K and 236 K, showing loss of lipid headgroup and acyl chain signals, respectively (down arrows). (i) 1D 1H spectrum at 257 K, with intense liquid water peak. (j) 1D 1H spectrum at 250 K, after freezing of solvent (down arrow). (k) 1H 1D at 236 K, showing a loss of acyl chain signals (down arrow). Data obtained at 600 MHz (1H) and 8.33 kHz MAS, and processed with a cosine-bell apodization function and zero-filling to twice the original size. Selected spectra are scaled as indicated. To see this figure in color, go online.
Figure 4.
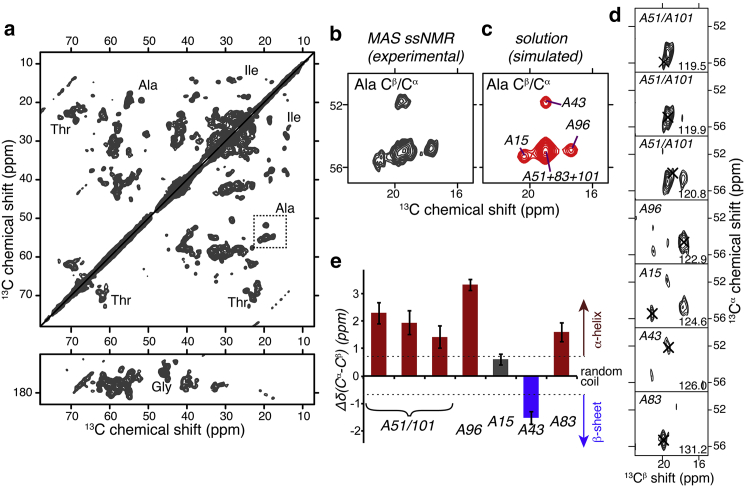
2D and 3D MAS NMR. (a) 2D 13C-13C spectrum on 1.7 mg oxidized 13C,15N-cyt-c bound to DOPC/TOCL LUVs, obtained with 10 ms DARR mixing. Selected residue types are indicated and the dashed box marks Ala Cα-Cβ crosspeaks. The spectrum was processed with fourfold zero-filling after 15 Hz Lorentzian line sharpening and 40 or 50 Hz Gaussian line broadening in the direct and indirect dimensions, respectively. (b) Enlargement of the Ala Cβ-Cα crosspeaks; (c) Analogous (simulated) peaks from solution NMR data (46) on the soluble protein (see text for details). (d) 2D slices from a 3D NCACX spectrum reveal distinct Ala signals (black X), at 15N shifts shown at bottom right. Tentative assignments based on correspondence to solution NMR shifts are indicated. (e) MAS NMR secondary chemical shifts Δδ(Cα-Cβ) deviate from random coil values (at zero) and indicate mostly α-helical structure for the Ala, in line with the native structure (Fig. 1a). Dashed lines at ±0.7 ppm indicate the typical boundaries to the random coil regime. Experiments performed at 233 K, 8.33 kHz MAS, and 600 MHz (for 1H). To see this figure in color, go online.
Optical spectroscopy
Quantitative analysis of the cyt-c oxidation state was performed based on the UV-VIS absorbance profiles for reduced and oxidized protein, measured at room temperature (64). The absorbance at 434 nm was employed to determine the protein concentration. Corrections were applied for baseline shifts due to any turbidity in lipid-containing samples, where needed. FTIR spectroscopy was performed at room temperature for both soluble cyt-c and for cyt-c bound to 1:4 (molar) TOCL/DOPC, using an MB series spectrophotometer (ABB Bomem, Quebec City, QC). The buffer signal was subtracted from each of the FTIR spectra using PROTA software (Biotools, Jupiter, FL). Peak deconvolution was then applied to the amide I region after using the Savitzky-Golay algorithm to apply a 2.9% smoothing and baseline correction. Second derivative analysis was used to determine peak positions, and peak fitting was performed using Gaussian line shapes in the PeakFit v4.12 program (Systat Software, San Jose, CA).
Activity measurements
Peroxidase activity measurements were performed based on previous reports (14, 15). Fluorescence measurements were performed at room temperature, using a Horiba Scientific Fluoromax-4 spectrofluorometer (Edison, NJ). Samples were prepared analogous to the NMR samples. LUVs were prepared by extrusion in 20 mM HEPES pH 7.4 buffer, which here included 100 μM of the chelating agent diethylene triamine pentaacetic acid. Stock solution of unlabeled cyt-c was added, followed by the substrate amplex red (AR) (ThermoFisher Scientific, Waltham, MA), to a 50 μM final AR concentration and a final protein to AR ratio of 1/100. A 5-min baseline was measured, after which H2O2 was added at 50 μM final concentration. Peroxidase activity was monitored based on the time-dependent increase in fluorescence from the oxidation product resorufin (65). Relative activity rates were determined by linear fitting.
Results
Peroxidase activity of LUV-bound 13C,15N-labeled cyt-c
Unilamellar vesicles were prepared from a mixture of TOCL and DOPC, to assure optimal access of the protein to the lipids. DLS measurements showed that the obtained vesicles were homogeneous in terms of their diameter (Fig. S1). The desired amount of protein stock solution was added to the LUVs at room temperature, resulting in CL-dependent binding. Membrane-binding was observed by pelleting the cyt-c/LUV samples by centrifugation, followed by UV-VIS measurements of the supernatant to measure residual unbound cyt-c, and further confirmed by monitoring the color of the pelleted LUVs. In the activity assays and all other experiments, the same variant of horse heart cyt-c was expressed in-house as described in the Materials and Methods section, and added to the vesicles at a 1:40 protein/lipid (P/L) molar ratio. This ratio is designed to mimic conditions during the early stages of the proapoptotic process. As discussed in the Introduction section, the CL asymmetry across the IMM becomes disrupted, which allows for the formation of CL-rich domains in the IMS-facing leaflet of the IMM. Our in vitro experiments are designed to probe the cyt-c peroxidase activity and molecular features of protein and lipid bilayer under these kinds of conditions. We measured the peroxidase activity of the LUV-bound cyt-c in fluorescence-based assays, performed at room temperature in the presence and absence of LUVs. Fig. 1 b shows the time-dependent generation of fluorescent product, and Fig. 1 c the corresponding activity rates for the different samples. Binding to CL-containing LUVs causes a significant increase in peroxidase activity, relative to the nascent activity of the protein, in line with previous reports (14, 15).
Lipid-bilayer-bound cyt-c experiences significant dynamics
Using the same protocols, uniformly 13C and 15N-labeled (U-13C,15N) cyt-c was bound to 4:1 (molar) DOPC/TOCL LUVs, and the membrane-bound cyt-c was pelleted into ceramic MAS sample holders using home-built ultracentrifugal sample packing tools. This packing method ensured the fast, straightforward, and highly reproducible preparation of LUV-based MAS NMR samples without risk of sample dehydration during transfer of membrane pellets. Analysis of the supernatant by UV-VIS showed that typically ∼75% of the U-13C,15N cyt-c was pelleted, in LUV-bound form, into the MAS rotor.
Fig. 2 a shows 1D 13C MAS NMR spectra acquired on such a sample featuring oxidized U-13C,15N-labeled cyt-c, using either direct 13C excitation (red) or 1H-13C CP (blue). The CP spectrum shows fewer peaks and much lower intensity peaks compared to the direct excitation data. The strongest 13C signals in the CP spectrum are lipid signals rather than those of the labeled protein. Similar results were obtained with the reduced form of the protein (Fig. 2 b), providing no direct evidence of excessive paramagnetic broadening or relaxation due to the heme. The spectra in Fig. 2, a and b), were obtained at 271 K internal sample temperature. Little change is seen at higher temperatures (up to room temperature), or down to 257 K (see Fig. 2, c, f, and i). Note that the water present in the typical densely packed, mm-sized MAS NMR samples have a much suppressed freezing temperature (55), causing the water to remain liquid down to −15 to −20°C. The (lack of) freezing of the water is conveniently probed by observation of the water 1H signals. The liquid water has a narrow and intense signal (e.g., Fig. 2 i), which is greatly reduced and broadened upon freezing (e.g., Fig. 2 j). Similarly, these 1H spectra will also reveal changes in the fluidity of lipid acyl chains (see below).
The initially low intensity in the CP spectra is due to the dynamics experienced by the protein and lipids. These protein and lipid dynamics occur on distinct timescales, as becomes more evident in other types of NMR spectra. In a 1H-13C INEPT spectrum (Fig. 2 f) we see again primarily lipid peaks (see also Fig. S2) and virtually no protein signals. The INEPT experiment is very sensitive to T2 relaxation, and suffers large intensity losses for all but the most mobile atomic sites. The lipid molecules have long T2 values as they undergo rapid diffusion and dynamics within the fluid bilayer. Cyt-c must be experiencing motion on a slower timescale than the surrounding lipids, leading to shorter T2 relaxation times (66) and a lack of INEPT signal. Thus, the protein is dynamic but its motion is constrained while it is associated with the membrane. The lack of cyt-c INEPT peaks and the distinct dynamics of the protein and lipids have another important implication. Any unfolded cyt-c segments would be expected to be highly flexible, such that they match the dynamic features of surrounding solvent or lipid molecules (66). Thus, the lack of INEPT peaks appears inconsistent with a large-scale unfolding of the membrane-bound protein.
Fig. 2 d shows that cooling of the sample results in a dramatic increase of the protein’s CP signal. At the same time, the lipid acyl chains still retain their mobility, as evidenced from their intense INEPT (Fig. 2 g) and 1H signals (Fig. 2 j). Immobilization of the protein is therefore coupled to the rigidification of the lipid headgroups (Fig. 2 g) and freezing of the solvent (Fig. 2 j), rather than the immobilization of the lipid acyl chains. The latter occurs at lower temperatures, when we observe a subsequent loss of the lipid acyl signals in the INEPT and 1H spectra (Fig. 2, h and k). This acyl chain rigidification does not translate into a significant increase in the protein’s CP signal (Fig. 2 e), further indicating cyt-c’s dynamics being coupled to the lipid headgroups rather than the hydrophobic membrane core. This strongly suggests the protein to be located outside the hydrophobic membrane core, in a peripherally bound state, and mirrors prior ssNMR studies of a different peripheral membrane protein (67).
Bound cyt-c retains a well-defined conformation
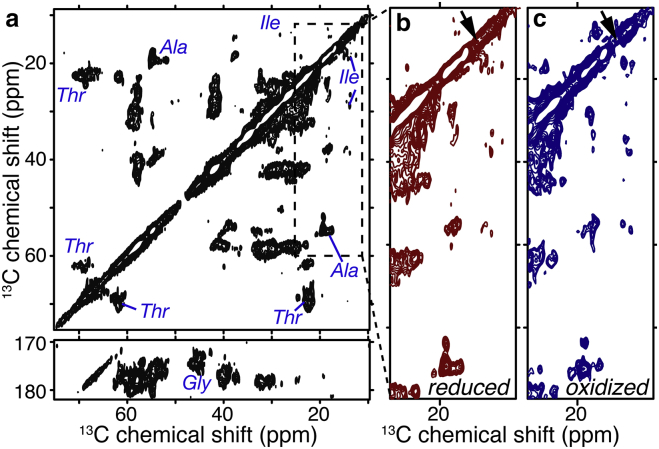
The enhanced CP efficiency at reduced temperatures enabled multidimensional MAS ssNMR studies of the labeled protein. This will allow site-specific insight into the structure of the immobilized protein, although it also implies the loss of direct insight into the dynamics that the protein experiences at the higher temperatures employed in the activity assays. Fig. 3 a shows a 2D 13C-13C MAS NMR spectrum that was obtained on the LUV-bound reduced U-13C,15N cyt-c. It features many protein-derived crosspeaks that can only be from the fully 13C-labeled protein. These peaks are sufficiently narrow to resolve many individual atomic sites in distinct residues, which then allows for site-specific insight into protein structure. Residues throughout a protein tend to have distinct NMR signals (high dispersion) in folded proteins, or quite similar NMR signals (low dispersion) in the absence of a well-defined fold. The observed resolution and dispersion of the membrane-bound cyt-c are not consistent with a disordered unfolded state, and show that the protein retains a high degree of structural order. The evidence supporting this conclusion will be examined in more detail below. Previous reports indicated differences in conformation, dynamics, and membrane-binding characteristics of cyt-c as a function of its oxidation state (39, 43, 68, 69, 70). We examined whether this translates into a detectable change in conformation of the LUV-bound protein. The reduced sample (Fig. 3, a and b), was recovered, treated with ferricyanide for in situ oxidation (71), and then repacked into its sample holder. The change in redox state was monitored by UV absorbance (Fig. S3) (64). The complete 2D 13C-13C spectra of the reduced and oxidized samples are shown in Fig. S4, and enlarged regions are shown in Fig. 3, b and c. Separately, another sample of U-13C,15N-cyt-c was oxidized before LUV binding and packed for MAS NMR. In both cases, 2D 13C-13C MAS ssNMR spectra resemble those of the reduced protein, in terms of resonance frequencies, line widths, and peak counts (Figs. 3 and 4 a). A few specific differences are visible in the spectral regions in Fig. 3, b and c, which will be examined in more detail in the Discussion section.
Figure 3.
2D MAS ssNMR on LUV-bound U-13C,15N cyt-c. (a) 13C-13C ssNMR spectrum on 0.75 mg reduced 13C,15N-cyt-c bound to DOPC/TOCL (4/1) LUVs at a P/L = 1:40. Off-diagonal crosspeaks are due to 13C-labeled protein, with selected residue types indicated. The dashed box indicates the region enlarged in (b). The spectrum was obtained using 15 ms DARR mixing, at 236 K, 8.33 kHz MAS, and 600 MHz (1H frequency). (b and c) Spectral regions before and after conversion of membrane-bound reduced cyt-c to the oxidized state. Minor chemical shift and intensity changes affect Ile and other methyl-bearing residues. Arrows indicate the Met-80 13Cε chemical shift for reduced cyt-c in solution (46). To see this figure in color, go online.
On the conformation of the membrane-bound oxidized cyt-c
These multidimensional MAS NMR spectra on the oxidized sample obtained at low temperatures provide, to our knowledge, new insight into the conformation of the membrane-bound cyt-c. First, we examine the easily identifiable crosspeaks from cyt-c’s Ala residues, which are distributed throughout the protein (Fig. 1) and thus should reveal both global and local structural changes. In the 2D spectrum four distinct Ala Cα-Cβ crosspeaks are seen with varying peak intensities (boxed in Fig. 4 a and enlarged in Fig. 4 b). As cyt-c has six Ala residues (see Fig. 1 a), this could indicate missing peaks due to unfolding-induced disorder or dynamics, peak overlap, or a combination thereof. To evaluate this, we also obtained a 3D NCACX spectrum on the sample, which offers the 15N resolution needed to reveal the existence of as many as seven Ala 15N/13Cα/13Cβ peaks (Fig. 4 d). The Cα and Cβ chemical shifts depend on the presence or absence of secondary structure, with opposite effects of α-helical or β-sheet structure. This is reflected clearly in the secondary shift ΔδCα-Cβ (the difference between Cα and Cβ shifts minus the corresponding random coil value), which is zero for random coil conformations and deviates in positive or negative directions, depending on the type of secondary structure (see Fig. 4 e) (72). The secondary shifts of the Ala seen in the 3D spectrum are plotted in Fig. 4 e, and indicate that most are α-helical in structure (red bars), rather than random coil. Similar analyses applied to other residue types also show that there are many peaks that are inconsistent with random coil structure. However, broad lines and extensive peak overlap in the 105-residue protein (with e.g., 10 Thr) limit this analysis for other residue types. A different kind of analysis proved more fruitful, as will be discussed next.
We used the NMR chemical shifts obtained in these low-temperature ssNMR experiments to compare the membrane-bound structure to that of the unbound protein. NMR shifts are highly sensitive to a protein’s conformation, and both the structure and the NMR signals of the soluble protein are known (46). Based on these solution NMR shifts, we created synthetic 13C-13C spectra as shown in Fig. S5, with the Ala peaks reproduced in Fig. 4 c. Comparison of Fig. 4, b and c, illustrates the remarkable similarity that characterizes most of the spectrum (Fig. S5). Clearly, the Ala peak pattern is perfectly reproduced, and even the individual 13C shifts show only minor differences. Based on this close correspondence, we were even able to make the Ala assignments indicated in Fig. 4, d and e. More generally, there is a good correspondence between the peak patterns of various easily identifiable residue types (Fig S5, g–i). That said, it does appear that a number of peaks are moved, missing, or attenuated. Although other Ile peaks are easily identified (Fig. 3), Ile-81 is for instance missing or showing up at a different chemical shift. This Ile is part of a proposed membrane-binding site (more below) and stands apart in the solution data by its particularly high-field 13Cδ shift, which is not seen in our ssNMR data. Based on observed variations in the peak intensities, it is also likely that a subset of peaks is affected by line broadening due to structural heterogeneity or residual dynamics, causing them to be hard to resolve or detect. Nonetheless, Fig. S5 shows that the degree of correspondence between the ssNMR and solution NMR signals is remarkable. This implies a strong preservation of the native conformation and is not consistent with the membrane-bound protein experiencing a large degree of unfolding or denaturation (20, 21, 22, 23, 24, 25). A more detailed analysis of the protein’s structure will require de novo ssNMR assignments and further structural measurements.
FTIR corroborates the preservation of secondary structure
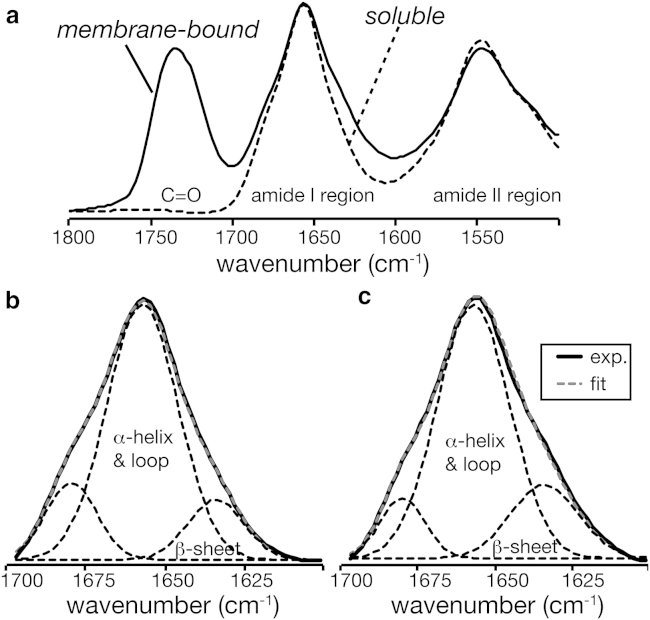
We also performed FTIR on membrane-bound and free cyt-c at room temperature, using analogous samples as used for ssNMR. The employed cyt-c was not isotopically labeled, but was otherwise obtained using the same construct and expression protocols as used for the NMR samples. We obtained analogous data for the wild-type protein, similar to prior studies (25, 73). Fig. 5 shows the experimental FTIR data, along with a deconvolution of the amide I region. The indicated C=O signals in the membrane-bound spectrum are dominated by the lipids (74). These room-temperature FTIR results corroborate the preservation of secondary structure in the membrane-bound peroxidase-active protein, consistent with various prior studies (e.g. (22, 23, 73)).
Figure 5.
FTIR on free and bound cyt-c. (a) FTIR on unlabeled cyt-c in buffer (dashed line), and bound at 1:40 P/L to DOPC/TOCL (4:1) LUVs (solid line) at room temperature. (b and c) Deconvolution of the amide I region of soluble and membrane-bound cyt-c, respectively. Individual fit components are shown as dashed black lines and their sum as a dashed gray line.
The cyt-c-bound vesicles retain a lipid bilayer conformation
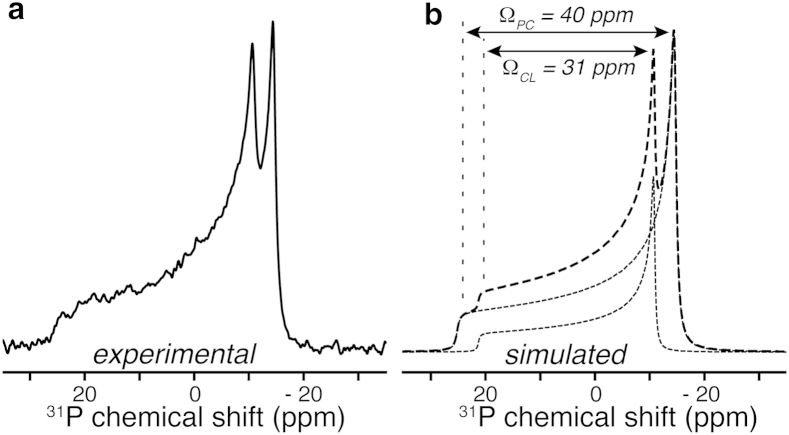
Several studies have emphasized the ability of cyt-c to cause nonbilayer formation in CL-rich membranes, with potential implications for membrane disruption and OMM permeation in mitochondrial apoptosis (75, 76). Static 31P ssNMR (without MAS) is an excellent tool to detect such bilayer disruption, because bilayer and nonbilayer lipid phases have distinct line shapes (36, 77). Briefly, lipid mobility in liquid crystalline bilayers results in uniaxial motional averaging of the phosphate’s 31P CSA. This results in characteristic spectra featuring a low-field, low-intensity shoulder and a high-field intense maximum, with an averaged CSA span of 30–40 ppm. Nonbilayer phases feature additional modes of motional averaging, resulting in much narrower line shapes. Cubic phases and (inverted) micellar structures allow for isotropic averaging, resulting in narrow isotropic peaks. Tubular inverted hexagonal (HII) phases have a narrowed, but still anisotropic line shape with a low-field maximum and a high-field shoulder (i.e., the opposite of bilayers). Fig. 6 shows the 1D 31P ssNMR spectrum for the sample studied by MAS NMR in Fig. 4. The spectrum features the characteristic line shape of lipids in a liquid crystalline bilayer. The 31P line shapes do display the typical features of slightly elongated, not-quite-spherical vesicles in high magnetic fields, as observed previously by us and others (33, 78, 79, 80). This manifests itself as a slight reduction in the low-intensity shoulder (lipids aligned with the field) and an increase in the high-intensity maximum that reflects the lipids oriented perpendicular to the field. The individual components of the CL and DOPC headgroups are visible, allowing extraction of their 31P CSA span values (59), which are found to be 31 and 40 ppm, for TOCL and DOPC, respectively, consistent with prior work (42). However, we observe no detectable nonbilayer phase signals. Thus, our FTIR and MAS ssNMR data reflect cyt-c as it interacts with the lipids in a bilayer state. This contrasts with prior 31P NMR studies that showed nonbilayer phases, seemingly due to the presence of higher amounts of cyt-c than employed here (21, 38, 40, 81).
Figure 6.
Static 31P ssNMR indicates a lipid bilayer structure. (a) 1D 31P static ssNMR spectrum on the oxidized 13C,15N-labeled cyt-c/LUV sample from Fig. 4. (b) Simulation of the spectral line shape (dashed line) based on two CSA components (dotted line). The magnitudes of the 31P CSA spans (Ω) for CL and PC are indicated.
Discussion
Disorder and dynamics of the membrane-bound cyt-c
FTIR, static ssNMR, and MAS NMR were used to probe cyt-c that is bound to LUVs featuring the mitochondrial lipid CL, which is the condition where it displays its proapoptotic peroxidase activity. Previous ssNMR studies suggested the bound protein to be dynamic and largely unfolded, but provided only partial insights due to the limited degree of isotopic labeling (20, 21, 40). We applied MAS ssNMR to U-13C,15N-labeled, membrane-bound cyt-c, representing one of the first MAS NMR studies of a fully labeled peripheral membrane protein (67). In line with the known affinity for CL (42, 82), cyt-c was found to associate tightly with CL-containing LUVs, causing the protein to gain significantly increased peroxidase activity (Fig. 1, b and c). The ssNMR spectra revealed extensive dynamics of the bound protein (Fig. 2). These dynamics occur on a ms timescale, explaining the initially low efficiency of both CP- and INEPT-based experiments. Increased dynamics were also noted in previous ssNMR on 13Cε-Met-labeled cyt-c (40), and explained as a full or partial unfolding induced by binding to CL-containing membranes. Pronounced protein dynamics have also been reported by studies that employ other spectroscopic methods (22, 23, 73, 83). Disordered or unstructured proteins or protein segments normally feature dynamics and relaxation behavior that match those of surrounding lipids (66, 84). However, we found that cyt-c exhibits dynamics that are significantly different from those of the lipids, as evidenced by the lack of protein peaks in the INEPT spectra (Fig. 2, f–h). This complete lack of INEPT signals is inconsistent with an extensively unfolded CL-bound cyt-c, and provides one piece of evidence that the protein dynamics are not necessarily representative of an unfolded state within the membrane. A similar behavior was noted in ssNMR studies of another folded but dynamic peripheral membrane protein (67).
Conformation of the membrane-bound protein
Our low-temperature-enabled 2D and 3D ssNMR spectra, as well as room-temperature FTIR, show that the membrane-bound cyt-c retains its secondary structure. In the NMR data, there is little evidence of global structural or dynamic disorder, which would have induced much more substantial line broadening and reduced spectral dispersion (84). The reproducible observation of these spectral features across multiple samples (in both redox states) shows that membrane-induced unfolding is not occurring under the sample conditions studied here. We do note that much of the ssNMR was performed at low temperature (unlike the activity measurements). Thus, it is possible that functionally relevant intramolecular dynamics are not represented in those spectra. The observed NMR peak positions are highly sensitive indicators of (changes in) the protein structure, because NMR resonance frequencies are directly correlated to the local protein structure. Comparisons between solution and solid-state shifts can be used to detect structural changes upon protein misfolding, aggregation, or membrane binding (85). The experimental ssNMR spectra closely match the peak patterns predicted from the known solution NMR structure (Fig. S5). This observation is not only inconsistent with the protein being unfolded within the membrane, but also indicates that the retained conformation is rather similar to that of the unbound, soluble state. We examined in particular detail the Ala residues that are distributed throughout the protein (Fig. 4). As illustrated in Fig. 1 a, cyt-c’s Ala are found at the end of N-helix (A15), in the partly α-helical loop before the 60’s helix (A43; A51), within the 71–85 Ω loop (A83), and within the C-helix (A96, A101) (86). Changes in their NMR signals should accompany any significant changes in structure throughout the molecule. Our 2D and 3D data (Fig. 4) revealed as many as seven Ala signals, with secondary (13Cα-13Cβ) chemical shifts (72) that differ from random coil values (Fig. 4 e) and are mostly α-helical as expected for the native protein (Fig. 1 a). The Ala 15N, 13Cα, and 13Cβ shifts are also strikingly similar to previously published solution NMR data for native cyt-c (46). There is some variation in peak intensities, which may be due to some variation in residue-specific dynamics along the length of the protein. Nonetheless, these data require that cyt-c retains a relatively well-ordered conformation.
Several key ssNMR spectra (Figs. 3 and 4) were obtained at low temperature, unlike the activity measurements. Suppression of the proteins’ dynamics was critical to obtain high quality multidimensional ssNMR spectra, as often seen in membrane protein ssNMR. Low-temperature measurements are commonly applied to enable structural studies, whether in x-ray crystallography, ssNMR, or other techniques. Naturally, this does modulate and abolish certain dynamical modes present at elevated temperatures. Indeed, the NMR data in Fig. 2 show clearly the presence of extensive dynamics, which may include some degree of intramolecular motion that could be functionally important. It seems however unlikely that the protein would be largely unfolded or denatured (20, 21, 22, 23, 24, 25). In prior ssNMR studies of highly disordered or unfolded membrane-bound proteins or peptides, the membrane-embedded polypeptides maintained a similar (lack of) structure and similar peak positions upon freezing of the membrane (84, 87). For cyt-c, prior FTIR and binding studies provide no evidence of a dramatic change in structure or membrane-binding between liquid crystalline and gel phase lipids (26, 73). As discussed previously, much (though maybe not all) of the dynamics experienced by cyt-c are likely due to its peripheral association with the dynamic membrane (67).
Location of membrane-bound cyt-c
The MAS NMR data in Fig. 2 examined the dynamical properties of the solvent, the lipid headgroups, lipid acyl chains, and the membrane-bound protein. It was found that changes in the protein’s motion did not correlate to the changes in the hydrophobic acyl chains. Instead, it accompanied the immobilization of lipid headgroups, and freezing of the solvent motion. These observations are most consistent with a shallow, peripheral interaction with the membrane. We note that the high degree of similarity of chemical shifts and tertiary structure to those of soluble cyt-c would also be hard to reconcile with the protein being embedded within the highly hydrophobic membrane core, and be more consistent with a relatively hydrophilic environment. Specific chemical shift differences observed for Met-80 and Ile-81 lend some support to prior suggestions of their involvement in the membrane facing side of the protein (88). Prior studies have also shown Met-80 to be dislodged upon membrane binding (7, 40, 82, 83, 88). Moreover, the binding of cyt-c to the negatively charged membranes is greatly reduced upon increasing salt concentrations (data not shown and reference (89)), which is a characteristic feature of an electrostatically driven peripheral interaction, rather than a hydrophobic interaction with the membrane core. Thus, our data indicate that in our samples cyt-c retains a native-like fold and interacts with the CL-containing membrane via a peripheral interaction with the lipid headgroups. Prior studies showed significant membrane disruption in the presence of cyt-c (21, 38, 40, 81). 31P ssNMR showed that, under the experimental conditions employed here, the lipid bilayer remains intact (Fig. 6). The previously mentioned membrane disruption was seen under different experimental conditions and most importantly with significantly higher P/L ratios. It is possible that the protein’s membrane impact would differ qualitatively under such conditions, because electron spin resonance, Raman, and other methods have shown membrane-bound cyt-c to undergo further structural changes at higher P/L ratios (beyond P/L ∼1/20) (88, 90).
Implications for cyt-c’s role as a peroxidase in apoptosis and mitophagy
The interest in cyt-c’s interactions with CL, and the lipid peroxidase activity that results, relates to the requirement for this cyt-c activity in mitochondrial apoptosis and mitophagy. It has long been argued that some kind of conformational change must underlie the formation of such a peroxidase-active cyt-c/CL complex. We observed the peroxidase activity for the membrane-bound protein and showed cyt-c to be highly dynamic. Our spectroscopic studies of the bound protein suggest that (at least under the employed experimental conditions) this motion is not due to a complete unfolding of the protein. Our data also indicate that cyt-c interacts primarily with the membrane interface without extensively perturbing the lipid bilayer or penetrating deeply into the hydrophobic membrane core. It is important to point out that the interaction of cyt-c with membranes is well known to depend on the experimental conditions. Although our measurements were performed at neutral pH, it has been shown that more acidic conditions enable a different binding mode (89). This could in part explain some of the differences in prior ssNMR studies that were performed at pH 6–6.3 (20, 40). As discussed previously, the binding affinity and binding mode are also expected to be sensitive to the P/L ratio, salt concentration, lipid composition, and other experimental conditions (88, 90, 91).
Our observations fit well with the generally accepted idea that electrostatically driven interactions between cyt-c and the negatively charged CL headgroups drive the initial protein-membrane interactions (26, 92, 93). The surface-bound cyt-c would bind specifically and tightly to multiple charged CL headgroups, while locally enriching the membrane in CL lipids. Our data suggest however that this initial interaction is not necessarily followed by a large-scale loss of structure within the membrane core. Such unfolding would facilitate interactions between the heme and the lipid acyl chains and thus rationalize the lipid peroxidation activity. However, one might expect penetration of the largely unfolded protein to at least partly decouple the disordered polypeptide chain (and heme) from the CL that facilitated the initial binding event. Accordingly, the freed heme could interact more easily with the diversity of acyl chains present within the membrane, predicting a somewhat indiscriminate peroxidation activity. However, as noted above and previously (17, 18, 19), the in vivo peroxidase activity displays remarkable selectivity, with specific types of lipids (in particular CL) and acyl chains being the preferred substrates for cyt-c’s peroxidase activity. Based on our data, we propose that this may be explained by a formation of a more compact, folded CL-cyt-c complex in which the protein retains a native-like fold and the heme remains largely sequestered within the protein. This would preserve a specific interaction with a cluster of CL lipids, thus enhancing access to CL and limiting access to other lipids. Second, the more limited access of the active site in this peripherally-bound state, would also place structural and dynamic constraints on the acyl chains, as is discussed next.
At first glance, one may wonder how an interfacial interaction would allow cyt-c to catalyze the efficient peroxidation of the acyl chain double bonds of CL, deep within the membrane core. The extended lipid anchoring model may suggest that a close interaction between the heme and an acyl chain necessitates the extraction of CL acyl chains from the bilayer (28, 29). We propose an alternative model in which dynamics of the lipid bilayer and dynamics experienced by the protein, possibly combined with dynamics within the protein, are responsible for transient interactions with the active site of bound cyt-c. This could occur without a permanent loss of the native fold, without deep penetration into the lipid bilayer core, or lipid acyl chain extraction. The peroxidase activity in vivo mostly targets CL that feature specific PUFA-based acyl chains, over other lipid types (19). A noteworthy feature of such PUFA chains is that they feature pronounced flexibility along the membrane normal, placing acyl chain segments closer to the bilayer surface than one may expect (94, 95). Simulation studies on linoleoyl acyl chains, which are preferentially oxidized by cyt-c (19), have noted the reduced density and high disorder of these acyl chains, in particular near the headgroups (96). This is dramatically different from what happens in saturated lipids. Thus, even in a superficial binding mode, the dynamic nature of both the PUFA-containing membranes and the membrane-associated protein may enable the necessary proximity between the protein’s active site and the PUFA double bonds. A relatively subtle or localized structural or dynamical change in the cyt-c would then be sufficient to allow lipid peroxidation to occur. Recent x-ray crystallographic studies show how cyt-c mutants can gain a much enhanced peroxidase activity (in absence of lipids) through such localized structural changes (30, 31).
Conclusions
We have used an array of experiments to probe the structure and function of the membrane-bound state of cyt-c. We found a CL-induced gain in peroxidase activity, but did not find evidence for a significant loss of the native structure, in contrast to various prior reports. Rather, our results suggest that a more modest change in the protein’s structure may be sufficient to cause peroxidase activation. Of course, it is possible that more extensive unfolding occurs under other conditions, e.g., in terms of the P/L ratio and/or lipid composition. The conditions employed here reflect limiting CL and ROS concentrations, as thought to be present early in the apoptotic process. Insights into the membrane-bound protein, and its effect on the lipid bilayer, were made possible by our use of complementary ssNMR measurements. We expect that these (and other) ssNMR methods will continue to be essential and powerful tools for ongoing and future efforts to elucidate the membrane interactions of cyt-c as well as other peripherally binding membrane proteins.
Author Contributions
A.M. and M.D. performed protein expression and purification; A.M., C.L.H., and R.K. made samples and performed spectroscopic (non-NMR) measurements; A.M., C.L.H., and P.v.d.W. performed ssNMR experiments; A.M. and P.v.d.W. analyzed data; A.M., V.E.K., J.A., and P.v.d.W. designed research; A.M. and P.v.d.W. wrote the article.
Acknowledgments
We thank Alexandr Kapralov for advice and training on the activity assays, Travis Wheeler at the Department of Cell Biology machine shop for construction of the packing tools, and Mike Delk for technical assistance. We thank Jun Li, Naveena Yanamala, and Judith Klein-Seetharaman for helpful discussions and the preparation of earlier ssNMR samples.
Funding support was from the University of Pittsburgh (J.A., P.v.d.W.), the National Institutes of Health (grants HL114453, U19 AIO68021, and ES 020693 to V.K.; T32 GM088119 to C.L.H.), National Institute for Occupation Safety and Health (NIOSH) grant OH008282 (V.K.), Human Frontier Science Program grant HFSP-RGP0013/2014 (V.K.), and grant UL1 RR024153 from the National Center for Research Resources (NCRR).
Editor: Mei Hong.
Footnotes
Five figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00953-4.
Supporting Material
References
- 1.Fulda S., Debatin K.-M. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 2.van Loo G., Saelens X., Vandenabeele P. The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ. 2002;9:1031–1042. doi: 10.1038/sj.cdd.4401088. [DOI] [PubMed] [Google Scholar]
- 3.Okouchi M., Ekshyyan O., Aw T.Y. Neuronal apoptosis in neurodegeneration. Antioxid. Redox Signal. 2007;9:1059–1096. doi: 10.1089/ars.2007.1511. [DOI] [PubMed] [Google Scholar]
- 4.Xun Z., Rivera-Sánchez S., McMurray C.T. Targeting of XJB-5-131 to mitochondria suppresses oxidative DNA damage and motor decline in a mouse model of Huntington’s disease. Cell Reports. 2012;2:1137–1142. doi: 10.1016/j.celrep.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schug Z.T., Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim. Biophys. Acta. 2009;1788:2022–2031. doi: 10.1016/j.bbamem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Osman C., Voelker D.R., Langer T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagan V.E., Tyurin V.A., Borisenko G.G. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 8.Garcia Fernandez M., Troiano L., Cossarizza A. Early changes in intramitochondrial cardiolipin distribution during apoptosis. Cell Growth Differ. 2002;13:449–455. [PubMed] [Google Scholar]
- 9.Scorrano L., Ashiya M., Korsmeyer S.J. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev. Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 10.Montero J., Marí M., Fernández-Checa J.C. Cholesterol and peroxidized cardiolipin in mitochondrial membrane properties, permeabilization and cell death. Biochim. Biophys. Acta. 2010;1797:1217–1224. doi: 10.1016/j.bbabio.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckmann J., Eckert S.H., Eckert G.P. Mitochondria: mitochondrial membranes in brain ageing and neurodegeneration. Int. J. Biochem. Cell Biol. 2013;45:76–80. doi: 10.1016/j.biocel.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Epand R.F., Martinou J.-C., Epand R.M. The apoptotic protein tBid promotes leakage by altering membrane curvature. J. Biol. Chem. 2002;277:32632–32639. doi: 10.1074/jbc.M202396200. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalvez F., Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 14.Kapralov A.A., Yanamala N., Kagan V.E. Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim. Biophys. Acta. 2011;1808:2147–2155. doi: 10.1016/j.bbamem.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe M., Niibayashi R., Miyoshi H. Molecular mechanisms for the induction of peroxidase activity of the cytochrome c-cardiolipin complex. Biochemistry. 2011;50:8383–8391. doi: 10.1021/bi2010202. [DOI] [PubMed] [Google Scholar]
- 16.Kagan V.E., Wipf P., Bayir H. Mitochondrial targeting of electron scavenging antioxidants: regulation of selective oxidation vs. random chain reactions. Adv. Drug Deliv. Rev. 2009;61:1375–1385. doi: 10.1016/j.addr.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown L.R., Wüthrich K. A spin label study of lipid oxidation catalyzed by heme proteins. Biochim. Biophys. Acta. 1977;464:356–369. doi: 10.1016/0005-2736(77)90010-4. [DOI] [PubMed] [Google Scholar]
- 18.Kagan V.E., Bayir H.A., Borisenko G. Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic. Biol. Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagan V.E., Tyurina Y.Y., Bayir H. Cardiolipin signaling mechanisms: collapse of asymmetry and oxidation. Antioxid. Redox Signal. 2015;22:1667–1680. doi: 10.1089/ars.2014.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spooner P.J.R., Watts A. Reversible unfolding of cytochrome c upon interaction with cardiolipin bilayers. 1. Evidence from deuterium NMR measurements. Biochemistry. 1991;30:3871–3879. doi: 10.1021/bi00230a010. [DOI] [PubMed] [Google Scholar]
- 21.Spooner P.J.R., Watts A. Reversible unfolding of cytochrome c upon interaction with cardiolipin bilayers. 2. Evidence from phosphorus-31 NMR measurements. Biochemistry. 1991;30:3880–3885. doi: 10.1021/bi00230a011. [DOI] [PubMed] [Google Scholar]
- 22.Hong Y., Muenzner J., Pletneva E.V. Origin of the conformational heterogeneity of cardiolipin-bound cytochrome C. J. Am. Chem. Soc. 2012;134:18713–18723. doi: 10.1021/ja307426k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanske J., Toffey J.R., Pletneva E.V. Conformational properties of cardiolipin-bound cytochrome c. Proc. Natl. Acad. Sci. USA. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernabeu A., Contreras L.M., Villalaín J. Two-dimensional infrared correlation spectroscopy study of the interaction of oxidized and reduced cytochrome c with phospholipid model membranes. Biochim. Biophys. Acta. 2007;1768:2409–2420. doi: 10.1016/j.bbamem.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Choi S., Swanson J.M. Interaction of cytochrome c with cardiolipin: an infrared spectroscopic study. Biochim. Biophys. Acta. 1995;54:271–278. doi: 10.1016/0301-4622(94)00151-9. [DOI] [PubMed] [Google Scholar]
- 26.Rytömaa M., Kinnunen P.K. Reversibility of the binding of cytochrome c to liposomes. Implications for lipid-protein interactions. J. Biol. Chem. 1995;270:3197–3202. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 27.Kalanxhi E., Wallace C.J.A. Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem. J. 2007;407:179–187. doi: 10.1042/BJ20070459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuominen E.K.J., Wallace C.J.A., Kinnunen P.K.J. Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. J. Biol. Chem. 2002;277:8822–8826. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- 29.Sinibaldi F., Howes B.D., Santucci R. Extended cardiolipin anchorage to cytochrome c: a model for protein-mitochondrial membrane binding. J. Biol. Inorg. Chem. 2010;15:689–700. doi: 10.1007/s00775-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 30.McClelland L.J., Mou T.-C., Bowler B.E. Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity. Proc. Natl. Acad. Sci. USA. 2014;111:6648–6653. doi: 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan W., Wang Z., Huang Z.-X. Structural basis for cytochrome c Y67H mutant to function as a peroxidase. PLoS One. 2014;9:e107305. doi: 10.1371/journal.pone.0107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watts A. Solid-state NMR approaches for studying the interaction of peptides and proteins with membranes. Biochim. Biophys. Acta. 1998;1376:297–318. doi: 10.1016/s0304-4157(98)00012-4. [DOI] [PubMed] [Google Scholar]
- 33.Hoop C.L., Sivanandam V.N., van der Wel P.C.A. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry. 2012;51:90–99. doi: 10.1021/bi201356v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong M., Zhang Y., Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annu. Rev. Phys. Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eddy M.T., Ong T.-C., Griffin R.G. Lipid dynamics and protein-lipid interactions in 2D crystals formed with the β-barrel integral membrane protein VDAC1. J. Am. Chem. Soc. 2012;134:6375–6387. doi: 10.1021/ja300347v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Wel P.C.A. Lipid dynamics and protein-lipid interactions in integral membrane proteins: insights from solid-state NMR. eMagRes. 2014;3:111–118. [Google Scholar]
- 37.Wylie B.J., Bhate M.P., McDermott A.E. Transmembrane allosteric coupling of the gates in a potassium channel. Proc. Natl. Acad. Sci. USA. 2014;111:185–190. doi: 10.1073/pnas.1319577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Kruijff B., Cullis P.R. Cytochrome c specifically induces non-bilayer structures in cardiolipin-containing model membranes. Biochim. Biophys. Acta. 1980;602:477–490. doi: 10.1016/0005-2736(80)90327-2. [DOI] [PubMed] [Google Scholar]
- 39.Waltham M.C., Cornell B.A., Smith R. Association of ferri- and ferro-cytochrome c with lipid multilayers: a 31P solid-state NMR study. Biochim. Biophys. Acta. 1986;862:451–456. doi: 10.1016/0005-2736(86)90250-6. [DOI] [PubMed] [Google Scholar]
- 40.Spooner P.J.R., Watts A. Cytochrome c interactions with cardiolipin in bilayers: a multinuclear magic-angle spinning NMR study. Biochemistry. 1992;31:10129–10138. doi: 10.1021/bi00156a037. [DOI] [PubMed] [Google Scholar]
- 41.Spooner P.J.R., Duralski A.A., Watts A. Dynamics in a protein-lipid complex: nuclear magnetic resonance measurements on the headgroup of cardiolipin when bound to cytochrome c. Biophys. J. 1993;65:106–112. doi: 10.1016/S0006-3495(93)81048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinheiro T.J., Watts A. Resolution of individual lipids in mixed phospholipid membranes and specific lipid-cytochrome c interactions by magic-angle spinning solid-state phosphorus-31 NMR. Biochemistry. 1994;33:2459–2467. doi: 10.1021/bi00175a014. [DOI] [PubMed] [Google Scholar]
- 43.Kim S.-m., Shin K., Akutsu H. The interactions of ferric and ferrous cytochrome c with cardiolipin in phospholipid membranes studied by solid-state H-2 and P-31 NMR. J. Mol. Struct. 1998;441:183–188. [Google Scholar]
- 44.Bushnell G.W., Louie G.V., Brayer G.D. High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 45.Rumbley J.N., Hoang L., Englander S.W. Recombinant equine cytochrome c in Escherichia coli: high-level expression, characterization, and folding and assembly mutants. Biochemistry. 2002;41:13894–13901. doi: 10.1021/bi026543y. [DOI] [PubMed] [Google Scholar]
- 46.Liu W., Rumbley J., Wand A.J. Backbone and side-chain heteronuclear resonance assignments and hyperfine NMR shifts in horse cytochrome c. Protein Sci. 2003;12:2104–2108. doi: 10.1110/ps.03211303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollock W.B., Rosell F.I., Mauk A.G. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys-72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 48.Bertini I., Engelke F., Ravera E. On the use of ultracentrifugal devices for sedimented solute NMR. J. Biomol. NMR. 2012;54:123–127. doi: 10.1007/s10858-012-9657-y. [DOI] [PubMed] [Google Scholar]
- 49.Metz G., Wu X., Smith S.O. Ramped-amplitude cross-polarization in magic-angle-spinning NMR. J. Magn. Reson. A. 1994;110:219–227. [Google Scholar]
- 50.Morris G.A., Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 1979;101:760–762. [Google Scholar]
- 51.Bennett A.E., Rienstra C.M., Griffin R.G. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 52.Takegoshi K., Nakamura S., Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 53.Dvinskikh S.V., Castro V., Sandström D. Heating caused by radiofrequency irradiation and sample rotation in 13C magic angle spinning NMR studies of lipid membranes. Magn. Reson. Chem. 2004;42:875–881. doi: 10.1002/mrc.1477. [DOI] [PubMed] [Google Scholar]
- 54.Thurber K.R., Tycko R. Measurement of sample temperatures under magic-angle spinning from the chemical shift and spin-lattice relaxation rate of 79Br in KBr powder. J. Magn. Reson. 2009;196:84–87. doi: 10.1016/j.jmr.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Böckmann A., Gardiennet C., Lesage A. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR. 2009;45:319–327. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 56.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Stevens T.J., Fogh R.H., Laue E.D. A software framework for analysing solid-state MAS NMR data. J. Biomol. NMR. 2011;51:437–447. doi: 10.1007/s10858-011-9569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 59.Harris R.K., Becker E.D., Zilm K.W., International Union of Pure and Applied Chemistry Physical and Biophysical Chemistry Division Further conventions for NMR shielding and chemical shifts (IUPAC Recommendations 2008) Magn. Reson. Chem. 2008;46:582–598. doi: 10.1002/mrc.2225. [DOI] [PubMed] [Google Scholar]
- 60.Zorin V., Ciesielski F., Bonev B.B. Heteronuclear chemical shift correlation and J-resolved MAS NMR spectroscopy of lipid membranes. Magn. Reson. Chem. 2010;48:925–934. doi: 10.1002/mrc.2690. [DOI] [PubMed] [Google Scholar]
- 61.Tang M., Waring A.J., Hong M. Effects of arginine density on the membrane-bound structure of a cationic antimicrobial peptide from solid-state NMR. Biochim. Biophys. Acta. 2009;1788:514–521. doi: 10.1016/j.bbamem.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morcombe C.R., Zilm K.W. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 63.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Margoliash E., Frohwirt N. Spectrum of horse-heart cytochrome c. Biochem. J. 1959;71:570–572. doi: 10.1042/bj0710570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou M., Diwu Z., Haugland R.P. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 66.Wang T., Cady S.D., Hong M. NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide. Biophys. J. 2012;102:787–794. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong L., Bamm V.V., Ladizhansky V. Solid-state NMR spectroscopy of 18.5 kDa myelin basic protein reconstituted with lipid vesicles: spectroscopic characterisation and spectral assignments of solvent-exposed protein fragments. Biochim. Biophys. Acta. 2007;1768:3193–3205. doi: 10.1016/j.bbamem.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berghuis A.M., Brayer G.D. Oxidation state-dependent conformational changes in cytochrome c. J. Mol. Biol. 1992;223:959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 69.Volkov A.N., Vanwetswinkel S., van Nuland N.A.J. Redox-dependent conformational changes in eukaryotic cytochromes revealed by paramagnetic NMR spectroscopy. J. Biomol. NMR. 2012;52:245–256. doi: 10.1007/s10858-012-9607-8. [DOI] [PubMed] [Google Scholar]
- 70.Letellier L., Shechter E. Correlations between structure and spectroscopic properties in membrane model system. Fluorescence and circular dichroism of the cytochrome c-cardiolipin system. Eur. J. Biochem. 1973;40:507–512. doi: 10.1111/j.1432-1033.1973.tb03220.x. [DOI] [PubMed] [Google Scholar]
- 71.Oldfield E., Allerhand A. Cytochrome c: observation of numerous single-carbon sites of the reduced and oxidized species by means of natural-abundance 13C nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA. 1973;70:3531–3535. doi: 10.1073/pnas.70.12.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luca S., Filippov D.V., Baldus M. Secondary chemical shifts in immobilized peptides and proteins: a qualitative basis for structure refinement under magic angle spinning. J. Biomol. NMR. 2001;20:325–331. doi: 10.1023/a:1011278317489. [DOI] [PubMed] [Google Scholar]
- 73.Heimburg T., Marsh D. Investigation of secondary and tertiary structural changes of cytochrome c in complexes with anionic lipids using amide hydrogen exchange measurements: an FTIR study. Biophys. J. 1993;65:2408–2417. doi: 10.1016/S0006-3495(93)81299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reis O., Winter R., Zerda T.W. The effect of high external pressure on DPPC-cholesterol multilamellar vesicles: a pressure-tuning Fourier transform infrared spectroscopy study. Biochim. Biophys. Acta. 1996;1279:5–16. doi: 10.1016/0005-2736(95)00233-2. [DOI] [PubMed] [Google Scholar]
- 75.Bergstrom C.L., Beales P.A., Groves J.T. Cytochrome c causes pore formation in cardiolipin-containing membranes. Proc. Natl. Acad. Sci. USA. 2013;110:6269–6274. doi: 10.1073/pnas.1303819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Antonov V.F., Puchkov M.N., Borodulin V. Soft perforation of cardiolipin-containing planar lipid bilayer membrane by cytochrome c and H(2)O(2) Eur. Biophys. J. 2014;43:469–476. doi: 10.1007/s00249-014-0977-6. [DOI] [PubMed] [Google Scholar]
- 77.Van der Wel P.C.A., Pott T., Killian J.A. Tryptophan-anchored transmembrane peptides promote formation of nonlamellar phases in phosphatidylethanolamine model membranes in a mismatch-dependent manner. Biochemistry. 2000;39:3124–3133. doi: 10.1021/bi9922594. [DOI] [PubMed] [Google Scholar]
- 78.Brumm T., Möps A., Bayerl T.M. Macroscopic orientation effects in broadline NMR-spectra of model membranes at high magnetic field strength: a method preventing such effects. Biophys. J. 1992;61:1018–1024. doi: 10.1016/S0006-3495(92)81909-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pott T., Dufourc E.J. Action of melittin on the DPPC-cholesterol liquid-ordered phase: a solid state 2H-and 31P-NMR study. Biophys. J. 1995;68:965–977. doi: 10.1016/S0006-3495(95)80272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Costello A.L., Alam T.M. Investigating the impact of cholesterol on magnetically aligned sphingomyelin/cholesterol multilamellar vesicles using static (31)P NMR. Chem. Phys. Lipids. 2010;163:506–513. doi: 10.1016/j.chemphyslip.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 81.Pinheiro T.J.T., Duer M.J., Watts A. Phospholipid headgroup dynamics in DOPG-d5-cytochrome c complexes as revealed by 2H and 31P NMR: the effects of a peripheral protein on collective lipid fluctuations. Solid State Nucl. Magn. Reson. 1997;8:55–64. doi: 10.1016/s0926-2040(96)01255-6. [DOI] [PubMed] [Google Scholar]
- 82.Belikova N.A., Vladimirov Y.A., Kagan V.E. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Jongh H.H., Ritsema T., Killian J.A. Lipid specificity for membrane mediated partial unfolding of cytochrome c. FEBS Lett. 1995;360:255–260. doi: 10.1016/0014-5793(95)00115-p. [DOI] [PubMed] [Google Scholar]
- 84.Su Y., Hong M. Conformational disorder of membrane peptides investigated from solid-state NMR line widths and line shapes. J. Phys. Chem. B. 2011;115:10758–10767. doi: 10.1021/jp205002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li J., Hoop C.L., van der Wel P.C.A. Amyloid-like fibrils from a domain-swapping protein feature a parallel, in-register conformation without native-like interactions. J. Biol. Chem. 2011;286:28988–28995. doi: 10.1074/jbc.M111.261750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krishna M.M.G., Lin Y., Walter Englander S. Cooperative omega loops in cytochrome c: role in folding and function. Biochim. Biophys. Acta. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 87.Su Y., Waring A.J., Hong M. Membrane-bound dynamic structure of an arginine-rich cell-penetrating peptide, the protein transduction domain of HIV TAT, from solid-state NMR. Biochemistry. 2010;49:6009–6020. doi: 10.1021/bi100642n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oellerich S., Lecomte S., Hildebrandt P. Peripheral and integral binding of cytochrome c to phospholipids vesicles. J. Phys. Chem. B. 2004;108:3871–3878. [Google Scholar]
- 89.Rytömaa M., Mustonen P., Kinnunen P.K. Reversible, nonionic, and pH-dependent association of cytochrome c with cardiolipin-phosphatidylcholine liposomes. J. Biol. Chem. 1992;267:22243–22248. [PubMed] [Google Scholar]
- 90.Heimburg T., Marsh D. Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes. Biophys. J. 1995;68:536–546. doi: 10.1016/S0006-3495(95)80215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rytömaa M., Kinnunen P.K. Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J. Biol. Chem. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 92.Pinheiro T.J., Elöve G.A., Roder H. Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry. 1997;36:13122–13132. doi: 10.1021/bi971235z. [DOI] [PubMed] [Google Scholar]
- 93.Zuckermann M.J., Heimburg T. Insertion and pore formation driven by adsorption of proteins onto lipid bilayer membrane-water interfaces. Biophys. J. 2001;81:2458–2472. doi: 10.1016/S0006-3495(01)75892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gawrisch K., Eldho N.V., Holte L.L. The structure of DHA in phospholipid membranes. Lipids. 2003;38:445–452. doi: 10.1007/s11745-003-1082-0. [DOI] [PubMed] [Google Scholar]
- 95.Feller S.E., Gawrisch K., MacKerell A.D., Jr. Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J. Am. Chem. Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 96.Bachar M., Brunelle P., Rauk A. Molecular dynamics simulation of a polyunsaturated lipid bilayer susceptible to lipid peroxidation. J. Phys. Chem. B. 2004;108:7170–7179. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.